Phenotypic and Genomic Characterization of Pseudomonas wuhanensis sp. nov., a Novel Species with Promising Features as a Potential Plant Growth-Promoting and Biocontrol Agent
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Bacterial Isolation
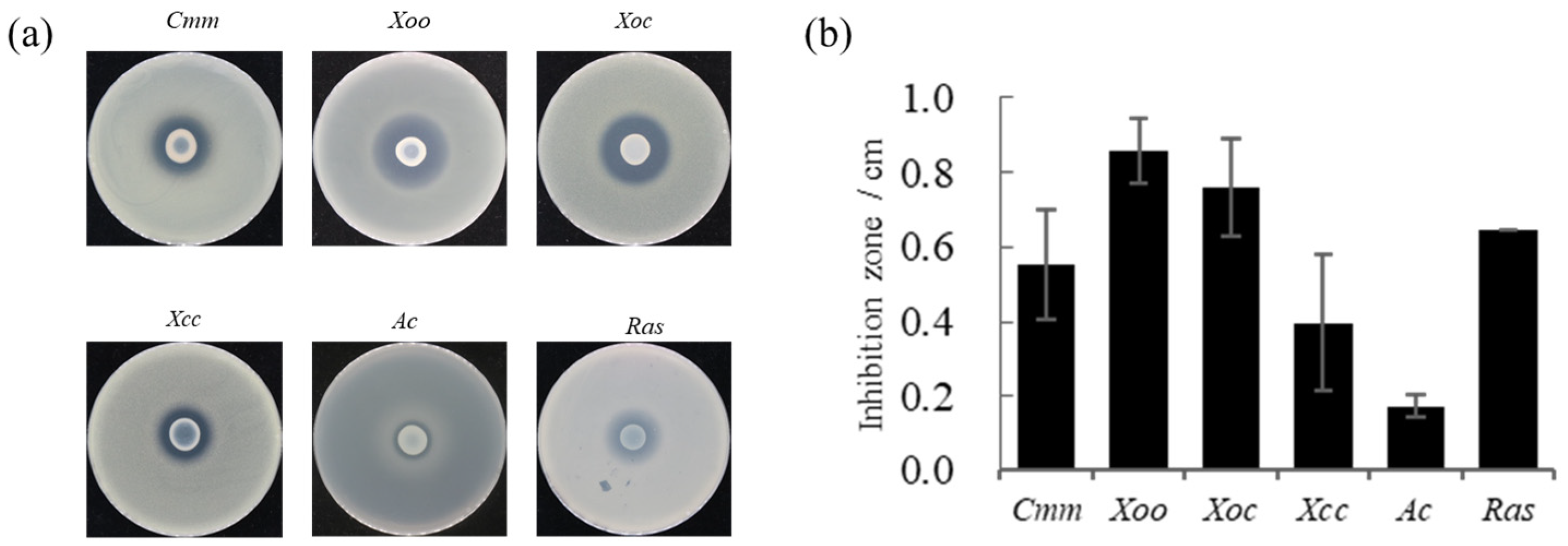
2.2. Antagonistic Test
2.3. Identification of Plant Growth-Promoting Traits
2.4. Physiology and Chemotaxonomic Characterization
2.5. Molecular Identification
2.6. Genome Sequencing, Annotation, and Comparative Genomic Analysis
2.7. Statistical Analysis
3. Results and Discussion
3.1. Antagonism against Phytopathogens
3.2. Plant Growth-Promoting Activities
3.3. 16S rRNA and MLSA Phylogenies
3.4. Physiology and Chemotaxonomic Characterization
3.5. Genomic Characterization
3.6. Description of P. wuhanensis sp. nov.
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Palleroni, N.J. Introduction to the family Pseudomonadaceae. In The Prokaryotes; Starr, M.P., Stolp, H., Trüper, H.G., Balows, A., Schlegel, H.G., Eds.; Springer: Berlin/Heidelberg, Germany, 1992; Volume 3, pp. 3074–3085. [Google Scholar]
- Kaminski, M.A.; Furmanczyk, E.M.; Sobczak, A.; Dziembowski, A.; Lipinski, L. Pseudomonas silesiensis sp. nov. strain A3T isolated from a biological pesticide sewage treatment plant and analysis of the complete genome sequence. Syst. Appl. Microbiol. 2018, 41, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Sawada, H.; Takeuchi, K.; Someya, N.; Morohoshi, T.; Satou, M. Pseudomonas solani sp. nov. isolated from the rhizosphere of eggplant in Japan. Int. J. Syst. Evol. Microbiol. 2023, 73, 005942. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Zhang, Y.; Chen, M.; Lu, W.; Chen, M.; Yan, Y.; Lin, M.; Zhang, W.; Zhou, Z. Pseudomonas nanhaiensis sp. nov., a lipase-producing bacterium isolated from deep-sea sediment of the South China Sea. Antonie van Leeuwenhoek 2021, 114, 1791–1804. [Google Scholar] [CrossRef] [PubMed]
- Mamtimin, T.; Anwar, N.; Abdurahman, M.; Kurban, M.; Rozahon, M.; Mamtimin, H.; Hamood, B.; Rahman, E.; Wu, M. Pseudomonas lopnurensis sp. nov., an endophytic bacterium isolated from Populus euphratica at the ancient Ugan river. Antonie van Leeuwenhoek 2021, 114, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Rao, Z.; Wu, S.; Peng, F.; Xie, Z.; Long, Y. Pseudomonas benzopyrenica sp. nov., isolated from soil, exhibiting high-efficiency degradation of benzo(a)pyrene. Int. J. Syst. Evol. Microbiol. 2023, 73, 006034. [Google Scholar] [CrossRef] [PubMed]
- Azhar, E.I.; Papadioti, A.; Bibi, F.; Ashshi, A.M.; Raoult, D.; Angelakis, E. ’Pseudomonas saudimassiliensis’ sp. nov. a new bacterial species isolated from air samples in the urban environment of Makkah, Saudi Arabia. New Microbes New Infect. 2017, 16, 43–44. [Google Scholar] [CrossRef] [PubMed]
- Busquets, A.; Gomila, M.; Beiki, F.; Mulet, M.; Rahimian, H.; García-Valdés, E.; Lalucat, J. Pseudomonas caspiana sp. nov., a citrus pathogen in the Pseudomonas syringae phylogenetic group. Syst. Appl. Microbiol. 2017, 40, 266–273. [Google Scholar] [CrossRef]
- Hunter, W.J.; Manter, D.K. Pseudomonas kuykendallii sp. nov.: A novel γ-proteo-bacteria isolated from a hexazinone degrading bioreactor. Curr. Microbiol. 2012, 65, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Ridgway, H.F.; Safarik, J.; Phipps, D.; Carl, P.; Clark, D. Identification and catabolic activity of well-derived gasoline-degrading bacteria from a contaminated aquifer. Appl. Environ. Microbiol. 1990, 56, 3565–3575. [Google Scholar] [CrossRef]
- Peix, A.; Ramírez-Bahena, M.-H.; Velázquez, E. The current status on the taxonomy of Pseudomonas revisited: An update. Infect. Genet. Evol. 2018, 57, 106–116. [Google Scholar] [CrossRef]
- Gross, H.; Loper, J.E. Genomics of secondary metabolite production by Pseudomonas spp. Nat. Prod. Rep. 2009, 26, 1408–1446. [Google Scholar] [CrossRef]
- Jiang, Y.; Huang, H.; Wu, M.; Yu, X.; Chen, Y.; Liu, P.; Li, X. Pseudomonas sp. LZ-Q continuously degrades phenanthrene under hypersaline and hyperalkaline condition in a membrane bioreactor system. Biophys. Rep. 2015, 1, 156–167. [Google Scholar] [CrossRef]
- Gu, Y.; Wang, J.; Xia, Z.; Wei, H. Characterization of a versatile plant growth-promoting rhizobacterium Pseudomonas mediterranea strain S58. Microorganisms 2020, 8, 334. [Google Scholar] [CrossRef]
- Widnyana, I.K.; Javandira, C. Activities Pseudomonas spp. and Bacillus sp. to stimulate germination and seedling growth of tomato plants. Agric. Agric. Sci. Procedia 2016, 9, 419–423. [Google Scholar] [CrossRef]
- Lyu, D.; Backer, R.; Robinson, W.G.; Smith, D.L. Plant growth-promoting rhizobacteria for cannabis production: Yield, cannabinoid profile and disease resistance. Front. Microbiol. 2019, 10, 1761. [Google Scholar] [CrossRef]
- Rosier, A.; Bishnoi, U.; Lakshmanan, V.; Sherrier, D.J.; Bais, H.P. A perspective on inter-kingdom signaling in plant-beneficial microbe interactions. Plant Mol. Biol. 2016, 90, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Lugtenberg, B.; Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Kujur, R.R.A.; Das, S.K. Pseudomonas phenolilytica sp. nov., a novel phenol-degrading bacterium. Arch. Microbiol. 2022, 204, 320. [Google Scholar] [CrossRef] [PubMed]
- Höfte, M.; Altier, N. Fluorescent pseudomonads as biocontrol agents for sustainable agricultural systems. Res. Microbiol. 2010, 161, 464–471. [Google Scholar] [CrossRef]
- de León, L.; Siverio, F.; López, M.M.; Rodríguez, A. Clavibacter michiganesis subsp. michiganensis, a seedborne tomato pathogen: Healthy seeds are still the goal. Plant Dis. 2011, 95, 1328–1338. [Google Scholar]
- Peng, C.; Chen, J.; Li, N.; Wang, R. Seedling Petri-dish inoculation method: A robust, easy-to-use and reliable assay for studying plant-Ralstonia solanacearum interactions. J. Integr. Agric. 2023, 22, 3709–3719. [Google Scholar]
- Niu, X.-N.; Wei, Z.-Q.; Zou, H.-F.; Xie, G.-G.; Wu, F.; Li, K.-J.; Jiang, W.; Tang, J.-L.; He, Y.-Q. Complete sequence and detailed analysis of the first indigenous plasmid from Xanthomonas oryzae pv. oryzicola. BMC Microbiol. 2015, 15, 233. [Google Scholar] [CrossRef]
- Wang, F.; Gao, S.; Niran, J.; Li, N.; Yin, Y.; Yu, C.; Jiao, C.; Yao, M. iTRAQ proteomics reveals the regulatory response to Xanthomonas campestris pv. vesicatoria in resistant vs. susceptible pepper genotypes. Hortic. Plant J. 2022, 8, 747–756. [Google Scholar] [CrossRef]
- He, Y.-W.; Wu, J.; Cha, J.-S.; Zhang, L.-H. Rice bacterial blight pathogen Xanthomonas oryzae pv. oryzae produces multiple DSF-family signals in regulation of virulence factor production. BMC Microbiol. 2010, 10, 187. [Google Scholar] [CrossRef] [PubMed]
- de Assunção, E.F.; da Conceicao, C.S.; Alexandre, E.R.; da Gama, M.A.S.; de Souza Nunes, G.H.; de Souza, E.B. New sources of melon accessions with resistance to bacterial fruit blotch at different phenological stages of melon growth and to multiple strains of Acidovorax citrulli. Euphytica 2021, 217, 79. [Google Scholar] [CrossRef]
- Bunsangiam, S.; Thongpae, N.; Limtong, S.; Srisuk, N. Large scale production of indole-3-acetic acid and evaluation of the inhibitory effect of indole-3-acetic acid on weed growth. Sci. Rep. 2021, 11, 13094. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Miranda, S.; Cabirol, N.; George-Téllez, R.; Zamudio-Rivera, L.; Fernández, F. O-CAS, a fast and universal method for siderophore detection. J. Microbiol. Methods 2007, 70, 127–131. [Google Scholar] [CrossRef]
- Minnikin, D.E.; O’Donnell, A.G.; Goodfellow, M.; Alderson, G.; Athalye, M.; Schaal, A.; Parlett, J.H. An integrated procedure for the extraction of bacterial isopre-noid quinones and polar lipids. J. Microbiol. Methods 1984, 2, 233–241. [Google Scholar] [CrossRef]
- Collins, M.D.; Jones, D. A note on the separation of natural mixtures of bacterial ubiquinones using reverse-phase partition thin-layer chromatography and high-performance liquid chromatography. J. Appl. Bacteriol. 1981, 51, 129–134. [Google Scholar] [CrossRef]
- Oyaizu, H.; Komagata, K. Grouping of Pseudomonas species on the basis of cellular fatty acid composition and the quinone system with special reference to the existence of 3-hydroxy fatty acids. J. Gen. Appl. Microbiol. 1983, 29, 17–40. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613. [Google Scholar] [CrossRef]
- Bennasar, A.; Mulet, M.; Lalucat, J.; García-Valdés, E. PseudoMLSA: A database for multigenic sequence analysis of Pseudomonas species. BMC Microbiol. 2010, 10, 118. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szcześniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, 46–50. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, 246–251. [Google Scholar] [CrossRef]
- Abby, S.S.; Rocha, E.P.C. Identification of protein secretion systems in bacterial genomes using MacSyFinder. In Bacterial Protein Secretion Systems; Journet, L., Cascales, E., Eds.; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2017; Volume 1615, pp. 1–21. [Google Scholar]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. UniProt: The universal protein knowledgebase in 2023. Nucleic Acids. Res. 2023, 51, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Girard, L.; Lood, C.; Höfte, M.; Vandamme, P.; Rokni-Zadeh, H.; van Noort, V.; Lavigne, R.; De Mot, R. The ever-expanding Pseudomonas genus: Description of 43 new species and partition of the Pseudomonas putida group. Microorganisms 2021, 9, 1766. [Google Scholar] [CrossRef] [PubMed]
- Durán, D.; Bernal, P.; Vazquez-Arias, D.; Blanco-Romero, E.; Garrido-Sanz, D.; Redondo-Nieto, M.; Rivilla, R.; Martín, M. Pseudomonas fluorescens F113 type VI secretion systems mediate bacterial killing and adaption to the rhizosphere microbiome. Sci. Rep. 2021, 11, 5772. [Google Scholar] [CrossRef] [PubMed]
- Kramer, J.; Özkaya, Ö.; Kümmerli, R. Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol. 2020, 18, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Drehe, I.; Simonetti, E.; Ruiz, J.A. Contribution of the siderophores pyoverdine and enantio-pyochelin to fitness in soil of Pseudomonas protegens Pf-5. Curr. Microbiol. 2018, 75, 1560–1565. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Yao, Y.; Yeon, S.K.; Seiple, I.B. Modular approaches to lankacidin antibiotics. J. Am. Chem. Soc. 2020, 142, 15116–15126. [Google Scholar] [CrossRef] [PubMed]
- Webster, G.; Mullins, A.J.; Petrova, Y.D.; Mahenthiralingam, E. Polyyne-producing Burkholderia suppress Globisporangium ultimum damping-off disease of Pisum sativum (pea). Front. Microbiol. 2023, 14, 1240206. [Google Scholar] [CrossRef]
- Lin, F.; Zhu, X.; Sun, J.; Meng, F.; Lu, Z.; Lu, Y. Bacillomycin D-C16 inhibits growth of Fusarium verticillioides and production of fumonisin B1 in maize kernels. Pestic. Biochem. Physiol. 2022, 181, 105015. [Google Scholar] [CrossRef]
- Fira, D.; Dimkić, I.; Berić, T.; Lozo, J.; Stanković, S. Biological control of plant pathogens by Bacillus species. J. Biotechnol. 2018, 285, 44–55. [Google Scholar] [CrossRef]
- Jenul, C.; Sieber, S.; Daeppen, C.; Mathew, A.; Lardi, M.; Pessi, G.; Hoepfner, D.; Neuburger, M.; Linden, A.; Gademann, K.; et al. Biosynthesis of fragin is controlled by a novel quorum sensing signal. Nat. Commun. 2018, 9, 1297. [Google Scholar] [CrossRef] [PubMed]
- Tohya, M.; Watanabe, S.; Teramoto, K.; Tada, T.; Kuwahara-Arai, K.; Mya, S.; Zin, K.N.; Kirikae, T.; Tin, H.H. Pseudomonas yangonensis sp. nov., isolated from wound samples of patients in a hospital in Myanmar. Int. J. Syst. Evol. Microbiol. 2020, 70, 3597–3605. [Google Scholar] [CrossRef] [PubMed]
- Haas, D.; Défago, G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 2005, 3, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Poblete-Morales, M.; Carvajal, D.; Almasia, R.; Michea, S.; Cantillana, C.; Levican, A.; Silva-Moreno, E. Pseudomonas atacamensis sp. nov., isolated from the rhizosphere of desert bloom plant in the region of Atacama, Chile. Antonie van Leeuwenhoek 2020, 113, 1201–1211. [Google Scholar] [CrossRef]
- Sun, J.; Wang, W.; Ying, Y.; Zhu, X.; Liu, J.; Hao, J. Pseudomonas profundi sp. nov., isolated from deep-sea water. Int. J. Syst. Evol. Microbiol. 2018, 68, 1776–1780. [Google Scholar] [CrossRef]
- Silby, M.W.; Winstanley, C.; Godfrey, S.A.C.; Levy, S.B.; Jackson, R.W. Pseudomonas genomes: Diverse and adaptable. FEMS Microbiol. Rev. 2011, 35, 652–680. [Google Scholar] [CrossRef]



| Species | dDDH (%) | ANI (%) |
|---|---|---|
| FP607T | FP607T | |
| Pseudomonas farris SWRI79T | 66.5 | 93.6 |
| Pseudomonas lini DSM 16768T | 54.2 | 94.0 |
| Pseudomonas frederiksbergensis LMG 19851T | 35.9 | 88.8 |
| Pseudomonas silesiensis A3T | 34.1 | 87.5 |
| Pseudomonas mandelii LMG 21607T | 34.1 | 88.1 |
| Pseudomonas arsenicoxydans CECT 7543T | 33.5 | 87.9 |
| Pseudomonas moorei DSM 12647T | 31.6 | 86.8 |
| Pseudomonas mohnii DSM 18327T | 31.5 | 86.7 |
| Pseudomonas jessenii LMG 21605T | 31.2 | 86.9 |
| Pseudomonas laurylsulfatiphila AP3 16T | 31.1 | 86.7 |
| Fatty Acid | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Summed features: * | |||||
| 3(C16:1 ω7c/C16:1 ω6c) | 7.1 | 28.0 | 31.2 | 34 | 36.9 |
| 8(C18:1 ω7c/C18:1 ω6c) | 1.5 | 8.4 | 4.8 | 11.9 | 18.3 |
| Saturated: | |||||
| C12:0 | 13.9 | 10.3 | 4.1 | 2.1 | 6.5 |
| C16:0 | 1.7 | 16.1 | 19.5 | 32.1 | 27.9 |
| Hydroxy: | |||||
| C10:0 3-OH | 39.0 | 7.6 | 8.8 | 3.8 | 3.7 |
| C12:0 2-OH | 11.4 | 5.4 | 8.9 | 4.1 | 1.0 |
| C12:0 3-OH | 18.1 | 6.7 | 7.4 | 4.3 | 3.1 |
| Region | Type | Most Similar Known Cluster | Location | BGC Type |
|---|---|---|---|---|
| 1 | redox-cofactor | Lankacidin | 603,022–625,194 | NRP + Polyketide |
| 2 | Ripp-like | - | 727,901–740,105 | - |
| 3 | lanthipeptide class II | - | 880,816–903,950 | - |
| 4 | Ripp-like | - | 982,561–994,438 | - |
| 5 | NAGGN | - | 1,814,162–1,829,030 | - |
| 6 | NRPS | Pf-5 pyoverdine | 1,969,442–2,022,425 | NRP |
| 7 | NRP-metallophore | histicorrugatin | 2,829,428–2,905,150 | NRP |
| 8 | HR-T2PKS | cepacin A | 2,933,224–2,975,466 | Polyketide |
| 9 | hydrogen-cyanide | - | 3,343,615–3,356,462 | - |
| 10 | isocyanide | pyoverdine DC3000 | 3,844,839–3,886,506 | NRP |
| 11 | Ripp-like | bacillomycin D | 3,926,086–3,936,925 | Polyketide + NRP: Lipopeptide |
| 12 | betalactone | fengycin | 4,157,666–4,180,849 | NRP |
| 13 | hydrogen-cyanide | - | 4,181,489–4,194,536 | - |
| 14 | NRPS-like | pyralomicin 1a | 4,454,343–4,509,652 | NRP + Polyketide: Modular type I polyketide |
| 15 | NRP-metallophore | Pf-5 pyoverdine | 4,547,832–4,633,873 | NRP |
| 16 | Ripp-like | - | 5,225,729–5,236,583 | - |
| 17 | Aryl polyene | APE Vf | 6,109,423–6,152,929 | Other |
| 18 | NRPS-like | fragin | 6,413,864–6,457,280 | NRP |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, J.; Liao, K.; Zhang, Y.-J.; Li, J.-Z.; Wei, H.-L. Phenotypic and Genomic Characterization of Pseudomonas wuhanensis sp. nov., a Novel Species with Promising Features as a Potential Plant Growth-Promoting and Biocontrol Agent. Microorganisms 2024, 12, 944. https://doi.org/10.3390/microorganisms12050944
Hou J, Liao K, Zhang Y-J, Li J-Z, Wei H-L. Phenotypic and Genomic Characterization of Pseudomonas wuhanensis sp. nov., a Novel Species with Promising Features as a Potential Plant Growth-Promoting and Biocontrol Agent. Microorganisms. 2024; 12(5):944. https://doi.org/10.3390/microorganisms12050944
Chicago/Turabian StyleHou, Jiawei, Kaiji Liao, Yong-Jie Zhang, Jun-Zhou Li, and Hai-Lei Wei. 2024. "Phenotypic and Genomic Characterization of Pseudomonas wuhanensis sp. nov., a Novel Species with Promising Features as a Potential Plant Growth-Promoting and Biocontrol Agent" Microorganisms 12, no. 5: 944. https://doi.org/10.3390/microorganisms12050944





