Antigenic Characterization of Circulating and Emerging SARS-CoV-2 Variants in the U.S. throughout the Delta to Omicron Waves
Abstract
:1. Introduction
2. Materials and Methods
2.1. Serum Samples
2.2. Biosafety Statement
2.3. Prevalence Analysis of SARS-CoV-2 Variants
2.4. Virus Isolation from Clinical Specimens
2.5. Focus Reduction Neutralization Test (FRNT)
2.6. Data Processing and Statistical Analysis
3. Results
3.1. Specimen Selection Process for Antigenic Characterization
3.2. Neutralization of SARS-CoV-2 Delta, Mu and Omicron Variants by Pooled Monovalent Vaccinee Sera Post the Primary Series
3.3. Neutralization of SARS-CoV-2 Omicron Variants by Individual Human Sera Collected from Post-Third-Dose Monovalent mRNA Vaccine Recipients
3.4. Neutralization of SARS-CoV-2 Omicron Variants by Pooled Human Sera Collected from Post-Third-Dose Monovalent mRNA Vaccine Recipients
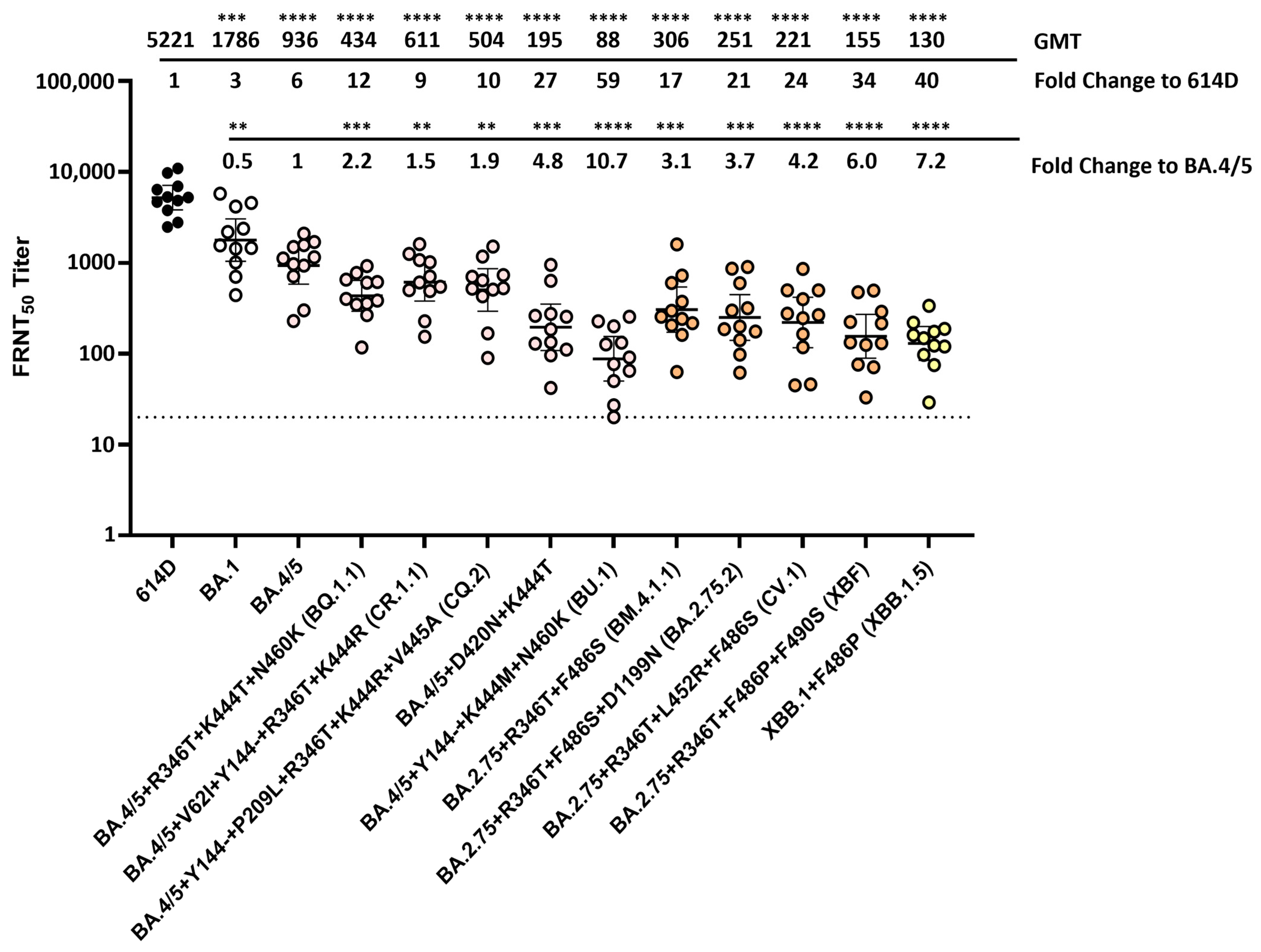
3.5. Neutralization of SARS-CoV-2 Omicron Variants by Individual Human Sera Collected from Vaccine Recipients Who Received the Bivalent mRNA Booster as the Fourth Dose
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Galloway, S.E.; Paul, P.; MacCannell, D.R.; Johansson, M.A.; Brooks, J.T.; MacNeil, A.; Slayton, R.B.; Tong, S.; Silk, B.J.; Armstrong, G.L.; et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage–United States, December 29, 2020–January 12, 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 95–99. [Google Scholar] [CrossRef]
- Zhou, B.; Thao, T.T.N.; Hoffmann, D.; Taddeo, A.; Ebert, N.; Labroussaa, F.; Pohlmann, A.; King, J.; Steiner, S.; Kelly, J.N.; et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature 2021, 592, 122–127. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.; France, A.M.; Aoki, Y.; Batra, D.; Biggerstaff, M.; Dugan, V.; Galloway, S.; Hall, A.J.; Johansson, M.A.; Kondor, R.J.; et al. Genomic Surveillance for SARS-CoV-2 Variants Circulating in the United States, December 2020–May 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 846–850. [Google Scholar] [CrossRef] [PubMed]
- Lambrou, A.S.; Shirk, P.; Steele, M.K.; Paul, P.; Paden, C.R.; Cadwell, B.; Reese, H.E.; Aoki, Y.; Hassell, N.; Zheng, X.Y.; et al. Genomic Surveillance for SARS-CoV-2 Variants: Predominance of the Delta (B.1.617.2) and Omicron (B.1.1.529) Variants—United States, June 2021–January 2022. MMWR Morb. Mortal. Wkly. Rep. 2022, 71, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Rahman, S.A.; Ehtesham, N.Z.; Hira, S.; Hasnain, S.E. SARS-CoV-2 variants of concern are emerging in India. Nat. Med. 2021, 27, 1131–1133. [Google Scholar] [CrossRef]
- Tian, D.; Sun, Y.; Zhou, J.; Ye, Q. The Global Epidemic of the SARS-CoV-2 Delta Variant, Key Spike Mutations and Immune Escape. Front. Immunol. 2021, 12, 751778. [Google Scholar] [CrossRef]
- Callaway, E. Heavily mutated Omicron variant puts scientists on alert. Nature 2021, 600, 21. [Google Scholar] [CrossRef]
- Viana, R.; Moyo, S.; Amoako, D.G.; Tegally, H.; Scheepers, C.; Althaus, C.L.; Anyaneji, U.J.; Bester, P.A.; Boni, M.F.; Chand, M.; et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 2022, 603, 679–686. [Google Scholar] [CrossRef]
- Ma, K.C.; Shirk, P.; Lambrou, A.S.; Hassell, N.; Zheng, X.Y.; Payne, A.B.; Ali, A.R.; Batra, D.; Caravas, J.; Chau, R.; et al. Genomic Surveillance for SARS-CoV-2 Variants: Circulation of Omicron Lineages—United States, January 2022–May 2023. MMWR Morb. Mortal. Wkly. Rep. 2023, 72, 651–656. [Google Scholar] [CrossRef]
- Chatterjee, S.; Bhattacharya, M.; Nag, S.; Dhama, K.; Chakraborty, C. A Detailed Overview of SARS-CoV-2 Omicron: Its Sub-Variants, Mutations and Pathophysiology, Clinical Characteristics, Immunological Landscape, Immune Escape, and Therapies. Viruses 2023, 15, 167. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, L.B.; Foster, C.; Rawlinson, W.; Tedla, N.; Bull, R.A. Evolution of the SARS-CoV-2 omicron variants BA.1 to BA.5: Implications for immune escape and transmission. Rev. Med. Virol. 2022, 32, e2381. [Google Scholar] [CrossRef] [PubMed]
- Rosenblum, H.G.; Wallace, M.; Godfrey, M.; Roper, L.E.; Hall, E.; Fleming-Dutra, K.E.; Link-Gelles, R.; Pilishvili, T.; Williams, J.; Moulia, D.L.; et al. Interim Recommendations from the Advisory Committee on Immunization Practices for the Use of Bivalent Booster Doses of COVID-19 Vaccines—United States, October 2022. MMWR Morb. Mortal. Wkly. Rep. 2022, 71, 1436–1441. [Google Scholar] [CrossRef]
- Focosi, D.; McConnell, S.; Casadevall, A.; Cappello, E.; Valdiserra, G.; Tuccori, M. Monoclonal antibody therapies against SARS-CoV-2. Lancet Infect. Dis. 2022, 22, e311–e326. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wilson, I.A. Protective neutralizing epitopes in SARS-CoV-2. Immunol. Rev. 2022, 310, 76–92. [Google Scholar] [CrossRef] [PubMed]
- Barnes, C.O.; Jette, C.A.; Abernathy, M.E.; Dam, K.A.; Esswein, S.R.; Gristick, H.B.; Malyutin, A.G.; Sharaf, N.G.; Huey-Tubman, K.E.; Lee, Y.E.; et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 2020, 588, 682–687. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, J.; Jian, F.; Xiao, T.; Song, W.; Yisimayi, A.; Huang, W.; Li, Q.; Wang, P.; An, R.; et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 2022, 602, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Yisimayi, A.; Jian, F.; Song, W.; Xiao, T.; Wang, L.; Du, S.; Wang, J.; Li, Q.; Chen, X.; et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature 2022, 608, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Song, W.; Wang, L.; Liu, P.; Yue, C.; Jian, F.; Yu, Y.; Yisimayi, A.; Wang, P.; Wang, Y.; et al. Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75. Cell Host Microbe 2022, 30, 1527–1539.e5. [Google Scholar] [CrossRef]
- McCallum, M.; De Marco, A.; Lempp, F.A.; Tortorici, M.A.; Pinto, D.; Walls, A.C.; Beltramello, M.; Chen, A.; Liu, Z.; Zatta, F.; et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 2021, 184, 2332–2347.e16. [Google Scholar] [CrossRef]
- Lok, S.M. An NTD supersite of attack. Cell Host Microbe 2021, 29, 744–746. [Google Scholar] [CrossRef]
- Wang, L.; Kainulainen, M.H.; Jiang, N.; Di, H.; Bonenfant, G.; Mills, L.; Currier, M.; Shrivastava-Ranjan, P.; Calderon, B.M.; Sheth, M.; et al. Differential neutralization and inhibition of SARS-CoV-2 variants by antibodies elicited by COVID-19 mRNA vaccines. Nat. Commun. 2022, 13, 4350. [Google Scholar] [CrossRef]
- Matsuyama, S.; Nao, N.; Shirato, K.; Kawase, M.; Saito, S.; Takayama, I.; Nagata, N.; Sekizuka, T.; Katoh, H.; Kato, F.; et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. USA 2020, 117, 7001–7003. [Google Scholar] [CrossRef]
- Lavezzo, E.; Pacenti, M.; Manuto, L.; Boldrin, C.; Cattai, M.; Grazioli, M.; Bianca, F.; Sartori, M.; Caldart, F.; Castelli, G.; et al. Neutralising reactivity against SARS-CoV-2 Delta and Omicron variants by vaccination and infection history. Genome Med. 2022, 14, 61. [Google Scholar] [CrossRef]
- Planas, D.; Veyer, D.; Baidaliuk, A.; Staropoli, I.; Guivel-Benhassine, F.; Rajah, M.M.; Planchais, C.; Porrot, F.; Robillard, N.; Puech, J.; et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 2021, 596, 276–280. [Google Scholar] [CrossRef]
- Cao, Y.; Jian, F.; Wang, J.; Yu, Y.; Song, W.; Yisimayi, A.; Wang, J.; An, R.; Chen, X.; Zhang, N.; et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature 2023, 614, 521–529. [Google Scholar] [CrossRef]
- Qu, P.; Evans, J.P.; Faraone, J.N.; Zheng, Y.-M.; Carlin, C.; Anghelina, M.; Stevens, P.; Fernandez, S.; Jones, D.; Lozanski, G.; et al. Enhanced neutralization resistance of SARS-CoV-2 Omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2. Cell Host Microbe 2023, 31, 9–17.e3. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Iketani, S.; Li, Z.; Guo, Y.; Yeh, A.Y.; Liu, M.; Yu, J.; Sheng, Z.; Huang, Y.; Liu, L.; et al. Antigenic characterization of the SARS-CoV-2 Omicron subvariant BA.2.75. Cell Host Microbe 2022, 30, 1512–1517.e4. [Google Scholar] [CrossRef]
- Suryadevara, N.; Shrihari, S.; Gilchuk, P.; VanBlargan, L.A.; Binshtein, E.; Zost, S.J.; Nargi, R.S.; Sutton, R.E.; Winkler, E.S.; Chen, E.C.; et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell 2021, 184, 2316–2331.e15. [Google Scholar] [CrossRef]
- Witte, L.; Baharani, V.A.; Schmidt, F.; Wang, Z.; Cho, A.; Raspe, R.; Guzman-Cardozo, C.; Muecksch, F.; Canis, M.; Park, D.J.; et al. Epistasis lowers the genetic barrier to SARS-CoV-2 neutralizing antibody escape. Nat. Commun. 2023, 14, 302. [Google Scholar] [CrossRef]
- Wang, Q.; Iketani, S.; Li, Z.; Liu, L.; Guo, Y.; Huang, Y.; Bowen, A.D.; Liu, M.; Wang, M.; Yu, J.; et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 2023, 186, 279–286.e8. [Google Scholar] [CrossRef]
- Wang, Q.; Bowen, A.; Valdez, R.; Gherasim, C.; Gordon, A.; Liu, L.; Ho, D.D. Antibody Response to Omicron BA.4–BA.5 Bivalent Booster. N. Engl. J. Med. 2023, 388, 567–569. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Kurhade, C.; Patel, S.; Kitchin, N.; Tompkins, K.; Cutler, M.; Cooper, D.; Yang, Q.; Cai, H.; Muik, A.; et al. Neutralization of BA.4–BA.5, BA.4.6, BA.2.75.2, BQ.1.1, and XBB.1 with Bivalent Vaccine. N. Engl. J. Med. 2023, 388, 854–857. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Bowen, A.; Tam, A.R.; Valdez, R.; Stoneman, E.; Mellis, I.A.; Gordon, A.; Liu, L.; Ho, D.D. SARS-CoV-2 neutralising antibodies after bivalent versus monovalent booster. Lancet Infect. Dis. 2023, 23, 527–528. [Google Scholar] [CrossRef] [PubMed]
- Regan, J.J.; Moulia, D.L.; Link-Gelles, R.; Godfrey, M.; Mak, J.; Najdowski, M.; Rosenblum, H.G.; Shah, M.M.; Twentyman, E.; Meyer, S.; et al. Use of Updated COVID-19 Vaccines 2023-2024 Formula for Persons Aged ≥ 6 Months: Recommendations of the Advisory Committee on Immunization Practices—United States, September 2023. MMWR Morb. Mortal. Wkly. Rep. 2023, 72, 1140–1146. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zou, J.; Kurhade, C.; Deng, X.; Chang, H.C.; Kim, D.K.; Shi, P.Y.; Ren, P.; Xie, X. Less neutralization evasion of SARS-CoV-2 BA.2.86 than XBB sublineages and CH.1.1. Emerg. Microbes Infect. 2023, 12, 2271089. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Kempf, A.; Nehlmeier, I.; Cossmann, A.; Dopfer-Jablonka, A.; Stankov, M.V.; Schulz, S.R.; Jäck, H.M.; Behrens, G.M.N.; Pöhlmann, S.; et al. Neutralisation sensitivity of SARS-CoV-2 lineages EG.5.1 and XBB.2.3. Lancet Infect. Dis. 2023, 23, e391–e392. [Google Scholar] [CrossRef] [PubMed]
- Yamasoba, D.; Uriu, K.; Plianchaisuk, A.; Kosugi, Y.; Pan, L.; Zahradnik, J.; Ito, J.; Sato, K. Virological characteristics of the SARS-CoV-2 omicron XBB.1.16 variant. Lancet Infect. Dis. 2023, 23, 655–656. [Google Scholar] [CrossRef] [PubMed]
- Planas, D.; Staropoli, I.; Michel, V.; Lemoine, F.; Donati, F.; Prot, M.; Porrot, F.; Guivel-Benhassine, F.; Jeyarajah, B.; Brisebarre, A.; et al. Distinct evolution of SARS-CoV-2 Omicron XBB and BA.2.86/JN.1 lineages combining increased fitness and antibody evasion. Nat. Commun. 2024, 15, 2254. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, Y.; Bowen, A.; Mellis, I.A.; Valdez, R.; Gherasim, C.; Gordon, A.; Liu, L.; Ho, D.D. XBB.1.5 monovalent mRNA vaccine booster elicits robust neutralizing antibodies against XBB subvariants and JN.1. Cell Host Microbe 2024, 32, 315–321.e3. [Google Scholar] [CrossRef]
- Lasrado, N.; Rössler, A.; Rowe, M.; Collier, A.-r.Y.; Barouch, D.H. Neutralization of SARS-CoV-2 Omicron subvariant BA.2.87.1. Vaccine 2024, 42, 2117–2121. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di, H.; Pusch, E.A.; Jones, J.; Kovacs, N.A.; Hassell, N.; Sheth, M.; Lynn, K.S.; Keller, M.W.; Wilson, M.M.; Keong, L.M.; et al. Antigenic Characterization of Circulating and Emerging SARS-CoV-2 Variants in the U.S. throughout the Delta to Omicron Waves. Vaccines 2024, 12, 505. https://doi.org/10.3390/vaccines12050505
Di H, Pusch EA, Jones J, Kovacs NA, Hassell N, Sheth M, Lynn KS, Keller MW, Wilson MM, Keong LM, et al. Antigenic Characterization of Circulating and Emerging SARS-CoV-2 Variants in the U.S. throughout the Delta to Omicron Waves. Vaccines. 2024; 12(5):505. https://doi.org/10.3390/vaccines12050505
Chicago/Turabian StyleDi, Han, Elizabeth A. Pusch, Joyce Jones, Nicholas A. Kovacs, Norman Hassell, Mili Sheth, Kelly Sabrina Lynn, Matthew W. Keller, Malania M. Wilson, Lisa M. Keong, and et al. 2024. "Antigenic Characterization of Circulating and Emerging SARS-CoV-2 Variants in the U.S. throughout the Delta to Omicron Waves" Vaccines 12, no. 5: 505. https://doi.org/10.3390/vaccines12050505




