High-Throughput Phenotyping for the Evaluation of Agronomic Potential and Root Quality in Tropical Carrot Using RGB Sensors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genetic Material, Experimental Site, and Flow
2.2. Evaluation of Agronomic Traits in the Field
2.3. Acquisition and Processing of Aerial Images
2.4. Statistical Analysis
2.5. Artificial Neural Intelligence for Grouping
2.6. Selection of the Best Entries Using the Genotype–Ideotype Distance Index
3. Results and Discussion
3.1. Germoplasm Evaluation
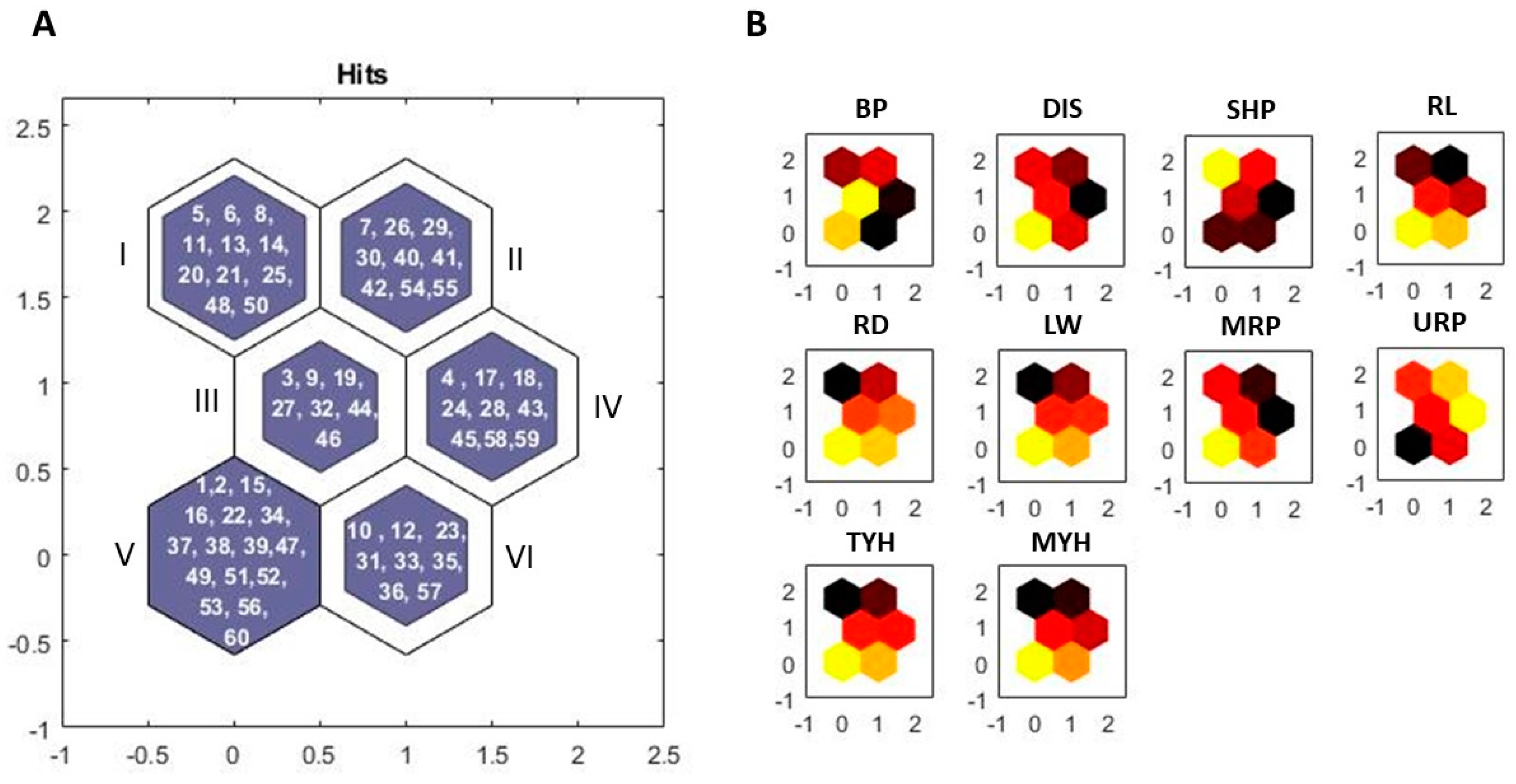
3.2. Grouping of Entries by Kohonen’s Self-Organizing Map
3.3. Validation of Image Phenotyping for Tropical Carrot
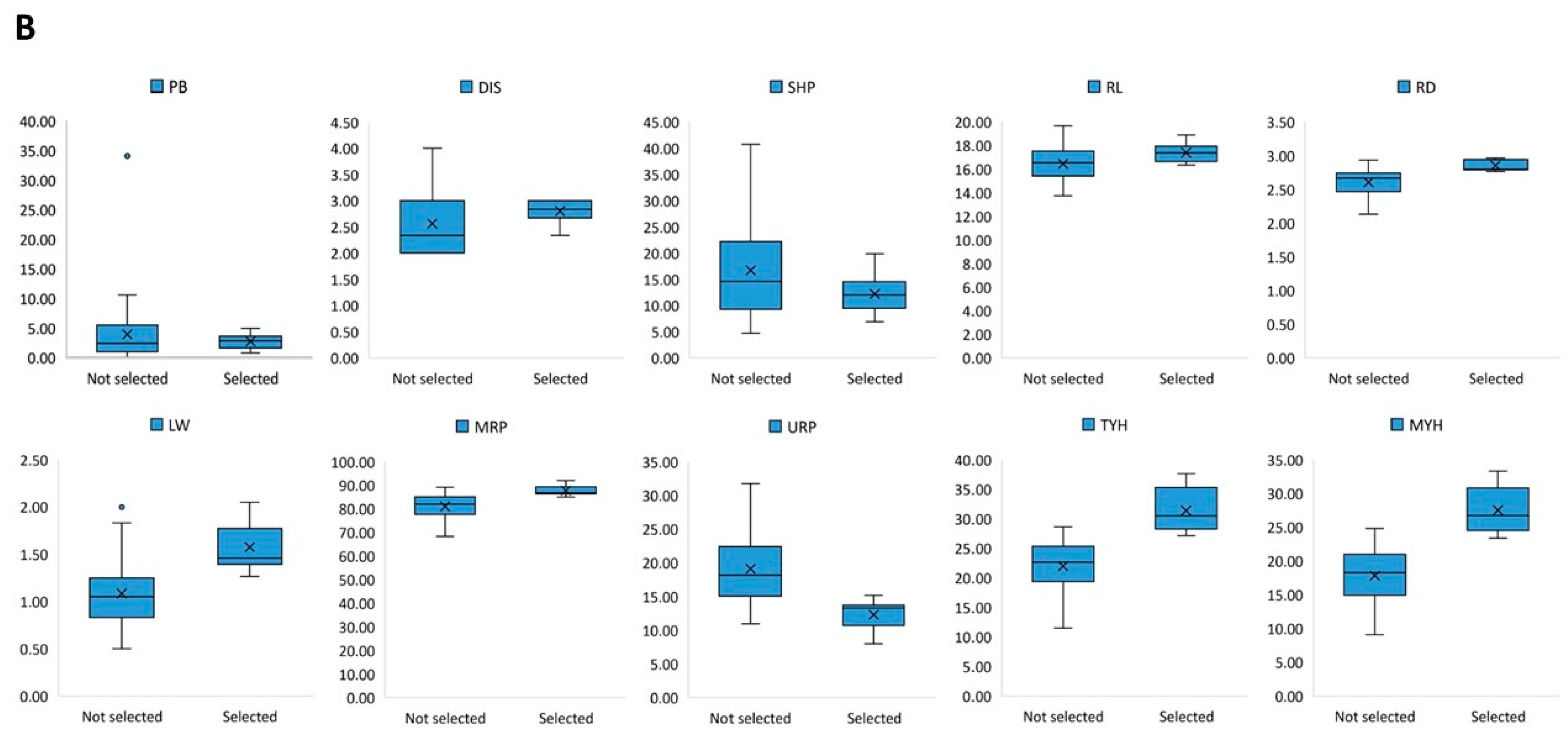
3.4. Selection of the Best Entries
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| RGB | Red–green–blue |
| UAV | Unmanned Aerial Vehicle |
| RPA | Remotely Piloted Aircraft |
| RCBD | Randomized complete block design |
| BP | Bolting percentage |
| DIS | Severity of leaf blight |
| LW | Fresh leaf weight (kg) |
| SHP | Percentage of purple or green shoulder |
| MRP | Percentage of marketable roots (%) |
| URP | Percentage of unmarketable roots (%) |
| MYH | Estimated yield of marketable roots (t ha−1) |
| TYH | Estimated total yield (t ha−1) |
| RD | Root diameter (cm) |
| RL | Root length (cm) |
| MRA | Marketable root area (m2) |
| URA | Unmarketable root area (m2) |
| TRA | Total root area (m2) |
| BI | Brightness Index |
| GLI | Green Leaf Index |
| HI | Primary Colors Hue Index |
| HUE | Overall Hue Index |
| NGRDI | Normalized Green Red Difference Index |
| SCI | Soil Color Index |
| SI | Spectral Slope Saturation Index |
| VARI | Visible Atmospherically Resistant Index |
| SOM | Self-Organizing Map |
References
- Abreu, D.J.M.D.; Lorenço, M.S.; Ferreira, A.N.; Scalice, H.K.; Vilas Boas, E.V.D.B.; Piccoli, R.H.; Carvalho, E.E.N. Artificial neural networks for the evaluation of physicochemical properties of carrots (Daucus carota L.) subjected to different cooking conditions as an alternative to traditional statistical methods. ACS Food Sci. Technol. 2021, 2, 143–150. [Google Scholar] [CrossRef]
- FAO. Food and Agriculture Organization of the United Nations Statistics Database. Available online: http://www.fao.org/faostat/en/#data (accessed on 2 May 2023).
- De Carvalho, A.D.F.; Pereira, G.E.; Da Silva, G.O. Estimates of genetic gains in the carrot using different selection indices. Rev. Agro@mbiente 2022, 16, 1–14. [Google Scholar] [CrossRef]
- Simon, P.W.; Freeman, R.E.; Vieira, J.V.; Boiteux, L.S.; Briard, M.; Nothnagel, T.; Michalik, B.; Kwon, Y.S. Carrot. In Vegetables II, 2nd ed.; Prohens, J., Nuez, F., Eds.; Springer: New York, NY, USA, 2008; Volume 2, pp. 327–357. [Google Scholar] [CrossRef]
- Walter, A.; Liebisch, F.; Hund, A. Plant phenotyping: From bean weighing to image analysis. Plant Methods 2015, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Maciel, G.M.; Gallis, R.B.A.; Barbosa, R.L.; Pereira, L.M.; Siquieroli, A.C.S.; Peixoto, J.V.M. Image phenotyping of inbred red lettuce lines with genetic diversity regarding carotenoid levels. Int. J. Appl. Earth Obs. Geoinf. 2019, 81, 154–160. [Google Scholar] [CrossRef]
- Dhondt, S.; Wuyts, N.; Inzé, D. Cell to whole-plant phenotyping: The best is yet to come. Trends Plant. Sci. 2013, 18, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Moeckel, T.; Dayananda, S.; Nidamanuri, R.R.; Nautiyal, S.; Hanumaiah, N.; Buerkert, A.; Wachendorf, M. Estimation of vegetable crop parameter by multi-temporal UAV-borne images. Remote Sens. 2018, 10, 805. [Google Scholar] [CrossRef]
- Herzig, P.; Borrmann, P.; Knauer, U.; Klück, H.-C.; Kilias, D.; Seiffert, U.; Pillen, K.; Maurer, A. Evaluation of RGB and Multispectral Unmanned Aerial Vehicle (UAV) Imagery for High-Throughput Phenotyping and Yield Prediction in Barley Breeding. Remote Sens. 2021, 13, 2670. [Google Scholar] [CrossRef]
- Xie, J.; Zhou, Z.; Zhang, H.; Zhang, L.; Li, M. Combining Canopy Coverage and Plant Height from UAV-Based RGB Images to Estimate Spraying Volume on Potato. Sustainability 2022, 14, 6473. [Google Scholar] [CrossRef]
- Ballesteros, R.; Ortega, J.F.; Hernandez, D.; Moreno, M.A. Onion biomass monitoring using UAV-based RGB imaging. Precis. Agric. 2018, 19, 840–857. [Google Scholar] [CrossRef]
- Ryu, J.H.; Na, S.I.; Cho, J. Inter-comparison of normalized difference vegetation index measured from different footprint sizes in cropland. Remote Sens. 2020, 12, 2980. [Google Scholar] [CrossRef]
- Ranđelović, P.; Đorđević, V.; Milić, S.; Balešević-Tubić, S.; Petrović, K.; Miladinović, J.; Đukić, V. Prediction of Soybean Plant Density Using a Machine Learning Model and Vegetation Indices Extracted from RGB Images Taken with a UAV. Agronomy 2020, 10, 1108. [Google Scholar] [CrossRef]
- Guo, Y.; Yin, G.; Sun, H.; Wang, H.; Chen, S.; Senthilnath, J.; Fu, Y. Scaling Effects on Chlorophyll Content Estimations with RGB Camera Mounted on a UAV Platform Using Machine-Learning Methods. Sensors 2020, 20, 5130. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.B.; De Carvalho, A.D.F.; Pinheiro, J.B.; Da Silva, G.O.; Vieira, J.V. Resistência de populações de cenoura à queima-das-folhas com diferentes níveis de germoplasma tropical. Hortic. Bras. 2012, 30, 489–493. [Google Scholar] [CrossRef]
- Richardson, A.J.; Wiegand, C. Distinguishing Vegetation from Soil Background Information. Photogramm. Eng. Remote Sens. 1977, 43, 1541–1552. [Google Scholar]
- Louhaichi, M.; Borman, M.M.; Johnson, D.E. Spatially Located Platform and Aerial Photography for Documentation of Grazing Impacts on Wheat. Geocarto Int. 2001, 16, 65–70. [Google Scholar] [CrossRef]
- Escadafal, R.; Belghith, A.; Bem, M.H. Indices spectraux pour la télédétection de la dégradation des milieux naturels en Tunisie aride. In Actes du Sixième Symposium International. Mes. Phys. Signat. Spectrales Télédétection 1994, 17–21. [Google Scholar]
- Tucker, C. Red and Photographic Infrared Linear Combinations for Monitoring Vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Mathieu, R.; Pouget, M.; Cervelle, B.; Escadafal, R. Relationships between satellite-based radiometric indices simulated using laboratory reflectance data and typic soil color of an arid environment. Remote Sens Environ 1998, 66, 17–28. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Stark, R.; Rundquist, D. Novel algorithms for remote estimation of vegetation fraction. Remote Sens. Environ. 2002, 80, 76–87. [Google Scholar] [CrossRef]
- Cruz, C.D. Genes: A software package for analysis in experimental statistics and quantitative genetics. Acta Sci. Agron. 2013, 35, 271–276. [Google Scholar] [CrossRef]
- De Carvalho, A.D.F.; Da Silva, G.O.; Pereira, G.E. Direct selection for phenotypic traits in carrot genotypes. Hortic. Bras. 2019, 37, 354–358. [Google Scholar] [CrossRef]
- De Carvalho, A.D.F.; Nogueira, M.T.M.; Silva, G.O.; Luz, J.M.Q.; Maciel, G.M.; Rabelo, P.G. Seleção de genótipos de cenoura para caracteres fenotípicos de raiz. Hortic. Bras. 2017, 35, 97–102. [Google Scholar] [CrossRef]
- Le Clerc, V.; Marques, S.; Suel, A.; Huet, S.; Hamama, L.; Voisine, L.; Auperpin, E.; Jourdan, M.; Barrot, L.; Prieur, R.; et al. QTL mapping of carrot resistance to leaf blight with connected populations: Stability across years and consequences for breeding. Theor. Appl. Genet. 2015, 128, 2177–2187. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho, A.D.F.; Silva, G.O. Divergência genética entre genótipos de cenoura através de caracteres agronômicos. Rev. Agro@mbiente 2017, 11, 137–144. [Google Scholar] [CrossRef]
- Janaszek, M.A.; Trajer, J.J. Method of evaluating diversity of carrot roots using a self-organizing map and image data. Comput. Electron. Agric. 2011, 79, 75–81. [Google Scholar] [CrossRef]
- Monyr, N.; Agoubi, B.; Berrada, M.; Abdelaziz, A.; El Hmaidi, A. Topological maps of Kohonen Self-Organization (SOM) applied to the study of sediments contaminated with heavy metals. Am. J. Eng. Res. 2016, 5, 99–105. [Google Scholar]
- Da Silva, G.O.; Vieira, J.V.; Vilela, M.S. Seleção de caracteres de cenoura cultivada em dois sistemas de produção agroecológicos no Distrito Federal. Rev. Ceres 2009, 56, 595–601. [Google Scholar]
- Neupane, K.; Baysal-Gurel, F. Automatic Identification and Monitoring of Plant Diseases Using Unmanned Aerial Vehi-cles: A Review. Remote Sens. 2021, 13, 3841. [Google Scholar] [CrossRef]
- Sweet, D.D.; Tirado, S.B.; Springer, N.M.; Hirsch, C.N.; Hirsch, C.D. Opportunities and challenges in phenotyping row crops using drone-based RGB imaging. Plant Phenome J. 2022, 5, e20044. [Google Scholar] [CrossRef]
- Lee, D.H.; Jeong, C.H.; Go, S.H.; Park, J.H. Evaluation of applicability of RGB image using support vector machine regression for estimation of leaf chlorophyll content of onion and garlic. Korean J. Remote Sens. 2021, 37, 1669–1683. [Google Scholar] [CrossRef]
- Olivoto, T.; Nardino, M. MGIDI: Toward an effective multivariate selection in biological experiments. Bioinformatics 2021, 37, 1383–1389. [Google Scholar] [CrossRef] [PubMed]






| Vegetation Indices | Equations ¹ | References |
|---|---|---|
| Brightness Index (BI) | √[(R2 + G2 + B2)/3] | [16] |
| Green Leaf Index (GLI) | (2G − R − B)/(2G + R + B) | [17] |
| Primary Colors Hue Index (HI) | (2R − G − B)/(G − B) | [18] |
| Overall Hue Index (HUE) | Atan [2(B − G − R)/30.5(G − R)] | [18] |
| Normalized Green Red Difference Index (NGRDI) | (G − R)(G + R) | [19] |
| Soil Color Index (SCI) | (R − G)/(R + G) | [20] |
| Spectral Slope Saturation Index (SI) | (R − B)/(R + B) | [18] |
| Visible Atmospherically Resistant Index (VARI) | (G − R)/(G + R − B) | [21] |
| areaporc | Plot area (cm2) |
| Mean Square | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SV | DF | BP | DIS | SHP | RL | RD | LW | MRP | URP | TYH | MYH |
| Block | 2 | 29.02 | 0.62 | 853.48 | 22.33 | 0.36 | 0.31 | 11.28 | 11.28 | 162.8 | 85.35 |
| Genotype | 59 | 72.99 ** | 0.81 ** | 219.81 ** | 5.69 ** | 0.14 ** | 0.4 ** | 83.27 * | 83.27 * | 88.99 ** | 81.52 ** |
| Residual | 118 | 7.69 | 0.34 | 60.47 | 1.38 | 0.02 | 0.05 | 52.58 | 52.58 | 9.66 | 9.64 |
| Mean | 3.72 | 2.6 | 15.96 | 16.6 | 2.65 | 1.16 | 82.04 | 17.96 | 23.53 | 19.44 | |
| CV | 74.58 | 22.42 | 48.73 | 7.06 | 5.6 | 19.69 | 8.84 | 40.37 | 13.21 | 15.97 | |
| Min | 0 | 1 | 1.54 | 11.8 | 1.9 | 0.35 | 51.39 | 2.65 | 7.9 | 5.37 | |
| Max | 37 | 5 | 59.26 | 21.3 | 3.2 | 2.43 | 97.35 | 48.61 | 42.4 | 38.43 | |
| F2 | 24.33 | 0.27 | 73.27 | 1.9 | 0.05 | 0.13 | 27.76 | 27.76 | 29.66 | 27.17 | |
| E2 | 2.56 | 0.11 | 20.15 | 0.46 | 0.01 | 0.02 | 17.53 | 17.53 | 3.22 | 3.21 | |
| G2 | 21.74 | 0.15 | 53.11 | 1.44 | 0.04 | 0.12 | 10.23 | 10.23 | 26.45 | 23.96 | |
| h2 | 89.46 | 55.11 | 72.48 | 75.8 | 83.96 | 87.01 | 36.86 | 36.86 | 89.15 | 88.17 | |
| CVg | 125.43 | 15.25 | 45.66 | 7.22 | 7.4 | 29.42 | 3.9 | 17.81 | 21.85 | 25.18 | |
| CVg/CVe | 1.68 | 0.68 | 0.94 | 1.02 | 1.32 | 1.49 | 0.44 | 0.44 | 1.65 | 1.58 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coelho, F.G.T.; Maciel, G.M.; Siquieroli, A.C.S.; Gallis, R.B.d.A.; Oliveira, C.S.d.; Ribeiro, A.L.A.; Pereira, L.M. High-Throughput Phenotyping for the Evaluation of Agronomic Potential and Root Quality in Tropical Carrot Using RGB Sensors. Agriculture 2024, 14, 710. https://doi.org/10.3390/agriculture14050710
Coelho FGT, Maciel GM, Siquieroli ACS, Gallis RBdA, Oliveira CSd, Ribeiro ALA, Pereira LM. High-Throughput Phenotyping for the Evaluation of Agronomic Potential and Root Quality in Tropical Carrot Using RGB Sensors. Agriculture. 2024; 14(5):710. https://doi.org/10.3390/agriculture14050710
Chicago/Turabian StyleCoelho, Fernanda Gabriela Teixeira, Gabriel Mascarenhas Maciel, Ana Carolina Silva Siquieroli, Rodrigo Bezerra de Araújo Gallis, Camila Soares de Oliveira, Ana Luisa Alves Ribeiro, and Lucas Medeiros Pereira. 2024. "High-Throughput Phenotyping for the Evaluation of Agronomic Potential and Root Quality in Tropical Carrot Using RGB Sensors" Agriculture 14, no. 5: 710. https://doi.org/10.3390/agriculture14050710





