Puzzle of Proteoform Variety—Where Is a Key?
Abstract
:1. Introduction
2. Standardization Aspects
3. Quantification of Proteoforms
4. Aspects of Generation of PTMs
5. Aspects of Proteoform Variety
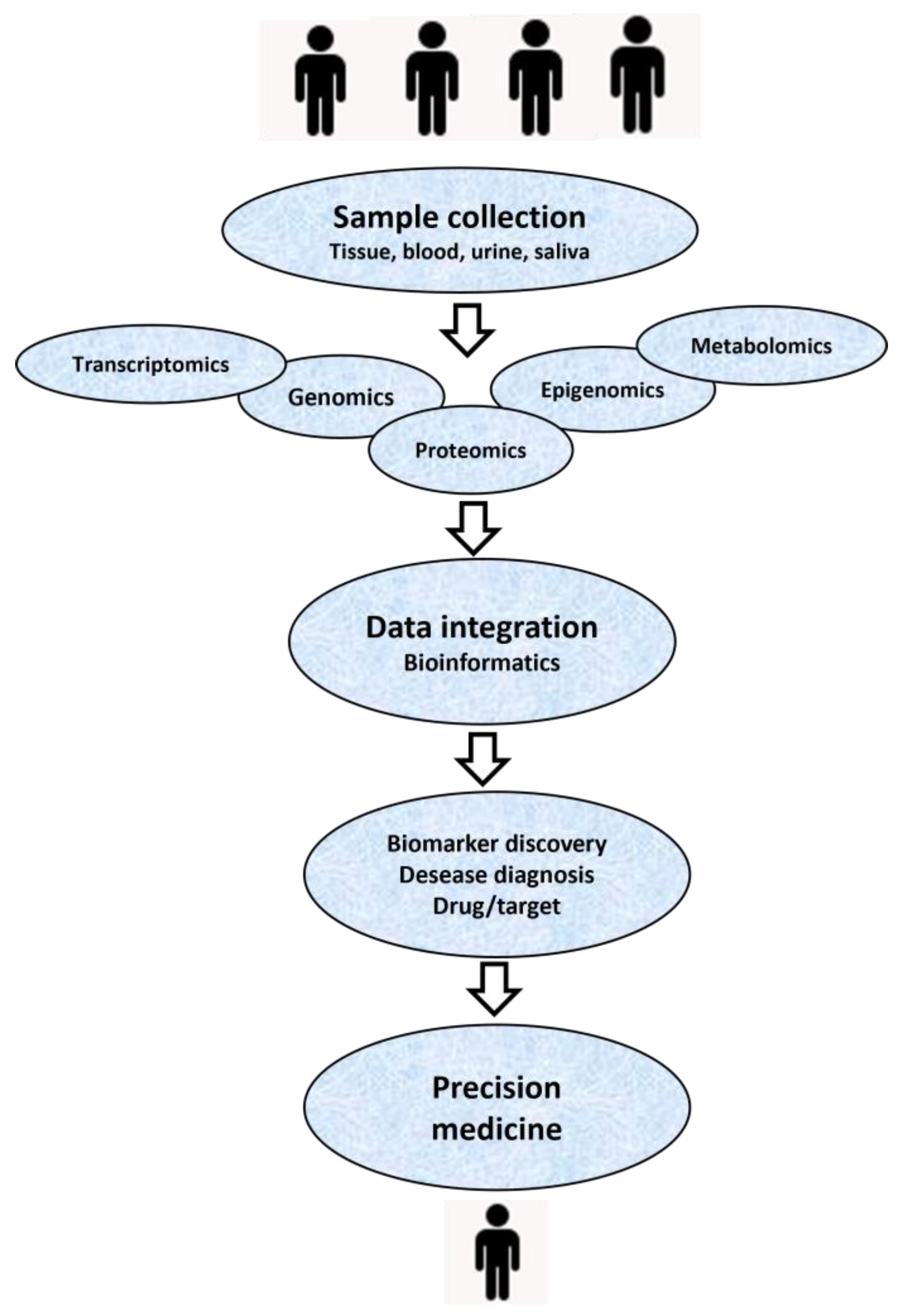
6. Role of Bioinformatics
7. Clinical Aspects of Proteoforms
8. Conclusions and Future Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Salzberg, S.L. Open questions: How many genes do we have? BMC Biol. 2018, 16, 94. [Google Scholar] [CrossRef] [PubMed]
- Aebersold, R.; Agar, J.N.; Amster, I.J.; Baker, M.S.; Bertozzi, C.R.; Boja, E.S.; Costello, C.E.; Cravatt, B.F.; Fenselau, C.; Garcia, B.A.; et al. How many human proteoforms are there? Nat. Chem. Biol. 2018, 14, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Marx, V. Inside the chase after those elusive proteoforms. Nat. Methods 2024, 21, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Schlüter, H.; Apweiler, R.; Holzhütter, H.-G.; Jungblut, P.R. Finding one’s way in proteomics: A protein species nomenclature. Chem. Cent. J. 2009, 3, 11. [Google Scholar] [CrossRef] [PubMed]
- Gunning, P.W. Protein Isoforms and Isozymes. In Encyclopedia of Life Sciences; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2006; ISBN 9780470015902. [Google Scholar]
- The UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res. 2012, 40, D71–D75. [Google Scholar] [CrossRef] [PubMed]
- Jungblut, P.; Thiede, B.; Zimmy-Arndt, U.; Muller, E.C.; Scheler, C.; Wittmann-Liebold, B.; Otto, A. Resolution power od 2-DE and identification of proteins from gels. Electrophoresis 1996, 17, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Jungblut, P.; Holzhütter, H.; Apweiler, R.; Schlüter, H. The speciation of the proteome. Chem. Cent. J. 2008, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Scheler, C.; Müller, E.-C.; Stahl, J.; Müller-Werdan, U.; Salnikow, J.; Jungblut, P. Identification and characterization of heat shock protein 27 protein species in human myocardial two-dimensional electrophoresis patterns. Electrophoresis 1997, 18, 2823–2831. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.M.; Kelleher, N.L. Proteoform: A single term describing protein complexity Lloyd. Nat. Methods 2013, 10, 186–187. [Google Scholar] [CrossRef] [PubMed]
- Carbonara, K.; Andonovski, M.; Coorssen, J.R. Proteomes Are of Proteoforms: Embracing the Complexity. Proteomes 2021, 9, 38. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Smith, L.M.; Agar, J.N.; Chamot-Rooke, J.; Danis, P.O.; Ge, Y.; Loo, J.A.; Paša-Tolić, L.; Tsybin, Y.O.; Kelleher, N.L. The Human Proteoform Project: Bringing Proteoforms to Life A Plan to Define the Human Proteome. Sci. Adv. 2020, 7, eabk0734. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.M.; Kelleher, N.L. Proteoforms as the next proteomics currency. Science 2018, 359, 1106–1107. [Google Scholar] [CrossRef] [PubMed]
- Fornelli, L.; Durbin, K.R.; Fellers, R.T.; Early, B.P.; Greer, J.B.; LeDuc, R.D.; Compton, P.D.; Kelleher, N.L. Advancing Top-down Analysis of the Human Proteome Using a Benchtop Quadrupole-Orbitrap Mass Spectrometer. J. Proteome Res. 2017, 16, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Tholey, A.; Schlüter, H. Top-down proteomics and proteoforms—Special issue. Proteomics 2024, 24, 2200375. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.M. Proteoforms and Proteoform Families: Past, Present, and Future. Methods Mol. Biol. 2022, 2500, 1–4. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Leduc, R.D.; Schwämmle, V.; Shortreed, M.R.; Cesnik, A.J.; Solntsev, S.K.; Shaw, J.B.; Martin, M.J.; Vizcaino, J.A.; Alpi, E.; Danis, P.; et al. ProForma: A Standard Proteoform Notation. J. Proteome Res. 2018, 17, 1321–1325. [Google Scholar] [CrossRef] [PubMed]
- LeDuc, R.D.; Deutsch, E.W.; Binz, P.-A.; Fellers, R.T.; Cesnik, A.J.; Klein, J.A.; Van Den Bossche, T.; Gabriels, R.; Yalavarthi, A.; Perez-Riverol, Y.; et al. Proteomics Standards Initiative’s ProForma 2.0: Unifying the Encoding of Proteoforms and Peptidoforms. J. Proteome Res. 2022, 21, 1189–1195. [Google Scholar] [CrossRef] [PubMed]
- Hollas, M.A.R.; Robey, M.T.; Fellers, R.T.; LeDuc, R.D.; Thomas, P.M.; Kelleher, N.L. The Human Proteoform Atlas: A FAIR community resource for experimentally derived proteoforms. Nucleic Acids Res. 2021, 50, D526–D533. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, L.V.; Millikin, R.J.; Shortreed, M.R.; Scalf, M.; Smith, L.M. Improving Proteoform Identifications in Complex Systems through Integration of Bottom-Up and Top-Down Data. J. Proteome Res. 2020, 19, 3510–3517. [Google Scholar] [CrossRef] [PubMed]
- Naryzhny, S. Inventory of proteoforms as a current challenge of proteomics: Some technical aspects. J. Proteom. 2019, 191, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Li, B.; Zhan, X.; Schlüter, H.; Jungblut, P.R.; Coorssen, J.R. Innovating the concept and practice of two-dimensional gel electrophoresis in the analysis of proteomes at the proteoform level. Proteomes 2019, 7, 36. [Google Scholar] [CrossRef]
- Naryzhny, S.N.; Maynskova, M.A.; Zgoda, V.G.; Ronzhina, N.L.; Kleyst, O.A.; Vakhrushev, I.V.; Archakov, A.I. Virtual-Experimental 2DE Approach in Chromosome-Centric Human Proteome Project. J. Proteome Res. 2016, 15, 525–530. [Google Scholar] [CrossRef]
- Naryzhny, S.N.; Zorina, E.S.; Kopylov, A.T.; Zgoda, V.G.; Kleyst, O.A.; Archakov, A.I. Next Steps on in Silico 2DE Analyses of Chromosome 18 Proteoforms. J. Proteome Res. 2018, 17, 4085–4096. [Google Scholar] [CrossRef] [PubMed]
- Petrenko, E.S.; Kopylov, A.T.; Kleist, O.A.; Legina, O.K.; Belyakova, N.V.; Pantina, R.A.; Naryzhny, S.N. Searching for Specific Markers of Glioblastoma: Analysis of Glioblastoma Cell Proteoforms. Cell Tissue Biol. 2018, 12, 455–459. [Google Scholar] [CrossRef]
- Naryzhny, S.; Zgoda, V.; Kopylov, A.; Petrenko, E.; Kleist, O.; Archakov, A. Variety and Dynamics of Proteoforms in the Human Proteome: Aspects of Markers for Hepatocellular Carcinoma. Proteomes 2017, 5, 33. [Google Scholar] [CrossRef] [PubMed]
- Naryzhny, S.; Klopov, N.; Ronzhina, N.; Zorina, E.; Zgoda, V.; Kleyst, O.; Belyakova, N.; Legina, O. A database for inventory of proteoform profiles: “2DE-pattern”. Electrophoresis 2020, 41, 1118–1124. [Google Scholar] [CrossRef]
- Marcus, K.; Lelong, C.; Rabilloud, T. What room for two-dimensional gel-based proteomics in a shotgun proteomics world? Proteomes 2020, 8, 17. [Google Scholar] [CrossRef] [PubMed]
- Rabilloud, T. Two-dimensional gel electrophoresis in proteomics: Old, old fashioned, but it still climbs up the mountains. Proteomics 2002, 2, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. Proteom. Protoc. Handb. 2005, 112, 571–607. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Rogers, L.D.; Overall, C.M. Proteolytic post-translational modification of proteins: Proteomic tools and methodology. Mol. Cell. Proteom. 2013, 12, 3532–3542. [Google Scholar] [CrossRef] [PubMed]
- Naryzhny, S.N.; Lee, H. Observation of multiple isoforms and specific proteolysis patterns of proliferating cell nuclear antigen in the context of cell cycle compartments and sample preparations. Proteomics 2003, 3, 930–936. [Google Scholar] [CrossRef] [PubMed]
- Rape, M.; Jentsch, S. Taking a bite: Proteasomal protein processing. Nat. Cell Biol. 2002, 4, E113–E116. [Google Scholar] [CrossRef] [PubMed]
- Sahu, I.; Glickman, M.H. Structural Insights into Substrate Recognition and Processing by the 20S Proteasome. Biomolecules 2021, 11, 148. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.W.; Petrera, A.; Schilling, O. The emerging role of the peptidome in biomarker discovery and degradome profiling. Biol. Chem. 2015, 396, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Jürgens, M.; Schrader, M. Peptidomic approaches in proteomic research. Curr. Opin. Mol. Ther. 2002, 4, 236–241. [Google Scholar] [PubMed]
- Wolf-Levy, H.; Javitt, A.; Eisenberg-Lerner, A.; Kacen, A.; Ulman, A.; Sheban, D.; Dassa, B.; Fishbain-Yoskovitz, V.; Carmona-Rivera, C.; Kramer, M.P.; et al. Revealing the cellular degradome by mass spectrometry analysis of proteasome-cleaved peptides. Nat. Biotechnol. 2018, 36, 1110–1116. [Google Scholar] [CrossRef] [PubMed]
- Puente, X.S.; Ordóñez, G.R.; López-Otín, C. Protease Genomics and the Cancer Degradome. In The Cancer Degradome: Proteases and Cancer Biology; Edwards, D., Høyer-Hansen, G., Blasi, F., Sloane, B.F., Eds.; Springer: New York, NY, USA, 2008; pp. 3–15. ISBN 978-0-387-69057-5. [Google Scholar]
- López-Otín, C.; Overall, C.M. Protease degradomics: A new challenge for proteomics. Nat. Rev. Mol. Cell Biol. 2002, 3, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Schrader, E.K.; Harstad, K.G.; Matouschek, A. Targeting proteins for degradation. Nat. Chem. Biol. 2009, 5, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Inobe, T.; Matouschek, A. Paradigms of protein degradation by the proteasome. Curr. Opin. Struct. Biol. 2014, 24, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Lyapina, I.; Ivanov, V.; Fesenko, I. Peptidome: Chaos or Inevitability. Int. J. Mol. Sci. 2021, 22, 3128. [Google Scholar] [CrossRef] [PubMed]
- Naryzhny, S. Quantitative Aspects of the Human Cell Proteome. Int. J. Mol. Sci. 2023, 24, 8524. [Google Scholar] [CrossRef] [PubMed]
- Deslignière, E.; Rolland, A.; Ebberink, E.H.T.M.; Yin, V.; Heck, A.J.R. Orbitrap-Based Mass and Charge Analysis of Single Molecules. Acc. Chem. Res. 2023, 56, 1458–1468. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chu, S.; Tan, S.; Yin, X.; Jiang, Y.; Dai, X.; Gong, X.; Fang, X.; Tian, D. Towards Higher Sensitivity of Mass Spectrometry: A Perspective From the Mass Analyzers. Front. Chem. 2021, 9, 813359. [Google Scholar] [CrossRef] [PubMed]
- Brunner, A.; Thielert, M.; Vasilopoulou, C.; Ammar, C.; Coscia, F.; Mund, A.; Hoerning, O.B.; Bache, N.; Apalategui, A.; Lubeck, M.; et al. Ultra-high sensitivity mass spectrometry quantifies single-cell proteome changes upon perturbation. Mol. Syst. Biol. 2022, 18, e10798. [Google Scholar] [CrossRef] [PubMed]
- Deslignière, E.; Yin, V.C.; Ebberink, E.H.T.M.; Rolland, A.D.; Barendregt, A.; Wörner, T.P.; Nagornov, K.O.; Kozhinov, A.N.; Fort, K.L.; Tsybin, Y.O.; et al. Ultralong transients enhance sensitivity and resolution in Orbitrap-based single-ion mass spectrometry. Nat. Methods 2024, 21, 619–622. [Google Scholar] [CrossRef] [PubMed]
- Thiede, B.; Koehler, C.J.; Strozynski, M.; Treumann, A.; Stein, R.; Zimny-Arndt, U.; Schmid, M.; Jungblut, P.R. High resolution quantitative proteomics of hela cells protein species using stable isotope labeling with amino acids in cell culture(SILAC), Two-dimensional gel electrophoresis(2DE) and nano-liquid chromatograpohy coupled to an LTQ-OrbitrapMass spectromet. Mol. Cell. Proteom. 2013, 12, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Zabel, C.; Klose, J. High-resolution large-gel 2DE. Methods Mol. Biol. 2009, 519, 311–338. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Li, N.; Zhan, X.; Qian, S. Revival of 2DE-LC/MS in Proteomics and Its Potential for Large-Scale Study of Human Proteoforms. Med One 2018, 3, e180008. [Google Scholar] [CrossRef]
- Ramazi, S.; Zahiri, J. Post-translational modifications in proteins: Resources, tools and prediction methods. Database 2021, 2021, baab012. [Google Scholar] [CrossRef] [PubMed]
- Naryzhny, S.N.; Maynskova, M.A.; Zgoda, V.G.; Ronzhina, N.; Novikova, S.E.; Belyakova, N.V.; Kleyst, O.A.; Legina, O.K.; Pantina, R.A.; Filatov, M.V. Proteomic Profiling of High-grade Glioblastoma Using Virtual experimental2DE. J. Proteom. Bioinform. 2016, 9, 158–165. [Google Scholar] [CrossRef]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef] [PubMed]
- Naryzhny, S.; Maynskova, M.; Zgoda, V.; Archakov, A. Zipf’s Law in Proteomics. J. Proteom. Bioinform. 2017, 10, 2–19. [Google Scholar] [CrossRef]
- Ebert, T.; Tran, N.; Schurgers, L.; Stenvinkel, P.; Shiels, P.G. Ageing—Oxidative stress, PTMs and disease. Mol. Aspects Med. 2022, 86, 101099. [Google Scholar] [CrossRef] [PubMed]
- Consortium, A.A. Aging Atlas: A multi-omics database for aging biology. Nucleic Acids Res. 2020, 49, D825–D830. [Google Scholar] [CrossRef] [PubMed]
- Melani, R.D.; Gerbasi, V.R.; Anderson, L.C.; Sikora, J.W.; Toby, T.K.; Hutton, J.E.; Butcher, D.S.; Negrão, F.; Seckler, H.S.; Srzentić, K.; et al. The Blood Proteoform Atlas: A reference map of proteoforms in human hematopoietic cells. Science 2022, 375, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.P.; Wiegers, T.C.; Johnson, R.J.; Sciaky, D.; Wiegers, J.; Mattingly, C.J. Comparative Toxicogenomics Database (CTD): Update 2023. Nucleic Acids Res. 2023, 51, D1257–D1262. [Google Scholar] [CrossRef] [PubMed]
- Murray-Zmijewski, F.; Slee, E.A.; Lu, X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat. Rev. Mol. Cell Biol. 2008, 9, 702–712. [Google Scholar] [CrossRef] [PubMed]
- Naryzhny, S.N.; Legina, O.K. Structural-functional diversity of p53 proteoforms. Biomeditsinskaya Khimiya 2019, 65, 263–276. [Google Scholar] [CrossRef] [PubMed]
- DeHart, C.J.; Chahal, J.S.; Flint, S.J.; Perlman, D.H. Extensive post-translational modification of active and inactivated forms of endogenous p53. Mol. Cell. Proteom. 2014, 13, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Sidoli, S.; Lin, S.; Karch, K.R.; Garcia, B.A. Bottom-up and middle-down proteomics have comparable accuracies in defining histone post-translational modification relative abundance and stoichiometry. Anal. Chem. 2015, 87, 3129–3133. [Google Scholar] [CrossRef] [PubMed]
- Garcia, B.A.; Pesavento, J.J.; Mizzen, C.A.; Kelleher, N.L. Pervasive combinatorial modification of histone H3 in human cells. Nat. Methods 2007, 4, 487–489. [Google Scholar] [CrossRef] [PubMed]
- Ronzhina, N.L.; Zorina, E.S.; Zavialova, M.G.; Legina, O.K.; Naryzhny, S.N. Variability of haptoglobin beta-chain proteoforms. Biomeditsinskaya Khimiya 2024, 70, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Vihinen, M. Bioinformatics in proteomics. Biomol. Eng. 2001, 18, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Domon, B.; Aebersold, R. Challenges and opportunities in proteomics data analysis. Mol. Cell. Proteom. 2006, 5, 1921–1926. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Forne, I.; Imhof, A. Bioinformatic analysis of proteomics data. BMC Syst. Biol. 2014, 8 (Suppl. S2), S3. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Guo, N.; Kejariwal, A.; Thomas, P.D. PANTHER version 6: Protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007, 35, D247–D252. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, C.F.; Anthony, K.; Krupa, S.; Buchoff, J.; Day, M.; Hannay, T.; Buetow, K.H. PID: The pathway interaction database. Nucleic Acids Res. 2009, 37, D674–D679. [Google Scholar] [CrossRef]
- Kandasamy, K.; Mohan, S.S.; Raju, R.; Keerthikumar, S.; Kumar, G.S.S.; Venugopal, A.K.; Telikicherla, D.; Navarro, J.D.; Mathivanan, S.; Pecquet, C.; et al. NetPath: A public resource of curated signal transduction pathways. Genome Biol. 2010, 11, R3. [Google Scholar] [CrossRef] [PubMed]
- Chatr-aryamontri, A.; Ceol, A.; Palazzi, L.M.; Nardelli, G.; Schneider, M.V.; Castagnoli, L.; Cesareni, G. MINT: The Molecular INTeraction database. Nucleic Acids Res. 2007, 35, D572–D574. [Google Scholar] [CrossRef] [PubMed]
- Del Toro, N.; Shrivastava, A.; Ragueneau, E.; Meldal, B.; Combe, C.; Barrera, E.; Perfetto, L.; How, K.; Ratan, P.; Shirodkar, G.; et al. The IntAct database: Efficient access to fine-grained molecular interaction data. Nucleic Acids Res. 2021, 50, D648–D653. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Al-Amrani, S.; Al-Jabri, Z.; Al-Zaabi, A.; Alshekaili, J.; Al-Khabori, M. Proteomics: Concepts and applications in human medicine. World J. Biol. Chem. 2021, 12, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X. Introductory Chapter: Proteoforms. In Proteoforms; Zhan, X., Ed.; IntechOpen: Rijeka, Croatia, 2020. [Google Scholar]
- Nice, E.C. The status of proteomics as we enter the 2020s: Towards personalised/precision medicine. Anal. Biochem. 2022, 644, 113840. [Google Scholar] [CrossRef] [PubMed]
- Apweiler, R.; Aslanidis, C.; Deufel, T.; Gerstner, A.; Hansen, J.; Hochstrasser, D.; Kellner, R.; Kubicek, M.; Lottspeich, F.; Maser, E.; et al. Approaching clinical proteomics: Current state and future fields of application in cellular proteomics. Cytom. Part A J. Int. Soc. Anal. Cytol. 2009, 75, 816–832. [Google Scholar] [CrossRef]
- Foster, J.B.; Koptyra, M.P.; Bagley, S.J. Recent Developments in Blood Biomarkers in Neuro-oncology. Curr. Neurol. Neurosci. Rep. 2023, 23, 857–867. [Google Scholar] [CrossRef] [PubMed]
- Ivanisevic, T.; Sewduth, R.N. Multi-Omics Integration for the Design of Novel Therapies and the Identification of Novel Biomarkers. Proteomes 2023, 11, 34. [Google Scholar] [CrossRef] [PubMed]
- Omenn, G.S.; Lane, L.; Overall, C.M.; Paik, Y.-K.; Cristea, I.M.; Corrales, F.J.; Lindskog, C.; Weintraub, S.; Roehrl, M.H.A.; Liu, S.; et al. Progress Identifying and Analyzing the Human Proteome: 2021 Metrics from the HUPO Human Proteome Project. J. Proteome Res. 2021, 20, 5227–5240. [Google Scholar] [CrossRef] [PubMed]
- Savaryn, J.P.; Catherman, A.D.; Thomas, P.M.; Abecassis, M.M.; Kelleher, N.L. The emergence of top-down proteomics in clinical research. Genome Med. 2013, 5, 53. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Yang, L.; Sun, Z.; Zhan, X. Personalized Drug Therapy: Innovative Concept Guided With Proteoformics. Mol. Cell. Proteom. 2024, 23, 100737. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, E.; Moriwaki, K.; Terao, N.; Tan, C.C.; Terao, M.; Nakagawa, T.; Matsumoto, H.; Shinzaki, S.; Kamada, Y. Fucosylation is a promising target for cancer diagnosis and therapy. Biomolecules 2012, 2, 34–45. [Google Scholar] [CrossRef] [PubMed]
- McGee, J.P.; Su, P.; Durbin, K.R.; Hollas, M.A.R.; Bateman, N.W.; Maxwell, G.L.; Conrads, T.P.; Fellers, R.T.; Melani, R.D.; Camarillo, J.M.; et al. Automated imaging and identification of proteoforms directly from ovarian cancer tissue. Nat. Commun. 2023, 14, 6478. [Google Scholar] [CrossRef] [PubMed]
- Forgrave, L.M.; Wang, M.; Yang, D.; DeMarco, M.L. Proteoforms and their expanding role in laboratory medicine. Pract. Lab. Med. 2022, 28, e00260. [Google Scholar] [CrossRef] [PubMed]
- Geyer, P.E.; Kulak, N.A.; Pichler, G.; Holdt, L.M.; Teupser, D.; Mann, M. Plasma Proteome Profiling to Assess Human Health and Disease. Cell Syst. 2016, 2, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Frantzi, M.; Latosinska, A.; Kontostathi, G.; Mischak, H. Clinical Proteomics: Closing the Gap from Discovery to Implementation. Proteomics 2018, 18, 1700463. [Google Scholar] [CrossRef] [PubMed]
- Verrills, N.M. Clinical proteomics: Present and future prospects. Clin. Biochem. Rev. 2006, 27, 99–116. [Google Scholar] [PubMed]
- Huang, C.-F.; Kline, J.T.; Negrão, F.; Robey, M.T.; Toby, T.K.; Durbin, K.R.; Fellers, R.T.; Friedewald, J.J.; Levitsky, J.; Abecassis, M.M.I.; et al. Targeted Quantification of Proteoforms in Complex Samples by Proteoform Reaction Monitoring. Anal. Chem. 2024, 96, 3578–3586. [Google Scholar] [CrossRef] [PubMed]

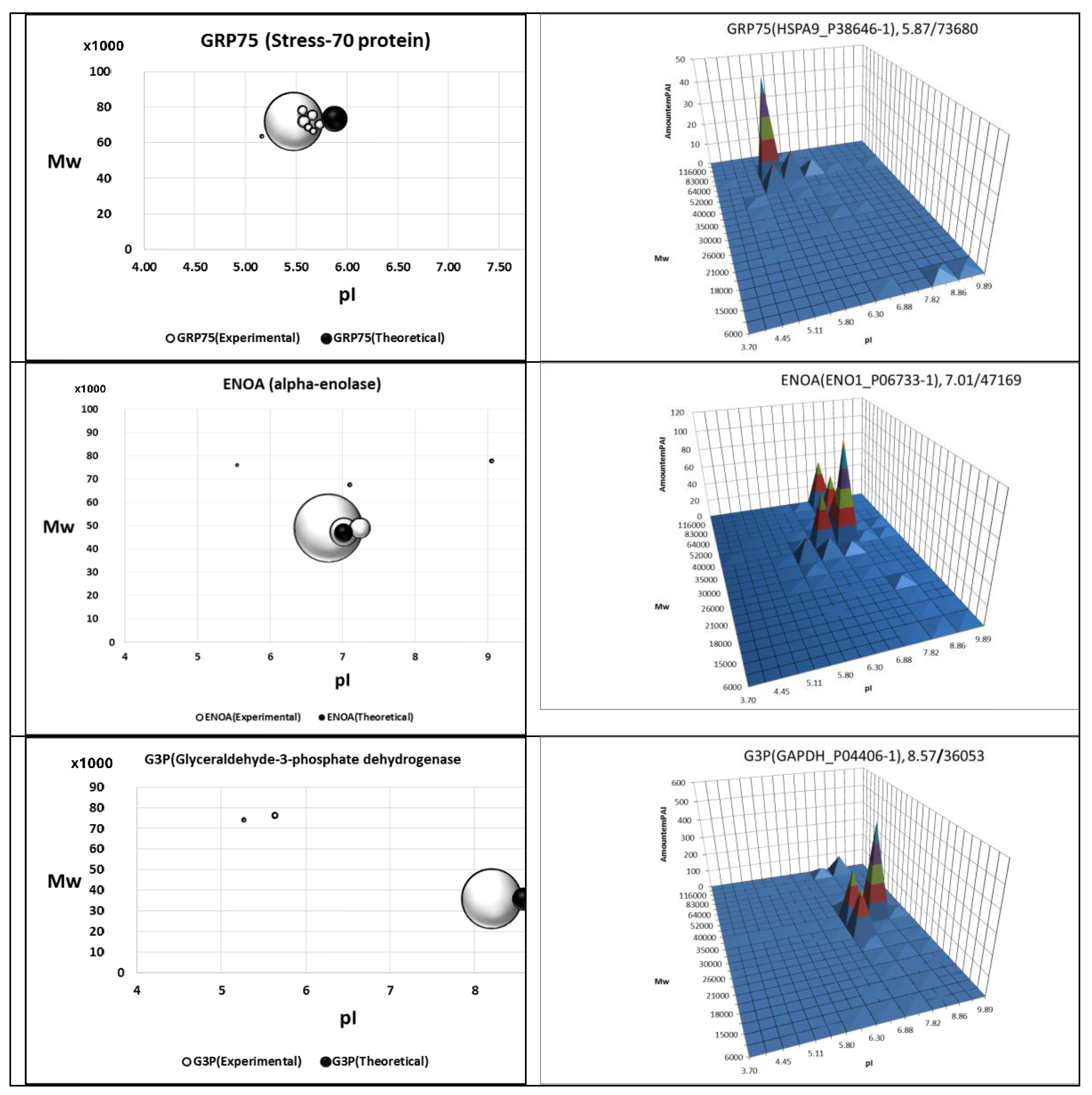



| Protein | Gene | Isoform Uniprot # | PTM Sites * | “2DE-Pattern” ** | “Proteoform Atlas” *** | Protein Class |
|---|---|---|---|---|---|---|
| H32 | HIST2H3A | Q71DI3-1 | 23 | 21 | 2979 | Chromatin |
| H4 | HIST1H4A | P62805-1 | 38 | 80 | 1113 | Chromatin |
| HS90B | HSP90AB1 | P08238-1 | 161 | 82 | 43 | Chaperone |
| CH60 | HSPD1 | P10809-1 | 157 | 48 | 91 | Chaperone |
| ENOA | ENO1 | P06733-1 | 111 | 78 | 302 | Metabolic |
| KPYM | PKM | P14618-1 | 132 | 77 | 124 | Metabolic |
| G3P | GAPDH | P04406-1 | 120 | 68 | 832 | Metabolic |
| PGK1 | PGK1 | P00558-1 | 97 | 53 | 104 | Metabolic |
| LDHA | LDHA | P00338-1 | 72 | 72 | 121 | Metabolic |
| EF1A1 | EEF1A1 | P68104-1 | 105 | 20 | 114 | Metabolic |
| HNRPK | HNRNPK | P61978-1 | 132 | 51 | 33 | RNA metabolism |
| TBB5 | TUBB | P07437-1 | 76 | 66 | 63 | Cytoskeleton |
| MYH9 | MYH9 | P35579-1 | 243 | 47 | 115 | Cytoskeleton |
| ACTB | ACTB | P60709-1 | 68 | 73 | 1014 | Cytoskeleton |
| VIME | VIM | P08670-1 | 139 | 65 | 261 | Cytoskeleton |
| FLNA | FLNA | P21333-1 | 323 | 57 | 139 | Cytoskeleton |
| RS27A | RPS27A | P62979-1 | 45 | 73 | 65 | Ribosomal |
| 1433Z | YWHAZ | P63104-1 | 64 | 34 | 125 | Scaffold/adaptor |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naryzhny, S. Puzzle of Proteoform Variety—Where Is a Key? Proteomes 2024, 12, 15. https://doi.org/10.3390/proteomes12020015
Naryzhny S. Puzzle of Proteoform Variety—Where Is a Key? Proteomes. 2024; 12(2):15. https://doi.org/10.3390/proteomes12020015
Chicago/Turabian StyleNaryzhny, Stanislav. 2024. "Puzzle of Proteoform Variety—Where Is a Key?" Proteomes 12, no. 2: 15. https://doi.org/10.3390/proteomes12020015








