Preharvest Sprouting in Quinoa: A New Screening Method Adapted to Panicles and GWAS Components
Abstract
:1. Introduction
2. Results
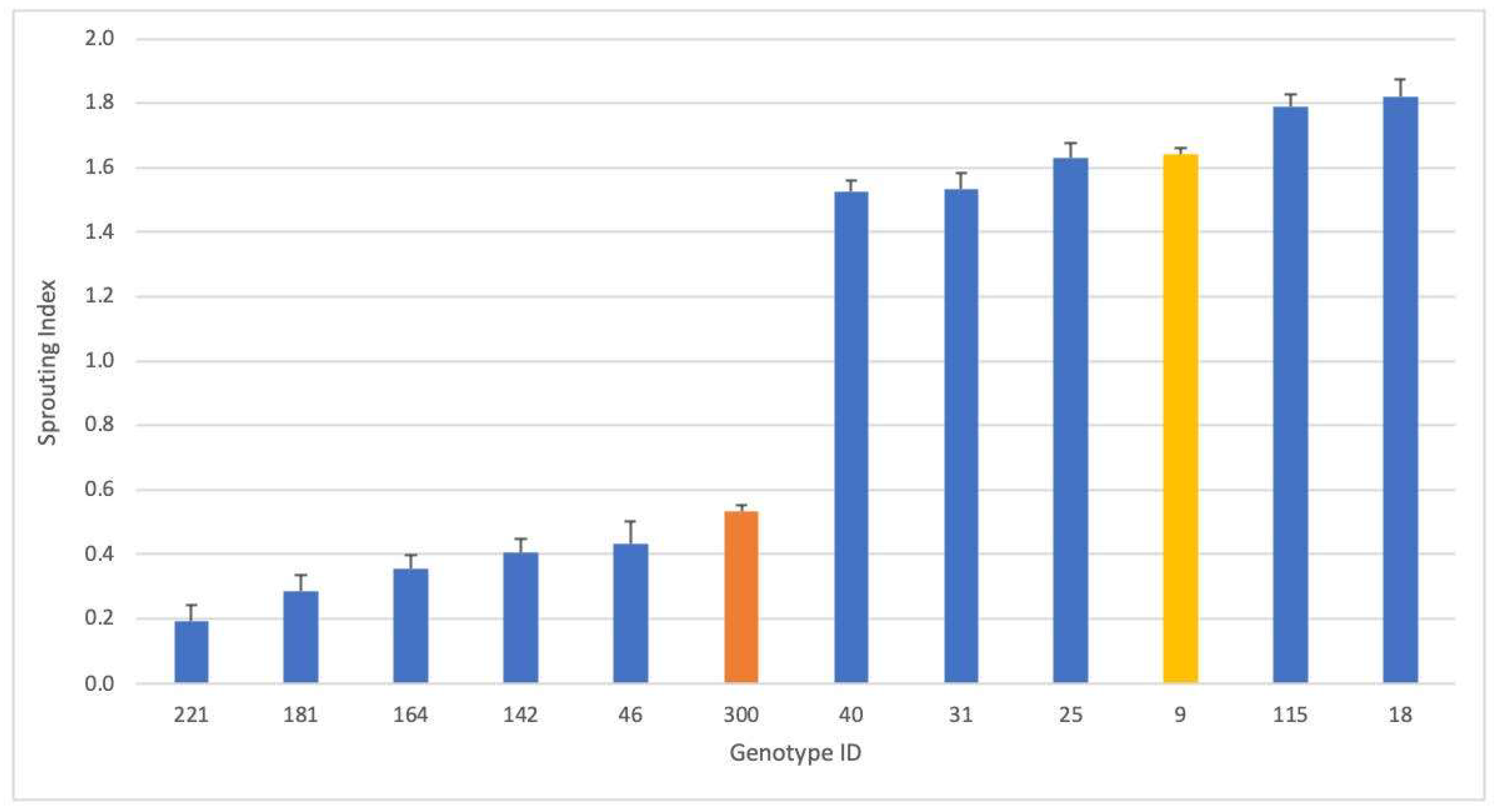
2.1. Genotype Rankings for PHS Tolerance
2.2. Addressing Population Stratification through PCA and an Indicator
2.3. Genome-Wide Association Study and Functional Annotations
3. Discussion
4. Materials and Methods
4.1. Germplasm
4.2. Experimental Design and Greenhouse Conditions
4.3. Development of the PHS Scoring Scale
4.4. Data Collection and Analysis
4.5. Phenotypic and Genotypic Data Integration for PHS Association Study
4.6. Population Structure Analysis
4.7. Association Study for Integrating Planting Date Effects Using an Additive–Additive (AA) Model
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bewley, D.J.; Black, M. Seeds: Physiology of Development and Germination; Springer: New York, NY, USA, 2013. [Google Scholar]
- Benech-Arnold, R.L. Bases of preharvest sprouting resistance in barley: Physiology, molecular biology and environmental control of dormancy in the barley grain in Barley science. In Recent Advances from Molecular Biology to Agronomy of Yield and Quality; Slafer, G.A., Molina-Cano, J.L., Savin, R., Araus, J.L., Romagosa, I., Eds.; Food Product Press: New York, NY, USA, 2001; pp. 481–502. [Google Scholar]
- Vetch, J.M.; Stougaard, R.N.; Martin, J.M.; Giroux, M.J. Revealing the genetic mechanisms of pre-harvest sprouting in hexaploid wheat (Triticum aestivum L.). Plant Sci. 2019, 281, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Ceccato, D.V.; Bertero, H.D.; Batlla, D. Environmental control of dormancy in quinoa (Chenopodium quinoa) seeds: Two potential genetic resources for pre-harvest sprouting tolerance. Seed Sci. Res. 2011, 21, 133–141. [Google Scholar] [CrossRef]
- Bazile, D.; Bertero, H.D.; Nieto, C. State of the Art Report on Quinoa around the World in 2013; FAO: Rome, Italy, 2015. [Google Scholar]
- Hinojosa, L.; González, J.A.; Barrios-Masias, F.H.; Fuentes, F.; Murphy, K.M. Quinoa abiotic stress responses: A review. Plants 2018, 7, 106. [Google Scholar] [CrossRef]
- McGinty, E.M.; Murphy, K.M.; Hauvermale, A.L. Seed dormancy and preharvest sprouting in quinoa (Chenopodium quinoa Willd). Plants 2021, 10, 458. [Google Scholar] [CrossRef]
- Paterson, A.H.; Sorrells, M.E.; Obendorf, R.L. Methods of evaluation for preharvest sprouting resistance in wheat breeding programs. Can. J. Plant Sci. 1989, 69, 681–689. [Google Scholar] [CrossRef]
- Chono, M.; Honda, I.; Shinoda, S.; Kushiro, T.; Kamiya, Y.; Nambara, E.; Watanabe, Y. Field studies on the regulation of abscisic acid content and germinability during grain development of barley: Molecular and chemical analysis of pre-harvest sprouting. J. Exp. Bot. 2006, 57, 2421–2434. [Google Scholar] [CrossRef]
- Cabral, A.L.; Jordan, M.C.; Mccartney, C.A.; You, F.M.; Humphreys, D.G.; Maclachlan, R.; Pozniak, C.J. Identification of candidate genes, regions and markers for pre-harvest sprouting resistance in wheat. BMC Plant Biol. 2014, 14, 340. [Google Scholar] [CrossRef]
- Gao, X.; Hu, C.; Li, H.; Yao, Y.; Meng, M.; Dong, J.; Li, X. Factors affecting pre-harvest sprouting resistance in wheat (Triticum aestivum L.): A review. J. Anim. Plant Sci. 2013, 23, 556–565. [Google Scholar]
- Ceccato, D.; Bertero, D.; Batlla, D.; Galati, B. Structural aspects of dormancy in quinoa (Chenopodium quinoa): Importance and possible action mechanisms of the seed coat. Seed Sci. Res. 2015, 25, 267–275. [Google Scholar] [CrossRef]
- Bertero, H.D.; Benech-Arnold, R. Release from dormancy during seed development in quinoa. Variation associated to cultivar origin. In Proceedings of the ASA (American Society of Agronomy), CSSA (Crop Science Society of America), SSSA (Soil Science Society of America) Meeting, Minneapolis, MN, USA, 10–13 November 2000. [Google Scholar]
- McGinty, E.M.; Craine, E.B.; Miller, N.D.; Ocana-Gallegos, C.; Spalding, E.P.; Murphy, K.M.; Hauvermale, A.L. Evaluating relationships between seed morphological traits and seed dormancy in Chenopodium quinoa Willd. Front. Plant Sci. 2023, 14, 1161165. [Google Scholar] [CrossRef]
- Sajeev, N.; Koornneef, M.; Bentsink, L. A commitment for life: Decades of unraveling the molecular mechanisms behind seed dormancy and germination. Plant Cell 2024, 36, 1358–1376. [Google Scholar] [CrossRef] [PubMed]
- Baskin, J.M.; Baskin, C.C. A classification system for seed dormancy. Seed Sci. Res. 2004, 14, 1–16. [Google Scholar] [CrossRef]
- Wu, Q.; Bai, X.; Wu, X.; Xiang, D.; Wan, Y.; Luo, Y.; Zhao, G. Transcriptome profiling identifies transcription factors and key homologs involved in seed dormancy and germination regulation of Chenopodium quinoa. Plant Physiol. Biochem. 2020, 151, 443–456. [Google Scholar] [CrossRef]
- Gubler, F.; Millar, A.A.; Jacobsen, J.V. Dormancy release, ABA and pre-harvest sprouting. Curr. Opin. Plant Biol. 2005, 8, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.; Reeves, W.; Ariizumi, T.; Steber, C. Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 2008, 59, 387–415. [Google Scholar] [CrossRef] [PubMed]
- Yan, A.; Chen, Z. The control of seed dormancy and germination by temperature, light and nitrate. Bot. Rev. 2020, 86, 39–75. [Google Scholar] [CrossRef]
- Tai, L.; Wang, H.J.; Xu, X.J.; Sun, W.H.; Ju, L.; Liu, W.T.; Chen, K.M. Pre-harvest sprouting in cereals: Genetic and biochemical mechanisms. J. Exp. Bot. 2021, 72, 2857–2876. [Google Scholar] [CrossRef] [PubMed]
- McMaster, G.J.; Derera, N.F. Methodology and sample preparation when screening for sprouting damage in cereals. Cereal Res. Commun. 1976, 4, 251–254. [Google Scholar]
- Perten, H. Application of the falling number method for evaluating alpha-amylase activity. Cereal Chem. 1964, 41, 127–140. [Google Scholar]
- He, Y.; Lin, Y.-L.; Chen, C.; Tsai, M.-H.; Lin, A.H.-M. Impacts of Starch and the Interactions between Starch and Other Macromolecules on Wheat Falling Number. Compr. Rev. Food Sci. Food Saf. 2019, 18, 641–654. [Google Scholar] [CrossRef]
- Hu, Y.; Sjoberg, S.M.; Chen, C.; Hauvermale, A.L.; Morris, C.F.; Delwiche, S.R.; Cannon, A.E.; Steber, C.M.; Zhang, Z. As the number falls, alternatives to the Hagberg-Perten falling number method: A review. Compr. Rev. Food Sci. Food Saf. 2022, 21, 2105–2117. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, K.; Nyanzi, F. Enzyme activities in quinoa (Chenopodium quinoa). Int. J. Food Sci. Technol. 1989, 24, 543–551. [Google Scholar] [CrossRef]
- Rosa, M.; Hilal, M.; González, J.A.; Prado, F.E. Changes in soluble carbohydrates and related enzymes induced by low temperature during early developmental stages of quinoa (Chenopodium quinoa) seedlings. J. Plant Physiol. 2004, 161, 683–689. [Google Scholar] [CrossRef] [PubMed]
- Hemalatha, P.; Bomzan, D.P.; Sathyendra Rao, B.V.; Sreerama, Y.N. Distribution of phenolic antioxidants in whole and milled fractions of quinoa and their inhibitory effects on α-amylase and α-glucosidase activities. Food Chem. 2016, 199, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Munkvold, J.D.; Tanaka, J.; Benscher, D.; Sorrells, M.E. Mapping quantitative trait loci for preharvest sprouting resistance in white wheat. Theor. Appl. Genet. 2009, 119, 1223–1235. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, M.V.; Barrero, J.M.; Corbineau, F.; Gubler, F.; Benech-Arnold, R.L. Dormancy in cereals (Not too much, not so little): About the mechanisms behind this trait. Seed Sci. Res. 2015, 25, 99–119. [Google Scholar]
- Depauw, R.; Mccaig, T. Components of variation, heritabilities and correlations for indices of sprouting tolerance and seed dormancy in Triticum spp. Euphytica 1991, 52, 221–229. [Google Scholar] [CrossRef]
- DePauw, R.M.; Knox, R.E.; Singh, A.K.; Fox, S.L.; Humphreys, D.G.; Hucl, P. Developing standardized methods for breeding preharvest sprouting resistant wheat, challenges and successes in Canadian wheat. Euphytica 2012, 188, 7–14. [Google Scholar] [CrossRef]
- Lopez-Marques, R.L.; Norrevang, A.F.; Ache, P.; Moog, M.; Visintainer, D.; Wendt, T.; Palmgren, M. Prospects for the accelerated improvement of the resilient crop quinoa. J. Exp. Bot. 2020, 71, 5333–5347. [Google Scholar] [CrossRef]
- Kulwal, P.L.; Kumar, N.; Gaur, A.; Khurana, P.; Khurana, J.P.; Tyagi, A.K.; Gupta, P.K. Mapping of a major QTL for pre-harvest sprouting tolerance on chromosome 3A in bread wheat. Theor. Appl. Genet. 2005, 111, 1052–1059. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Z. GAPIT version 3: Boosting power and accuracy for genomic association and prediction. Genom. Proteom. Bioinform. 2021, 19, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Liu, X.; Zhou, Y.; Summers, R.M.; Zhang, Z. BLINK: A package for the next level of genome-wide association studies with both individuals and markers in the millions. GigaScience 2018, 8, giy154. [Google Scholar] [CrossRef] [PubMed]
- Patiranage, D.S.; Rey, E.; Emrani, N.; Wellman, G.; Schmid, K.; Schmöckel, S.M.; Tester, M. Genome-wide association study in quinoa reveals selection pattern typical for crops with a short breeding history. eLife 2022, 11, e66873. [Google Scholar] [CrossRef] [PubMed]
- Peterson, A.J.; Murphy, K.M. Quinoa cultivation for temperate North America: Considerations and areas for investigation. In Quinoa: Improvement and Sustainable Production; Bazile, C., Bertero, M., Nieto, F., Eds.; Wiley & Sons: Hoboken, NJ, USA, 2015. [Google Scholar]
- Rasul, G.; Humphreys, G.D.; Wu, J.; Brûlé-Babel, A.; Fofana, B.; Glover, K.D. Evaluation of preharvest sprouting traits in a collection of spring wheat germplasm using genotype and genotype x environment interaction model. Plant Breed. 2012, 131, 244–251. [Google Scholar] [CrossRef]
- Xiang, Y.; Tang, N.; Du, H.; Ye, H.; Xiong, L. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 2008, 148, 1938–1952. [Google Scholar] [CrossRef] [PubMed]
- Mei, C.; Jiang, S.C.; Lu, Y.F.; Wu, F.Q.; Yu, Y.T.; Liang, S.; Zhang, D.P. Arabidopsis pentatricopeptide repeat protein SOAR1 plays a critical role in abscisic acid signaling. J. Exp. Bot. 2014, 65, 5317–5330. [Google Scholar] [CrossRef]
- Zhao, Y.; Cai, M.; Zhang, X.; Li, Y.; Zhang, J.; Zhao, H.; Qiu, F. Genome-wide identification, evolution and expression analysis of mTERF gene family in maize. PLoS ONE 2014, 9, e94126. [Google Scholar] [CrossRef] [PubMed]
- Gianinetti, A.; Finocchiaro, F.; Bagnaresi, P.; Zechini, A.; Faccioli, P.; Cattivelli, L.; Biselli, C. Seed dormancy involves a transcriptional program that supports early plastid functionality during imbibition. Plants 2018, 7, 35. [Google Scholar] [CrossRef]
- Li, T.; Pan, W.; Yuan, Y.; Liu, Y.; Li, Y.; Wu, X.; Cui, L. Identification, characterization, and expression profile Analysis of the mTERF gene family and its role in the response to abiotic stress in barley (Hordeum vulgare L.). Front. Plant Sci. 2021, 12, 684619. [Google Scholar] [CrossRef]
- Robles, P.; Quesada, V. Research progress in the molecular functions of plant mTERF proteins. Cells 2021, 10, 205. [Google Scholar] [CrossRef]
- Kanai, M.; Nishimura, M.; Hayashi, M. A peroxisomal ABC transporter promotes seed germination by inducing pectin degradation under the control of ABI5. Plant J. 2010, 62, 936–947. [Google Scholar] [CrossRef] [PubMed]
- Kebrom, T.H.; Mullet, J.E. Transcriptome profiling of tiller buds provides new insights into PhyB regulation of tillering and indeterminate growth in sorghum. Plant Physiol. 2016, 170, 2232–2250. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Wu, Y.; Guo, J.; Du, B.; Chen, R.; Zhu, L.; He, G. A rice lectin receptor-like kinase that is involved in innate immune responses also contributes to seed germination. Plant J. 2013, 76, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Ühlken, C.; Horvath, B.; Stadler, R.; Sauer, N.; Weingartner, M. MAIN-LIKE 1 is a crucial factor for correct cell division and differentiation in Arabidopsis thaliana. Plant J. 2014, 78, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Martinez, S.A.; Godoy, J.; Huang, M.; Zhang, Z.; Carter, A.H.; Garland Campbell, K.A.; Steber, C.M. Genome-wide association mapping for tolerance to preharvest sprouting and low falling numbers in wheat. Front. Plant Sci. 2018, 9, 141. [Google Scholar] [CrossRef] [PubMed]
- Sosa-Zuniga, V.; Brito, V.; Fuentes, F.; Steinfort, U. Phenological growth stages of quinoa (Chenopodium quinoa) based on the BBCH scale. Ann. Appl. Biol. 2017, 171, 117–124. [Google Scholar] [CrossRef]
- Stanschewski, C.S.; Rey, E.; Fiene, G.; Craine, E.B.; Wellman, G.; Melino, V.J.; Quinoa Phenotyping Consortium. Quinoa phenotyping methodologies: An international consensus. Plants 2021, 10, 1759. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: http://www.R-project.org/ (accessed on 15 March 2024).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Yutani, H. Welcome to the tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Tierney, N.; Cook, D.; McBain, M.; Fay, C. naniar: Data Structures, Summaries, and Visualisations for Missing Data. R Package Version 0.6.1.2021. 2021. Available online: https://CRAN.R-project.org/package=naniar (accessed on 15 March 2024).
- Tierney, N. Visdat: Visualising Whole Data Frames. J. Open Source Softw. 2017, 2, 355. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. Available online: https://ggplot2.tidyverse.org (accessed on 15 March 2024).
- Sarkar, D. Lattice: Multivariate Data Visualization with R; Springer: New York, NY, USA, 2008; ISBN 978-0-387-75968-5. [Google Scholar]
- McCulloch, W.S.; Pitts, W. A logical calculus of the ideas immanent in nervous activity. Bull. Math. Biophys. 1943, 5, 115–133. [Google Scholar] [CrossRef]
- Mares, D.J. Preservation of dormancy in freshly harvested wheat grain. Aust. J. Agric. Res. 1983, 34, 33–38. [Google Scholar] [CrossRef]
- Romero, G.; Heredia, A.; Chaparro-Zambrano, H.N. Germinative potential in quinoa (Chenopodium quinoa Willd.) seeds stored under cool conditions. Rev. UDCA Actual. Divulg. Científica 2018, 21, 341–350. [Google Scholar]
- Nelson, S.K.; Kanno, Y.; Seo, M.; Steber, C.M. Seed dormancy loss from dry after-ripening is associated with increasing gibberellin hormone levels in Arabidopsis thaliana. Front. Plant Sci. 2023, 14, 1145414. [Google Scholar] [CrossRef] [PubMed]







| SNP | Locus | Gene List (DAVID) |
|---|---|---|
| Cq1B_66402375 | XM_021904375 | basic leucine zipper 23-like (LOC110724880) |
| Cq2A_11615394 | XM_021877804 | uncharacterized mitochondrial protein AtMg00810-like (LOC110700265) |
| Cq2B_60857052 | XM_021883400 | glutamyl-tRNA(Gln) amidotransferase subunit C, chloroplastic/mitochondrial-like (LOC110705514) |
| Cq2B_9387424 | XM_021895300 | putative calcium-transporting ATPase 11, plasma membrane-type (LOC110716652) |
| Cq3A_10231545 | XM_021874234 | transcription termination factor MTERF2, chloroplastic-like (LOC110696907) |
| Cq3A_14217269 | XM_021883755 | uncharacterized LOC110705822 (LOC110705822) |
| Cq4B_4014100 | XR_002507314 | uncharacterized LOC110701120 (LOC110701120) |
| Cq4B_56717106 | XM_021880529 | katanin p80 WD40 repeat-containing subunit B1 homolog (LOC110702783) |
| Cq5A_582429 | XM_021887486 | protein MAIN-LIKE 1-like (LOC110709270) |
| Cq6A_8709044 | XM_021911740 | uncharacterized LOC110731845 (LOC110731845) |
| Cq6B_83520732 | XM_021858518 | ABC transporter G family member 11-like (LOC110682235) |
| Cq6B_9876457 | XR_002509647 | uncharacterized LOC110712666 (LOC110712666) |
| Cq7A_17313829 | XM_021919715 | peter Pan-like protein (LOC110739250) |
| Cq7A_17874020 | XM_021919734 | heavy metal-associated isoprenylated plant protein 36-like (LOC110739269) |
| Cq8A_9702538 | XM_021910407 | pentatricopeptide repeat-containing protein At1g71490-like (LOC110730586) |
| Cq9B_12163476 | XM_021894216 | G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 (LOC110715629) |
| Cq9B_24343108 | XR_002510671 | uncharacterized LOC110717110 (LOC110717110) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ocaña-Gallegos, C.; Liang, M.; McGinty, E.; Zhang, Z.; Murphy, K.M.; Hauvermale, A.L. Preharvest Sprouting in Quinoa: A New Screening Method Adapted to Panicles and GWAS Components. Plants 2024, 13, 1297. https://doi.org/10.3390/plants13101297
Ocaña-Gallegos C, Liang M, McGinty E, Zhang Z, Murphy KM, Hauvermale AL. Preharvest Sprouting in Quinoa: A New Screening Method Adapted to Panicles and GWAS Components. Plants. 2024; 13(10):1297. https://doi.org/10.3390/plants13101297
Chicago/Turabian StyleOcaña-Gallegos, Cristina, Meijing Liang, Emma McGinty, Zhiwu Zhang, Kevin M. Murphy, and Amber L. Hauvermale. 2024. "Preharvest Sprouting in Quinoa: A New Screening Method Adapted to Panicles and GWAS Components" Plants 13, no. 10: 1297. https://doi.org/10.3390/plants13101297







