Genome-Wide Identification of Members of the Soybean CBL Gene Family and Characterization of the Functional Role of GmCBL1 in Responses to Saline and Alkaline Stress
Abstract
:1. Introduction
2. Results
2.1. Identification of the G. max CBL Gene Family
2.2. Phylogenetic Analyses and Classification of CBL Genes
2.3. GmCBL Genes Syntenic Relationship, Gene Structure, and Protein Structure Analyses
2.4. GmCBL Chromosomal Distribution and Motif Analyses
2.5. Cis-Acting Element Analyses of GmCBL Gene Promoters
2.6. Multiple Sequence Alignment
2.7. Tissue-Specific Expression Profiling of GmCBLs
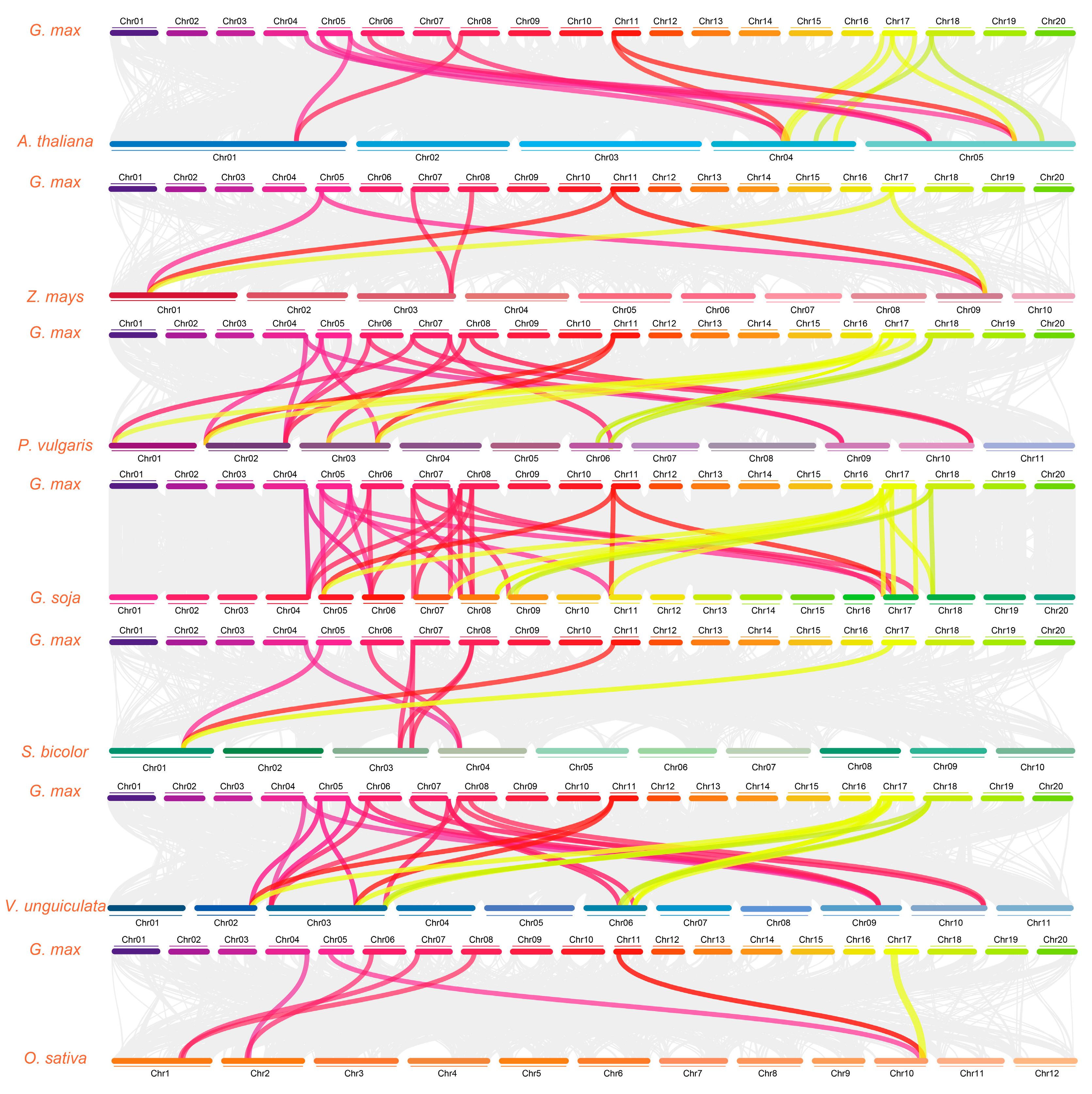
2.8. CBL Gene Synteny Analysis
2.9. Characterization of GmCBL Transcriptional Responses to Abiotic Stress and Hormone Stimulation
2.10. GmCBL1 Subcellular Localization and GmCBL1 Overexpression Enhances Soybean Saline–Alkaline Tolerance
3. Materials and Methods
3.1. Plant Materials and Treatment Conditions
3.2. Soybean CBL Gene Family Identification
3.3. Evolutionary, Tertiary Structure, Gene Structure, Promoter, Conserved Domain, and Synteny Analyses of GmCBLs
3.4. GmCBL Expression Analyses
3.5. qPCR
3.6. GmCBL1 Overexpression in Soybean Hairy Roots and Subcellular Localization Assays
3.7. Statistical Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Pagano, M.; Miransari, M. The Importance of Soybean Production Worldwide; Academic Press: New York, NY, USA, 2016; pp. 1–26. [Google Scholar]
- Cao, D.; Li, Y.; Liu, B.; Kong, F.; Tran, L.-S.P. Adaptive Mechanisms of Soybean Grown on Salt-Affected Soils. Land. Degrad. Dev. 2018, 29, 1054–1064. [Google Scholar] [CrossRef]
- Joshi, S.; Nath, J.; Singh, A.K.; Pareek, A.; Joshi, R. Ion transporters and their regulatory signal transduction mechanisms for salinity tolerance in plants. Physiol. Plant. 2022, 174, e13702. [Google Scholar] [CrossRef]
- Choi, W.-G.; Toyota, M.; Kim, S.-H.; Hilleary, R.; Gilroy, S. Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. USA 2014, 111, 6497–6502. [Google Scholar] [CrossRef]
- Knight, H.; Trewavas, A.J.; Knight, M.R. Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J. 1997, 12, 1067–1078. [Google Scholar] [CrossRef]
- Lecourieux, D.; Ranjeva, R.; Pugin, A. Calcium in plant defence-signalling pathways. New Phytol. 2006, 171, 249–269. [Google Scholar] [CrossRef]
- Hepler, P.K. Calcium: A Central Regulator of Plant Growth and Development. Plant Cell 2005, 17, 2142–2155. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhou, X.; Tao, M. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca(2+) influx. Nature 2019, 572, 341–346. [Google Scholar] [CrossRef]
- Aldon, D.; Mbengue, M.; Mazars, C.; Galaud, J.-P. Calcium Signalling in Plant Biotic Interactions. Int. J. Mol. Sci. 2018, 19, 665. [Google Scholar] [CrossRef]
- Zhu, K.; Chen, F.; Liu, J.; Chen, X.; Hewezi, T.; Cheng, Z.-M. Evolution of an intron-poor cluster of the CIPK gene family and expression in response to drought stress in soybean. Sci. Rep. 2016, 6, 28225. [Google Scholar] [CrossRef]
- Shi, J.; Kim, K.N.; Ritz, O.; Albrecht, V.; Gupta, R.; Harter, K.; Luan, S.; Kudla, J. Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell 1999, 11, 2393–2405. [Google Scholar]
- Kudla, J.; Xu, Q.; Harter, K.; Gruissem, W.; Luan, S. Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc. Natl. Acad. Sci. USA 1999, 96, 4718–4723. [Google Scholar] [CrossRef]
- Nagae, M.; Nozawa, A.; Koizumi, N.; Sano, H.; Hashimoto, H.; Sato, M.; Shimizu, T. The crystal structure of the novel calcium-binding protein AtCBL2 from Arabidopsis thaliana. J. Biol. Chem. 2003, 278, 42240–42246. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Huang, Y.; Xiong, L. Characterization of Stress-Responsive CIPK Genes in Rice for Stress Tolerance Improvement. Plant Physiol. 2007, 144, 1416–1428. [Google Scholar] [CrossRef]
- Gifford, J.L.; Walsh, M.P.; Vogel, H.J. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem. J. 2007, 405, 199–221. [Google Scholar] [CrossRef]
- Du, W.; Lin, H.; Chen, S.; Wu, Y.; Zhang, J.; Fuglsang, A.T.; Palmgren, M.G.; Wu, W.; Guo, Y. Phosphorylation of SOS3-Like Calcium-Binding Proteins by Their Interacting SOS2-Like Protein Kinases Is a Common Regulatory Mechanism in Arabidopsis. Plant Physiol. 2011, 156, 2235–2243. [Google Scholar] [CrossRef] [PubMed]
- Ishitani, M.; Liu, J.; Halfter, U.; Kim, C.-S.; Shi, W.; Zhu, J.-K. SOS3 Function in Plant Salt Tolerance Requires N-Myristoylation and Calcium Binding. Plant Cell 2000, 12, 1667–1677. [Google Scholar] [CrossRef]
- Ma, X.; Li, Q.-H.; Yu, Y.-N.; Qiao, Y.-M.; Haq, S.u.; Gong, Z.-H. The CBL–CIPK Pathway in Plant Response to Stress Signals. Int. J. Mol. Sci. 2020, 21, 5668. [Google Scholar] [CrossRef] [PubMed]
- Luan, S. The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 2009, 14, 37–42. [Google Scholar] [CrossRef]
- Albrecht, V.; Ritz, O.; Linder, S.; Harter, K.; Kudla, J. The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J. 2001, 20, 1051–1063. [Google Scholar] [CrossRef] [PubMed]
- Kolukisaoglu, U.N.; Weinl, S.; Blazevic, D.; Batistic, O.; Kudla, J.R. Calcium Sensors and Their Interacting Protein Kinases: Genomics of the Arabidopsis and Rice CBL-CIPK Signaling Networks. Plant Physiol. 2004, 134, 43–58. [Google Scholar] [CrossRef]
- Qiu, Q.S.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J.K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436–8441. [Google Scholar] [CrossRef]
- Halfter, U.; Ishitani, M.; Zhu, J.K. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 2000, 97, 3735–3740. [Google Scholar] [CrossRef]
- Quintero, F.J.; Ohta, M.; Shi, H.; Zhu, J.K.; Pardo, J.M. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. USA 2002, 99, 9061–9066. [Google Scholar] [CrossRef]
- Cheong, Y.H.; Kim, K.-N.; Pandey, G.K.; Gupta, R.; Grant, J.J.; Luan, S. CBL1, a Calcium Sensor That Differentially Regulates Salt, Drought, and Cold Responses in Arabidopsis. Plant Cell 2003, 15, 1833–1845. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, V.; Weinl, S.; Blazevic, D.; D’Angelo, C.; Batistic, O.; Kolukisaoglu, Ü.; Bock, R.; Schulz, B.; Harter, K.; Kudla, J. The calcium sensor CBL1 integrates plant responses to abiotic stresses. Plant J. 2003, 36, 457–470. [Google Scholar] [CrossRef]
- Cheong, Y.H.; Sung, S.J.; Kim, B.-G.; Pandey, G.K.; Cho, J.-S.; Kim, K.-N.; Luan, S. Constitutive overexpression of the calcium sensor CBL5 confers osmotic or drought stress tolerance in Arabidopsis. Mol. Cells 2010, 29, 159–165. [Google Scholar] [CrossRef]
- Zhang, F.; Li, L.; Jiao, Z.; Chen, Y.; Liu, H.; Chen, X.; Fu, J.; Wang, G.; Zheng, J. Characterization of the calcineurin B-Like (CBL) gene family in maize and functional analysis of ZmCBL9 under abscisic acid and abiotic stress treatments. Plant Sci. 2016, 253, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Plasencia, F.A.; Estrada, Y.; Flores, F.B.; Ortíz-Atienza, A.; Lozano, R.; Egea, I. The Ca2+ Sensor Calcineurin B–Like Protein 10 in Plants: Emerging New Crucial Roles for Plant Abiotic Stress Tolerance. Front. Plant Sci. 2021, 11, 599944. [Google Scholar] [CrossRef]
- Mähs, A.; Steinhorst, L.; Han, J.P.; Shen, L.K.; Wang, Y.; Kudla, J. The calcineurin B-like Ca2+ sensors CBL1 and CBL9 function in pollen germination and pollen tube growth in Arabidopsis. Mol. Plant 2013, 6, 1149–1162. [Google Scholar] [CrossRef]
- Gu, Z.; Meng, D.; Yang, Q.; Yuan, H.; Wang, A.; Li, W.; Chen, Q.; Zhang, Y.; Wang, D.; Li, T. A CBL gene, MdCBL5, controls the calcium signal and influences pollen tube growth in apple. Tree Genet. Genomes 2015, 11, 27. [Google Scholar] [CrossRef]
- Tang, R.-J.; Liu, H.; Yang, Y.; Yang, L.; Gao, X.-S.; Garcia, V.J.; Luan, S.; Zhang, H.-X. Tonoplast calcium sensors CBL2 and CBL3 control plant growth and ion homeostasis through regulating V-ATPase activity in Arabidopsis. Cell Res. 2012, 22, 1650–1665. [Google Scholar] [CrossRef]
- Eckert, C.; Offenborn, J.N.; Heinz, T.; Armarego-Marriott, T.; Schültke, S.; Zhang, C.; Hillmer, S.; Heilmann, M.; Schumacher, K.; Bock, R.; et al. The vacuolar calcium sensors CBL2 and CBL3 affect seed size and embryonic development in Arabidopsis thaliana. Plant J. 2014, 78, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, B.; Liu, W.-Z.; Li, H.; Wang, L.; Wang, B.; Deng, M.; Liang, W.; Deyholos, M.K.; Jiang, Y.-Q. Identification and characterization of CBL and CIPK gene families in canola (Brassica napus L.). BMC Plant Biol. 2014, 14, 8. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Liu, J.; Dong, C.; Cheng, Z.-M. The CBL and CIPK Gene Family in Grapevine (Vitis vinifera): Genome-Wide Analysis and Expression Profiles in Response to Various Abiotic Stresses. Front. Plant Sci. 2017, 8, 978. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef]
- Li, R.; Zhang, J.; Wei, J.; Wang, H.; Wang, Y.; Ma, R. Functions and Mechanisms of the Cbl–Cipk Signaling System in Plant Response to Abiotic Stress. Prog. Nat. Sci. 2009, 19, 667–676. [Google Scholar] [CrossRef]
- Chen, L.; Wang, Q.-Q.; Zhou, L.; Ren, F.; Li, D.-D.; Li, X.-B. Arabidopsis Cbl-Interacting Protein Kinase (Cipk6) Is Involved in Plant Response to Salt/Osmotic Stress and Aba. Mol. Biol. Rep. 2013, 40, 4759–4767. [Google Scholar] [CrossRef]
- Sánchez-Barrena, M.J.; Martínez-Ripoll, M.; Albert, A. Structural Biology of a Major Signaling Network That Regulates Plant Abiotic Stress: The Cbl-Cipk Mediated Pathway. Int. J. Mol. Sci. 2013, 14, 5734–5749. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.-K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant Hormone Regulation of Abiotic Stress Responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple Sequence Alignment Using Clustalw and Clustalx. Curr. Protoc. Bioinform. 2002, 2–3. [Google Scholar] [CrossRef]
- Edgar, R.C. Muscle: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. Meme Suite: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. Tbtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, Z.; Zhou, R.; Cheng, P.; Li, H.; Wang, Z.; Liu, Y.; Li, M.; Zhao, Z.; Hu, Z.; et al. Genome-Wide Analysis of Soybean Dnaja-Family Genes and Functional Characterization of Gmdnaja6 Responses to Saline and Alkaline Stress. Crop J. 2023, 11, 1230–1241. [Google Scholar] [CrossRef]
- Li, M.; Hou, L.; Liu, S.; Zhang, C.; Yang, W.; Pang, X.; Li, Y. Genome-Wide Identification and Expression Analysis of Nac Transcription Factors in Ziziphus jujuba Mill. Reveal Their Putative Regulatory Effects on Tissue Senescence and Abiotic Stress Responses. Ind. Crops Prod. 2021, 173, 114093. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative Pcr and the 2−Δδct Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Tóth, K.; Batek, J.; Stacey, G. Generation of Soybean (Glycine max) Transient Transgenic Roots. Curr. Protoc. Plant Biol. 2016, 1, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ginestet, C. Ggplot2: Elegant Graphics for Data Analysis. J. R. Stat. Soc. Ser. A Stat. Soc. 2011, 174, 245–246. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, R.; Gao, Z.; Chen, C.; Jiang, Z.; Shu, H. A Genome-Wide Analysis of the Expansin Genes in Malus × domestica. Mol. Genet. Genom. 2014, 289, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Fan, P.; Liu, H.; Tan, P.; Ma, W.; Mo, Z.; Zhao, J.; Chu, G.; Peng, F. Insight into the Cbl and Cipk Gene Families in Pecan (Carya illinoinensis): Identification, Evolution and Expression Patterns in Drought Response. BMC Plant Biol. 2022, 22, 221. [Google Scholar] [CrossRef]
- Ma, X.; Gai, W.-X.; Qiao, Y.-M.; Ali, M.; Wei, A.-M.; Luo, D.-X.; Li, Q.-H.; Gong, Z.-H. Identification of CBL and CIPK gene families and functional characterization of CaCIPK1 under Phytophthora capsici in pepper (Capsicum annuum L.). BMC Genom. 2019, 20, 775. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Zhang, G.; Sun, L.; Wang, J.; Hao, F. Genome-wide identification of CBL family and expression analysis of CBLs in response to potassium deficiency in cotton. PeerJ 2017, 5, e3653. [Google Scholar] [CrossRef]
- Villao-Uzho, L.; Chávez-Navarrete, T.; Pacheco-Coello, R.; Sánchez-Timm, E.; Santos-Ordóñez, E. Plant Promoters: Their Identification, Characterization, and Role in Gene Regulation. Genes 2023, 14, 1226. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Wang, Y.; Wang, M.; Li, T.; Zhou, Y.; Wang, X.; Wei, S.; He, G.; Yang, G. Identification and comprehensive analyses of the CBL and CIPK gene families in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 269. [Google Scholar] [CrossRef]
- Jiang, M.; Zhao, C.; Zhao, M.; Li, Y.; Wen, G. Phylogeny and Evolution of Calcineurin B-Like (CBL) Gene Family in Grass and Functional Analyses of Rice CBLs. J. Plant Biol. 2020, 63, 117–130. [Google Scholar] [CrossRef]
- Du, W.; Yang, J.; Ma, L.; Su, Q.; Pang, Y. Identification and Characterization of Abiotic Stress Responsive CBL-CIPK Family Genes in Medicago. Int. J. Mol. Sci. 2021, 22, 4634. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Yang, L.; Liu, Y.; Zhao, Y.; Han, T.; Miao, X.; Zhang, A. Calcineurin B-like protein 5 (SiCBL5) in Setaria italica enhances salt tolerance by regulating Na+ homeostasis. Crop J. 2022, 10, 234–242. [Google Scholar] [CrossRef]
- Kumar Meena, M.; Kumar Vishwakarma, N.; Tripathi, V.; Chattopadhyay, D. CBL-interacting protein kinase 25 contributes to root meristem development. J. Exp. Bot. 2018, 70, 133–147. [Google Scholar] [CrossRef]
- Pandey, G.K.; Grant, J.J.; Cheong, Y.H.; Kim, B.G.; Li, G.; Luan, S. Calcineurin-B-like protein CBL9 interacts with target kinase CIPK3 in the regulation of ABA response in seed germination. Mol. Plant 2008, 1, 238–248. [Google Scholar] [CrossRef]
- Wang, C.; Yuan, Z.; Li, S.; Wang, W.; Xue, R.; Tai, F. Characterization of eight CBL genes expressions in maize early seeding development. Acta Physiol. Plant. 2014, 36, 3307–3314. [Google Scholar] [CrossRef]
- Ketehouli, T.; Idrice Carther, K.F.; Noman, M.; Wang, F.-W.; Li, X.-W.; Li, H.-Y. Adaptation of Plants to Salt Stress: Characterization of Na+ and K+ Transporters and Role of CBL Gene Family in Regulating Salt Stress Response. Agronomy 2019, 9, 687. [Google Scholar] [CrossRef]
- Song, Z.; Dong, B.; Yang, Q.; Niu, L.; Li, H.; Cao, H.; Amin, R.; Meng, D.; Yujie, F. Screening of CBL genes in pigeon pea with focus on the functional analysis of CBL4 in abiotic stress tolerance and flavonoid biosynthesis. Environ. Exp. Bot. 2020, 177, 104102. [Google Scholar] [CrossRef]
- Mao, J.; Yuan, G.; Han, K.; Xiang, H.; Zeng, W.; Visser, R.G.F.; Bai, Y.; van der Linden, C.G.; Liu, H.; Wang, Q. Genome-wide identification of CBL family genes in Nicotiana tabacum and the functional analysis of NtCBL4A-1 under salt stress. Environ. Exp. Bot. 2023, 209, 105311. [Google Scholar] [CrossRef]
- Steinhorst, L.; Kudla, J. Calcium and Reactive Oxygen Species Rule the Waves of Signaling. Plant Physiol. 2013, 163, 471–485. [Google Scholar] [CrossRef]
- Shi, H.; Lee, B.-h.; Wu, S.-J.; Zhu, J.-K. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol. 2003, 21, 81–85. [Google Scholar] [CrossRef]









| Gene Name | Gene ID | Gene Location | ORF Length (bp) | Protein Length | Isoelectric Point | Molecular Weight (kDa) | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| GmCBL1 | Glyma.04G235900 | Chr4 50399334…50405613 | 672 | 224 | 4.4 | 25.68 | Cytoplasmic |
| GmCBL2 | Glyma.05G066800 | Chr5 6731396…6740875 | 642 | 214 | 4.49 | 24.22 | Nuclear |
| GmCBL3 | Glyma.05G217700 | Chr5 39770219…39774659 | 693 | 231 | 4.59 | 26.1 | Nuclear |
| GmCBL4 | Glyma.06G128700 | Chr6 10581579…10588773 | 672 | 224 | 4.42 | 25.55 | Cytoplasmic |
| GmCBL5 | Glyma.07G010800 | Chr7 824696…827823 | 792 | 264 | 4.76 | 30.44 | Cytoplasmic |
| GmCBL6 | Glyma.07G269200 | Chr7 44230392…44236986 | 681 | 227 | 4.39 | 25.78 | Extracellular matrix |
| GmCBL7 | Glyma.08G023700 | Chr8 1884590…1887532 | 708 | 236 | 4.53 | 26.73 | Nuclear |
| GmCBL8 | Glyma.08G194000 | Chr8 15628395…15631841 | 798 | 266 | 4.76 | 30.65 | Cytoplasmic |
| GmCBL9 | Glyma.08G332800 | Chr8 44992808…44997458 | 681 | 227 | 4.45 | 26.04 | Cytoplasmic |
| GmCBL10 | Glyma.09G016000 | Chr9 1233158…1239867 | 681 | 227 | 4.64 | 25.93 | Nuclear |
| GmCBL11 | Glyma.11G038900 | Chr11 2773817…2777295 | 639 | 213 | 4.31 | 24.40 | Nuclear |
| GmCBL12 | Glyma.17G004400 | Chr17 434853…441609 | 681 | 227 | 4.40 | 25.80 | Cytoplasmic |
| GmCBL13 | Glyma.17G149000 | Chr17 12354772…12359193 | 660 | 220 | 4.72 | 25.24 | Cytoplasmic |
| GmCBL14 | Glyma.17G229500 | Chr17 38443770…38448625 | 777 | 259 | 4.44 | 29.26 | Chloroplast |
| GmCBL15 | Glyma.18G074200 | Chr18 7033194…7037977 | 681 | 227 | 4.50 | 26.07 | Cytoplasmic |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, F.; Zhang, D.; Chen, Y.; Wu, J. Genome-Wide Identification of Members of the Soybean CBL Gene Family and Characterization of the Functional Role of GmCBL1 in Responses to Saline and Alkaline Stress. Plants 2024, 13, 1304. https://doi.org/10.3390/plants13101304
Jiao F, Zhang D, Chen Y, Wu J. Genome-Wide Identification of Members of the Soybean CBL Gene Family and Characterization of the Functional Role of GmCBL1 in Responses to Saline and Alkaline Stress. Plants. 2024; 13(10):1304. https://doi.org/10.3390/plants13101304
Chicago/Turabian StyleJiao, Feng, Dongdong Zhang, Yang Chen, and Jinhua Wu. 2024. "Genome-Wide Identification of Members of the Soybean CBL Gene Family and Characterization of the Functional Role of GmCBL1 in Responses to Saline and Alkaline Stress" Plants 13, no. 10: 1304. https://doi.org/10.3390/plants13101304




