lncRNA Biomarkers of Glioblastoma Multiforme †
Abstract
:1. Introduction
2. Methodology
3. Genome Localisation and Expression of lncRNA
4. Molecular Mechanisms Underlying lncRNA Functions
- Signalling, where lncRNAs are transcribed at a specific site and time in a cell type-specific manner, inducing or governing an active signalling event (Figure 2).
- Decoying, where lncRNAs serve as decoys for target proteins. The lncRNA molecules occupy the binding site, and the proteins cannot interact with DNA (Figure 3a). In this way, lncRNA can interact with transcription factors, repressors, chromatin modifiers, and other proteins. Within this regulatory mode, lncRNAs can also interact with miRNAs (Figure 3b). Specific lncRNAs act as sponges for some miRNAs, i.e., the lncRNA binds to the miRNA, which then cannot perform its function.
- Guiding, where lncRNA molecules control the placement of ribonucleoprotein complexes at specific target sites, with precision effects (Figure 4).
- Scaffolding, where transcripts act as scaffolds for other molecules that can bind to a given lncRNA to form a ribonucleoprotein complex (Figure 5).
5. Functions of lncRNAs in Cancer
6. lncRNAs and GBM
7. lncRNA Biomarker in Diagnostic and Clinical Use
8. Emerging lncRNA Biomarkers of GBM
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Fernandes, J.C.; Acuña, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Non-Coding RNA 2019, 5, 17. [Google Scholar] [CrossRef]
- Taft, R.J.; Pheasant, M.; Mattick, J.S. The relationship between non-protein-coding DNA and eukaryotic complexity. BioEssays 2007, 29, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Lagarrigue, S.; Lorthiois, M.; Degalez, F.; Gilot, D.; Derrien, T. LncRNAs in domesticated animals: From dog to livestock species. Mamm. Genome 2022, 33, 248–270. [Google Scholar] [CrossRef] [PubMed]
- Ward, M.; McEwan, C.; Mills, J.D.; Janitz, M. Conservation and tissue-specific transcription patterns of long noncoding RNAs. J. Hum. Transcr. 2015, 1, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Murillo-Maldonado, J.M.; Riesgo-Escovar, J.R. The various and shared roles of lncRNAs during development. Dev. Dyn. 2019, 248, 1059–1069. [Google Scholar] [CrossRef]
- GENCODE. 2024. Available online: https://www.gencodegenes.org/human/stats.html (accessed on 31 January 2024).
- Kazimierczyk, M.; Kasprowicz, M.K.; Kasprzyk, M.E.; Wrzesinski, J. Human Long Noncoding RNA Interactome: Detection, Characterization and Function. Int. J. Mol. Sci. 2020, 21, 1027. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Seal, R.L.; Chen, L.; Griffiths-Jones, S.; Lowe, T.M.; Mathews, M.B.; O’Reilly, D.; Pierce, A.J.; Stadler, P.F.; Ulitsky, I.; Wolin, S.L.; et al. A guide to naming human non-coding RNA genes. EMBO J. 2020, 39, e103777. [Google Scholar] [CrossRef]
- Kanderi, T.; Gupta, V. Glioblastoma Multiforme. In Glioblastoma Multiforme; StatPearls Publishing: St. Petersburg, FL, USA, 2022; pp. 1–20. Available online: https://www.ncbi.nlm.nih.gov/books/NBK558954/ (accessed on 8 April 2023).
- Melhem, J.M.; Detsky, J.; Lim-Fat, M.J.; Perry, J.R. Updates in IDH-Wildtype Glioblastoma. Neurotherapeutics 2022, 19, 1705–1723. [Google Scholar] [CrossRef]
- Rezaei, O.; Tamizkar, K.H.; Sharifi, G.; Taheri, M.; Ghafouri-Fard, S. Emerging Role of Long Non-Coding RNAs in the Pathobiology of Glioblastoma. Front. Oncol. 2021, 10, 625884. [Google Scholar] [CrossRef]
- Yadav, B.; Pal, S.; Rubstov, Y.; Goel, A.; Garg, M.; Pavlyukov, M.; Pandey, A.K. LncRNAs associated with glioblastoma: From transcriptional noise to novel regulators with a promising role in therapeutics. Mol. Ther.-Nucleic Acids 2021, 24, 728–742. [Google Scholar] [CrossRef] [PubMed]
- Akindona, F.A.; Frederico, S.C.; Hancock, J.C.; Gilbert, M.R. Exploring the origin of the cancer stem cell niche and its role in anti-angiogenic treatment for glioblastoma. Front. Oncol. 2022, 12, 947634. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, R. Potential of long non-coding RNAs as a therapeutic target and molecular markers in glioblastoma pathogenesis. Heliyon 2021, 7, e06502. [Google Scholar] [CrossRef] [PubMed]
- Paul, Y.; Thomas, S.; Patil, V.; Kumar, N.; Mondal, B.; Hegde, A.S.; Arivazhagan, A.; Santosh, V.; Mahalingam, K.; Somasundaram, K. Genetic landscape of long noncoding RNA (lncRNAs) in glioblastoma: Identification of complex lncRNA regulatory networks and clinically relevant lncRNAs in glioblastoma. Oncotarget 2018, 9, 29548–29564. [Google Scholar] [CrossRef] [PubMed]
- Bhan, A.; Soleimani, M.; Mandal, S.S. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017, 77, 3965–3981. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Chan, J.J.; Tay, Y. Noncoding RNA: RNA Regulatory Networks in Cancer. Int. J. Mol. Sci. 2018, 19, 1310. [Google Scholar] [CrossRef] [PubMed]
- Ray, I.; Goswami, S. Circadian rhythm genes in cancer: Insight into their functions and regulation involving noncoding RNAs. Chronobiol. Int. 2021, 38, 1231–1243. [Google Scholar] [CrossRef]
- O’Leary, V.B.; Ovsepian, S.V.; Smida, J.; Atkinson, M.J. PARTICLE−The RNA podium for genomic silencers. J. Cell. Physiol. 2019, 234, 19464–19470. [Google Scholar] [CrossRef]
- Bugnon, L.A.; A Edera, A.; Prochetto, S.; Gerard, M.; Raad, J.; Fenoy, E.; Rubiolo, M.; Chorostecki, U.; Gabaldón, T.; Ariel, F.; et al. Secondary structure prediction of long noncoding RNA: Review and experimental comparison of existing approaches. Brief. Bioinform. 2022, 23, 4. [Google Scholar] [CrossRef] [PubMed]
- Kwok, Z.H.; Tay, Y. Long noncoding RNAs: Lincs between human health and disease. Biochem. Soc. Trans. 2017, 45, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; Li, Y.; Li, J.; Gao, Z.; Yang, Z.; Li, Y.; Liu, H.; Fan, T. Long Non-Coding RNAs: The Regulatory Mechanisms, Research Strategies, and Future Directions in Cancers. Front. Oncol. 2020, 10, 598817. [Google Scholar] [CrossRef] [PubMed]
- Beylerli, O.; Gareev, I.; Sufianov, A.; Ilyasova, T.; Guang, Y. Long noncoding RNAs as promising biomarkers in cancer. Non-Coding RNA Res. 2022, 7, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; He, P.; Bian, Z. Long Noncoding RNAs in Neurodegenerative Diseases: Pathogenesis and Potential Implications as Clinical Biomarkers. Front. Mol. Neurosci. 2021, 14, 685143. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.-Z.; Chen, Q.; Wang, L.; Tao, G.; Gan, H.; Deng, L.-J.; Huang, J.-Q.; Chen, J.-X. Emerging roles of long non-coding RNA in depression. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2022, 115, 110515. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Li, Y.; Zhang, E.; Feillet, F.; Zhang, S.; Blau, N. Importance of the long non-coding RNA (lncRNA) transcript HULC for the regulation of phenylalanine hydroxylase and treatment of phenylketonuria. Mol. Genet. Metab. 2022, 135, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Boussios, S.; Devo, P.; Goodall, I.C.A.; Sirlantzis, K.; Ghose, A.; Shinde, S.D.; Papadopoulos, V.; Sanchez, E.; Rassy, E.; Ovsepian, S.V. Exosomes in the Diagnosis and Treatment of Renal Cell Cancer. Int. J. Mol. Sci. 2023, 24, 4356. [Google Scholar] [CrossRef] [PubMed]
- Sideris, N.; Dama, P.; Bayraktar, S.; Stiff, T.; Castellano, L. LncRNAs in breast cancer: A link to future approaches. Cancer Gene Ther. 2022, 29, 1866–1877. [Google Scholar] [CrossRef]
- Pokorná, M.; Kútna, V.; Ovsepian, S.V.; Matěj, R.; Černá, M.; O’leary, V.B. Biomolecules to Biomarkers? U87MG Marker Evaluation on the Path towards Glioblastoma Multiforme Pathogenesis. Pharmaceutics 2024, 16, 123. [Google Scholar] [CrossRef]
- Ren, S.; Xu, Y. AC016405.3, a novel long noncoding RNA, acts as a tumor suppressor through modulation of TET2 by microRNA-19a-5p sponging in glioblastoma. Cancer Sci. 2019, 110, 1621–1632. [Google Scholar] [CrossRef]
- Wei, M.; Wang, J.; He, Q.; Liu, L.; Wang, Z. AC016405.3 functions as an oncogenic long non-coding RNA by regulating ERBB3 via sponging miR-22-3p in breast cancer. J. Clin. Lab. Anal. 2021, 35, e23952. [Google Scholar] [CrossRef] [PubMed]
- LncRNAfunc: Database. Available online: https://ccsm.uth.edu/lncRNAfunc/index.html (accessed on 24 March 2023).
- Stackhouse, C.T.; Gillespie, G.Y.; Willey, C.D. Exploring the Roles of lncRNAs in GBM Pathophysiology and Their Therapeutic Potential. Cells 2020, 9, 2369. [Google Scholar] [CrossRef]
- Amirmahani, F.; Vallian, S.; Asadi, M.H. The LncRNA MIAT is identified as a regulator of stemness-associated transcript in glioma. Mol. Biol. Rep. 2023, 50, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Xu, X.-K.; Li, J.-L.; Kong, K.-K.; Li, H.; Chen, C.; He, J.; Wang, F.; Li, P.; Ge, X.-S.; et al. MALAT1 is a prognostic factor in glioblastoma multiforme and induces chemoresistance to temozolomide through suppressing miR-203 and promoting thymidylate synthase expression. Oncotarget 2017, 8, 22783–22799. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Wang, H.; Chen, E.; Bian, E.; Xu, Y.; Ji, X.; Yang, Z.; Hua, X.; Zhang, Y.; Zhao, B. LncRNA-ATB promotes TGF-β-induced glioma cells invasion through NF-κB and P38/MAPK pathway. J. Cell. Physiol. 2019, 234, 23302–23314. [Google Scholar] [CrossRef]
- Ma, C.-C.; Xiong, Z.; Zhu, G.-N.; Wang, C.; Zong, G.; Wang, H.-L.; Bian, E.-B.; Zhao, B. Long non-coding RNA ATB promotes glioma malignancy by negatively regulating miR-200a. J. Exp. Clin. Cancer Res. 2016, 35, 1–13. [Google Scholar] [CrossRef]
- Yin, T.; Wu, J.; Hu, Y.; Zhang, M.; He, J. Long non-coding RNA HULC stimulates the epithelial–mesenchymal transition process and vasculogenic mimicry in human glioblastoma. Cancer Med. 2021, 10, 5270–5282. [Google Scholar] [CrossRef]
- ZZhang, P.; Liu, Y.; Fu, C.; Wang, C.; Duan, X.; Zou, W.; Zhao, T. Knockdown of long non-coding RNA PCAT1 in glioma stem cells promotes radiation sensitivity. Med. Mol. Morphol. 2019, 52, 114–122. [Google Scholar] [CrossRef]
- Gong, X.; Liao, X.; Huang, M. LncRNA CASC7 inhibits the progression of glioma via regulating Wnt/β-catenin signaling pathway. Pathol.-Res. Pract. 2019, 215, 564–570. [Google Scholar] [CrossRef]
- Liu, D.; Wan, Y.; Qu, N.; Fu, Q.; Liang, C.; Zeng, L.; Yang, Y. LncRNA-FAM66C Was Identified as a Key Regulator for Modulating Tumor Microenvironment and Hypoxia-Related Pathways in Glioblastoma. Front. Public Health 2022, 10, 898270. [Google Scholar] [CrossRef] [PubMed]
- Ho, K.; Shih, C.; Liu, A.; Chen, K. Hypoxia-inducible lncRNA MIR210HG interacting with OCT1 is involved in glioblastoma multiforme malignancy. Cancer Sci. 2022, 113, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Katsushima, K.; Natsume, A.; Ohka, F.; Shinjo, K.; Hatanaka, A.; Ichimura, N.; Sato, S.; Takahashi, S.; Kimura, H.; Totoki, Y.; et al. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat. Commun. 2016, 7, 13616. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Ma, J.; Yu, B.; Zhu, Z.; Hu, Y. Long Noncoding RNA TUNAR Represses Growth, Migration, and Invasion of Human Glioma Cells Through Regulating miR-200a and Rac1. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2018, 27, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Cai, J.; Chen, Q.; Han, B.; Meng, X.; Li, Y.; Li, Z.; Wang, R.; Lin, L.; Duan, C.; et al. Lnc-TALC promotes O6-methylguanine-DNA methyltransferase expression via regulating the c-Met pathway by competitively binding with miR-20b-3p. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Xu, Z.; Chen, X.; Wang, X.; Zeng, S.; Zhao, Z.; Qian, L.; Li, Z.; Wei, J.; Huo, L.; et al. Novel Function of lncRNA ADAMTS9-AS2 in Promoting Temozolomide Resistance in Glioblastoma via Upregulating the FUS/MDM2 Ubiquitination Axis. Front. Cell Dev. Biol. 2019, 7, 217. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ji, X.; Wang, H. Targeting Long Noncoding RNA HMMR-AS1 Suppresses and Radiosensitizes Glioblastoma. Neoplasia 2018, 20, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xiao, X.; Han, X.; Yao, L.; Lan, W. Natural flavonoids alleviate glioblastoma multiforme by regulating long non-coding RNA. Biomed. Pharmacother. 2023, 161, 114477. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Lu, S.; Xu, Y.; Zheng, J. Long non-coding RNA AGAP2-AS1 promotes the proliferation of glioma cells by sponging miR-15a/b-5p to upregulate the expression of HDGF and activating Wnt/β-catenin signaling pathway. Int. J. Biol. Macromol. 2019, 128, 521–530. [Google Scholar] [CrossRef]
- Dai, X.; Liao, K.; Zhuang, Z.; Chen, B.; Zhou, Z.; Zhou, S.; Lin, G.; Zhang, F.; Lin, Y.; Miao, Y.; et al. AHIF promotes glioblastoma progression and radioresistance via exosomes. Int. J. Oncol. 2018, 54, 261–270. [Google Scholar] [CrossRef]
- Sun, Y.; Jing, Y.; Zhang, Y. Serum lncRNA-ANRIL and SOX9 expression levels in glioma patients and their relationship with poor prognosis. World J. Surg. Oncol. 2021, 19, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Skiriute, D.; Stakaitis, R.; Steponaitis, G.; Tamasauskas, A.; Vaitkiene, P. The Role of CASC2 and miR-21 Interplay in Glioma Malignancy and Patient Outcome. Int. J. Mol. Sci. 2020, 21, 7962. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, C.; Yang, J.; Sun, Y.; Zhang, S.; Yang, J.; Yang, L.; Wang, Y.; Jiao, B. Long noncoding RNA CASC9/miR-519d/STAT3 positive feedback loop facilitate the glioma tumourigenesis. J. Cell. Mol. Med. 2018, 22, 6338–6344. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wei, D.-L.; Wan, L.; Yan, S.-F.; Sun, Y.-H. Highly expressed lncRNA CCND2-AS1 promotes glioma cell proliferation through Wnt/β-catenin signaling. Biochem. Biophys. Res. Commun. 2017, 482, 1219–1225. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Liu, M.; Long, W.; Yuan, J.; Li, H.; Zhang, C.; Tang, G.; Jiang, W.; Yuan, X.; Wu, M.; et al. Knockdown lncRNA CRNDE enhances temozolomide chemosensitivity by regulating autophagy in glioblastoma. Cancer Cell Int. 2021, 21, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Yao, Y.; Hu, X.; Zhu, Y. LncRNA DCST1-AS1 downregulates miR-29b through methylation in glioblastoma (GBM) to promote cancer cell proliferation. Clin. Transl. Oncol. 2020, 22, 2230–2235. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Long, J.; Yang, C.; Gong, B.; Cheng, M.; Wang, Q.; Tang, J. LncRNA DGCR5 plays a tumor-suppressive role in glioma via the miR-21/Smad7 and miR-23a/PTEN axes. Aging 2020, 12, 20285–20307. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.-L.; Wang, L.-C.; Li, D.-C.; Lin, Q.-X.; Shen, X.-L.; Liu, H.-Y.; Li, M.; Ji, Y.-L.; Qin, C.-Z.; Chen, S.-H. Knockdown lncRNA DLEU1 Inhibits Gliomas Progression and Promotes Temozolomide Chemosensitivity by Regulating Autophagy. Front. Pharmacol. 2020, 11, 560543. [Google Scholar] [CrossRef]
- Deguchi, S.; Katsushima, K.; Hatanaka, A.; Shinjo, K.; Ohka, F.; Wakabayashi, T.; Zong, H.; Natsume, A.; Kondo, Y. Oncogenic effects of evolutionarily conserved noncoding RNA ECONEXIN on gliomagenesis. Oncogene 2017, 36, 4629–4640. [Google Scholar] [CrossRef]
- Li, G.; Cai, Y.; Wang, C.; Huang, M.; Chen, J. LncRNA GAS5 regulates the proliferation, migration, invasion and apoptosis of brain glioma cells through targeting GSTM3 expression. The effect of LncRNA GAS5 on glioma cells. J. Neuro-Oncol. 2019, 143, 525–536. [Google Scholar] [CrossRef]
- Wang, G.; Lin, X.; Han, H.; Zhang, H.; Li, X.; Feng, M.; Jiang, C. LncRNA H19 promotes glioblastoma multiforme development by activating autophagy by sponging miR-491-5p. Bioengineered 2022, 13, 11440–11455. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Dong, C.; Cui, J.; Wang, Y.; Hong, X. Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. J. Exp. Clin. Cancer Res. 2018, 37, 265. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.K.; Pastori, C.; Penas, C.; Komotar, R.J.; Ivan, M.E.; Wahlestedt, C.; Ayad, N.G. Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Mol. Cancer 2018, 17, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ahmadov, U.; Picard, D.; Bartl, J.; Silginer, M.; Trajkovic-Arsic, M.; Qin, N.; Blümel, L.; Wolter, M.; Lim, J.K.M.; Pauck, D.; et al. The long non-coding RNA HOTAIRM1 promotes tumor aggressiveness and radiotherapy resistance in glioblastoma. Cell Death Dis. 2021, 12, 885. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yu, H.; Liu, Y.; Liu, X.; Zheng, J.; Ma, J.; Gong, W.; Chen, J.; Zhao, L.; Tian, Y.; et al. Long Non-Coding RNA HOXA-AS2 Regulates Malignant Glioma Behaviors and Vasculogenic Mimicry Formation via the MiR-373/EGFR Axis. Cell. Physiol. Biochem. 2018, 45, 131–147. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Kuang, W.; Lu, S.; Guo, H.; Wu, M.; Ye, M.; Wu, L. Long noncoding RNA HOXB 13- AS 1 regulates HOXB 13 gene methylation by interacting with EZH 2 in glioma. Cancer Med. 2018, 7, 4718–4728. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.-M.; Chen, L.; Li, F.; Zhang, R.; Li, Z.-Y.; Chen, F.-F.; Jiang, X.-D. Over-expression of the long non-coding RNA HOTTIP inhibits glioma cell growth by BRE. J. Exp. Clin. Cancer Res. 2016, 35, 162. [Google Scholar] [CrossRef]
- Mu, Y.; Tang, Q.; Feng, H.; Zhu, L.; Wang, Y. LncRNA KTN1 AS1 promotes glioma cell proliferation and invasion by negatively regulating miR 505 3p. Oncol. Rep. 2020, 44, 2645–2655. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, X.; Wu, Z.; Hu, D.; Jia, J.; Guo, J.; Tang, T.; Yao, J.; Liu, H.; Tang, H. Long Noncoding RNA LINC00467 Promotes Glioma Progression through Inhibiting P53 Expression via Binding to DNMT1. J. Cancer 2020, 11, 2935–2944. [Google Scholar] [CrossRef]
- Amer, R.G.; El Arab, L.R.E.; El Ghany, D.A.; Saad, A.S.; Bahie-Eldin, N.; Swellam, M. Prognostic utility of lncRNAs (LINC00565 and LINC00641) as molecular markers in glioblastoma multiforme (GBM). J. Neuro-Oncol. 2022, 158, 435–444. [Google Scholar] [CrossRef]
- Li, D.; Hu, J.; Li, S.; Zhou, C.; Feng, M.; Li, L.; Gao, Y.; Chen, X.; Wu, X.; Cao, Y.; et al. LINC01393, a Novel Long Non-Coding RNA, Promotes the Cell Proliferation, Migration and Invasion through MiR-128-3p/NUSAP1 Axis in Glioblastoma. Int. J. Mol. Sci. 2023, 24, 5878. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Tang, Z.; Su, Z. Long non-coding RNA LINC01426 facilitates glioblastoma progression via sponging miR-345-3p and upregulation of VAMP8. Cancer Cell Int. 2020, 20, 327. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, Q.; Wang, F.; Zhang, X.; Tang, Y.; Wang, S. LncRNA LINC01446 promotes glioblastoma progression by modulating miR-489-3p/TPT1 axis. Biochem. Biophys. Res. Commun. 2018, 503, 1484–1490. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Hu, G.; Wei, B.; Wang, L.; Liu, N. PlncRNA LINC01494 Promotes Proliferation, Migration and Invasion in Glioma Through miR-122-5p/CCNG1 Axis/p. OncoTargets Ther. 2019, 12, 7655–7662. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.; Jiang, J.; Xiao, M.; Zeng, M.; Liu, X.; Zhao, B.; Chen, F. The transcript ENST00000444125 of lncRNA LINC01503 promotes cancer stem cell properties of glioblastoma cells via reducing FBXW1 mediated GLI2 degradation. Exp. Cell Res. 2022, 412, 113009. [Google Scholar] [CrossRef] [PubMed]
- Shree, B.; Sengar, S.; Tripathi, S.; Sharma, V. TRIPATHI, Shraddha and SHARMA, Vivek. LINC01711 promotes transforming growth factor-beta (TGF-β) induced invasion in glioblastoma multiforme (GBM) by acting as a competing endogenous RNA for miR-34a and promoting ZEB1 expression. Neurosci. Lett. 2023, 792, 136937. [Google Scholar] [CrossRef] [PubMed]
- Goenka, A.; Song, X.; Tiek, D.; Iglesia, R.P.; Lu, M.; Zeng, C.; Horbinski, C.; Zhang, W.; Hu, B.; Cheng, S.-Y. Oncogenic long noncoding RNA LINC02283 enhances PDGF receptor A-mediated signaling and drives glioblastoma tumorigenesis. Neuro-Oncol. 2023, 25, 1592–1604. [Google Scholar] [CrossRef] [PubMed]
- Kovalenko, T.F.; Yadav, B.; Anufrieva, K.S.; Rubtsov, Y.P.; Zatsepin, T.S.; Shcherbinina, E.Y.; Solyus, E.M.; Staroverov, D.B.; Larionova, T.D.; Latyshev, Y.A.; et al. Functions of long non-coding RNA ROR in patient-derived glioblastoma cells. Biochimie 2022, 200, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Li, J.; Yan, X.; Bian, X. LncRNA MAFG-AS1 Suppresses the Maturation of miR-34a to Promote Glioblastoma Cell Proliferation. Cancer Manag. Res. 2021, 13, 3493–3501. [Google Scholar] [CrossRef]
- Yue, H.; Zhu, J.; Xie, S.; Li, F.; Xu, Q. MDC1-AS, an antisense long noncoding RNA, regulates cell proliferation of glioma. Biomed. Pharmacother. 2016, 81, 203–209. [Google Scholar] [CrossRef]
- Zhang, S.; Guo, W. Long non coding RNA MEG3 suppresses the growth of glioma cells by regulating the miR 96 5p/MTSS1 signaling pathway. Mol. Med. Rep. 2019, 20, 4215–4225. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Xu, Y.; Wang, J.; Yang, X.; Wen, L.; Feng, J. LncRNA MNX1-AS1 Promotes Glioblastoma Progression through Inhibition of miR-4443. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2019, 27, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Cheng, Y.; Yuan, Z.; Wang, F.; Yang, L.; Zhao, H. NCK1-AS1 Increases Drug Resistance of Glioma Cells to Temozolomide by Modulating miR-137/TRIM24. Cancer Biother. Radiopharm. 2020, 35, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Zhang, C.; Yao, H.; Zhang, X.; Zhou, Y.; Che, Y.; Huang, Y. Knockdown of long non-coding RNA NEAT1 inhibits glioma cell migration and invasion via modulation of SOX2 targeted by miR-132. Mol. Cancer 2018, 17, 105. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Piao, L.; Sun, G.; Lv, C.; Jing, Y.; Jin, R. Long Non-Coding RNA PART1 Exerts Tumor Suppressive Functions in Glioma via Sponging miR-190a-3p and Inactivation of PTEN/AKT Pathway/p. OncoTargets Ther. 2020, 13, 1073–1086. [Google Scholar] [CrossRef] [PubMed]
- Pokorná, M.; Hudec, M.; Juříčková, I.; Vácha, M.; Polívková, Z.; Kútna, V.; Pala, J.; Ovsepian, S.V.; Černá, M.; O’leary, V.B. All-Trans Retinoic Acid Fosters the Multifarious U87MG Cell Line as a Model of Glioblastoma. Brain Sci. 2021, 11, 812. [Google Scholar] [CrossRef]
- Lv, T.; Jin, Y.; Miao, Y.; Xu, T.; Jia, F.; Feng, H.; Zhang, X. LncRNA PVT1 promotes tumorigenesis of glioblastoma by recruiting COPS5 to deubiquitinate and stabilize TRIM24. Mol. Ther.-Nucleic Acids 2022, 27, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cui, Y.; Ma, W.; Wang, M.; Cai, Y.; Jiang, Y. LncRNA RBPMS-AS1 promotes NRGN transcription to enhance the radiosensitivity of glioblastoma through the microRNA-301a-3p/CAMTA1 axis. Transl. Oncol. 2022, 15, 101282. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Guo, X.; Lv, W.; Li, Y.; Zhang, L.; Dong, C.; Zhang, J.; Cheng, G. PLncRNA RPSAP52 Upregulates TGF-β1 to Increase Cancer Cell Stemness and Predict Postoperative Survival in Glioblastoma/p. Cancer Manag. Res. 2020, 12, 2541–2547. [Google Scholar] [CrossRef]
- Wu, Z. MiR-195 connects lncRNA RUNX1-IT1 and cyclin D1 to regulate the proliferation of glioblastoma cells. Int. J. Neurosci. 2023, 133, 13–18. [Google Scholar] [CrossRef]
- Ni, H.; Wang, K.; Xie, P.; Zuo, J.; Liu, W.; Liu, C. LncRNA SAMMSON Knockdown Inhibits the Malignancy of Glioblastoma Cells by Inactivation of the PI3K/Akt Pathway. Cell. Mol. Neurobiol. 2021, 41, 79–90. [Google Scholar] [CrossRef]
- Brodie, S.; Lee, H.K.; Jiang, W.; Cazacu, S.; Xiang, C.; Poisson, L.M.; Datta, I.; Kalkanis, S.; Ginsberg, D.; Brodie, C. Correction: The novel long non-coding RNA TALNEC2, regulates tumor cell growth and the stemness and radiation response of glioma stem cells. Oncotarget 2021, 12, 2546–2547. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, Q.; Wu, B.; Zhang, S.; Li, L.; Jin, K.; Li, S.; Li, K.; Wang, Z.; Lu, Y.; et al. Long non-coding RNA TP73-AS1 is a potential immune related prognostic biomarker for glioma. Aging 2021, 13, 5638–5649. [Google Scholar] [CrossRef]
- Qin, X.; Yao, J.; Geng, P.; Fu, X.; Xue, J.; Zhang, Z. LncRNA TSLC1-AS1 is a novel tumor suppressor in glioma. Int. J. Clin. Exp. Pathol. 2014, 7, 3065–3072. [Google Scholar]
- Shang, C.; Tang, W.; Pan, C.; Hu, X.; Hong, Y. Long non-coding RNA TUSC7 inhibits temozolomide resistance by targeting miR-10a in glioblastoma. Cancer Chemother. Pharmacol. 2018, 81, 671–678. [Google Scholar] [CrossRef]
- Cao, Y.; Chai, W.; Wang, Y.; Tang, D.; Shao, D.; Song, H.; Long, J. LncRNA TUG1 inhibits the cancer stem cell like properties of temozolomide resistant glioma cells by interacting with EZH2. Mol. Med. Rep. 2021, 24, 533. [Google Scholar] [CrossRef]
- Xin, H.; Liu, N.; Xu, X.; Zhang, J.; Li, Y.; Ma, Y.; Li, G.; Liang, J. Knockdown of lncRNA-UCA1 inhibits cell viability and migration of human glioma cells by miR-193a-mediated downregulation of CDK6. J. Cell. Biochem. 2019, 120, 15157–15169. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Lin, C.; Peng, M.; Ren, J.; Jing, Y.; Lei, L.; Tao, Y.; Huang, J.; Yang, J.; Sun, M.; et al. Circulating plasma exosomal long non-coding RNAs LINC00265, LINC00467, UCA1, and SNHG1 as biomarkers for diagnosis and treatment monitoring of acute myeloid leukemia. Front. Oncol. 2022, 12, 1033143. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Li, Z.; Ma, K.; Li, X.; Tian, N.; Duan, J.; Xiao, X.; Wang, Y. Long Non-coding RNA XIST Promotes Glioma Tumorigenicity and Angiogenesis by Acting as a Molecular Sponge of miR-429. J. Cancer 2017, 8, 4106–4116. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Askari, A.; Moghadam, K.B.; Hussen, B.M.; Taheri, M.; Samadian, M. A review on the role of ZEB1-AS1 in human disorders. Pathol.-Res. Pract. 2023, 245, 154486. [Google Scholar] [CrossRef]
- Dong, J.; Peng, Y.; Zhong, M.; Xie, Z.; Jiang, Z.; Wang, K.; Wu, Y. Implication of lncRNA ZBED3-AS1 downregulation in acquired resistance to Temozolomide and glycolysis in glioblastoma. Eur. J. Pharmacol. 2023, 938, 175444. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hong, R.; Chen, W.; Xu, M.; Wang, L. The role of long noncoding RNA in major human disease. Bioorganic Chem. 2019, 92, 103214. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Luo, H.; Li, D.; Wang, S.; Xuan, L.; Sun, L. Research advances on circulating long noncoding RNAs as biomarkers of cardiovascular diseases. Int. J. Cardiol. 2022, 353, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Kabzinski, J.; Kucharska-Lusina, A.; Majsterek, I. RNA-Based Liquid Biopsy in Head and Neck Cancer. Cells 2023, 12, 1916. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Chen, K.K.; Zhang, J.; Xiao, B.; Huang, Z.; Ju, C.; Sun, J.; Zhang, F.; Lv, X.-B.; Huang, G. The decade of exosomal long RNA species: An emerging cancer antagonist. Mol. Cancer 2018, 17, 75. [Google Scholar] [CrossRef] [PubMed]
- Badowski, C.; He, B.; Garmire, L.X. Blood-derived lncRNAs as biomarkers for cancer diagnosis: The Good, the Bad and the Beauty. Npj Precis. Oncol. 2022, 6, 1. [Google Scholar] [CrossRef]
- Turner, A.W.; Wong, D.; Khan, M.D.; Dreisbach, C.N.; Palmore, M.; Miller, C.L. Multi-Omics Approaches to Study Long Non-coding RNA Function in Atherosclerosis. Front. Cardiovasc. Med. 2019, 6, 9. [Google Scholar] [CrossRef]
- Feng, N.; Wang, Z.; Wu, Y.; Zheng, H.; Jiang, X.; Wang, Z.; Qu, F.; Zhang, Z. ADAMTS9-AS2 Promotes Angiogenesis of Brain Microvascular Endothelial Cells Through Regulating miR-185-5p/IGFBP-2 Axis in Ischemic Stroke. Mol. Neurobiol. 2022, 59, 2593–2604. [Google Scholar] [CrossRef]
- Abdul-Maksoud, R.S.; Rashad, N.M.; Elsayed, W.S.H.; Elsayed, R.S.; Sherif, M.M.; Abbas, A.; El Shabrawy, M. The diagnostic significance of circulating lncRNA ADAMTS9-AS2 tumor biomarker in non-small cell lung cancer among the Egyptian population. J. Gene Med. 2021, 23, e3381. [Google Scholar] [CrossRef]
- Alkhathami, A.G.; Hadi, A.; Alfaifi, M.; Alshahrani, M.Y.; Verma, A.K.; Beg, M.M.A. Serum-Based lncRNA ANRIL, TUG1, UCA1, and HIT Expressions in Breast Cancer Patients. Dis. Markers 2022, 2022, 9997212. [Google Scholar] [CrossRef]
- Hu, Y.; Hu, J. Diagnostic value of circulating lncRNA ANRIL and its correlation with coronary artery disease parameters. Braz. J. Med. Biol. Res. 2019, 52, e8309. [Google Scholar] [CrossRef] [PubMed]
- Rahni, Z.; Hosseini, S.M.; Shahrokh, S.; Niasar, M.S.; Shoraka, S.; Mirjalali, H.; Nazemalhosseini-Mojarad, E.; Rostami-Nejad, M.; Malekpour, H.; Zali, M.R.; et al. Long non-coding RNAs ANRIL, THRIL, and NEAT1 as potential circulating biomarkers of SARS-CoV-2 infection and disease severity. Virus Res. 2023, 336, 199214. [Google Scholar] [CrossRef] [PubMed]
- Erfan, R.; Shaker, O.G.; Khalil, M.A.F.; Mahmoud, F.A.M.; Gomaa, M.S.; Abu-El-Azayem, A.K.; Zaki, O.M.; Ahmed, A.M.; Samy, A.; Mohammed, A. Circulating miR-199a and long noncoding-RNA ANRIL as Promising Diagnostic Biomarkers for Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2024. [CrossRef]
- Biswas, S.; Coyle, A.; Chen, S.; Gostimir, M.; Gonder, J.; Chakrabarti, S. Expressions of Serum lncRNAs in Diabetic Retinopathy—A Potential Diagnostic Tool. Front. Endocrinol. 2022, 13, 851967. [Google Scholar] [CrossRef] [PubMed]
- Permuth, J.B.; Chen, D.-T.; Yoder, S.J.; Li, J.; Smith, A.T.; Choi, J.W.; Kim, J.; Balagurunathan, Y.; Jiang, K.; Coppola, D.; et al. Linc-ing Circulating Long Non-coding RNAs to the Diagnosis and Malignant Prediction of Intraductal Papillary Mucinous Neoplasms of the Pancreas. Sci. Rep. 2017, 7, 10484. [Google Scholar] [CrossRef]
- Zhang, K.; Qi, M.; Yang, Y.; Xu, P.; Zhua, Y.; Zhang, J. Circulating lncRNA ANRIL in the Serum of Patients with Ischemic Stroke. Clin. Lab. 2019, 65, 1459–1465. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Jin, J. The correlation of serum long non-coding RNA ANRIL with risk factors, functional outcome, and prognosis in atrial fibrillation patients with ischemic stroke. J. Clin. Lab. Anal. 2020, 34, e23352. [Google Scholar] [CrossRef]
- Yin, Y.; Yang, W.; Zhang, L.; Liu, K.; Luo, Z. Long non-coding RNA ANRIL and its target microRNAs (microRNA-34a, microRNA-125a and microRNA-186) relate to risk stratification and prognosis in multiple myeloma. Hematology 2021, 26, 160–169. [Google Scholar] [CrossRef]
- AbdAllah, N.B.; Al Ageeli, E.; Shbeer, A.; A Abdulhakim, J.; A Toraih, E.; O Salman, D.; Fawzy, M.S.; Nassar, S.S. Long Non-Coding RNAs ANRIL and HOTAIR Upregulation is Associated with Survival in Neonates with Sepsis in a Neonatal Intensive Care Unit. Int. J. Gen. Med. 2022, 15, 6237–6247. [Google Scholar] [CrossRef]
- Xie, Y.; Zhang, Y.; Du, L.; Jiang, X.; Yan, S.; Duan, W.; Li, J.; Zhan, Y.; Wang, L.; Zhang, S.; et al. Circulating long noncoding RNA act as potential novel biomarkers for diagnosis and prognosis of non-small cell lung cancer. Mol. Oncol. 2018, 12, 648–658. [Google Scholar] [CrossRef]
- Hu, X.; Bao, J.; Wang, Z.; Zhang, Z.; Gu, P.; Tao, F.; Cui, D.; Jiang, W. The plasma lncRNA acting as fingerprint in non-small-cell lung cancer. Tumor Biol. 2016, 37, 3497–3504. [Google Scholar] [CrossRef] [PubMed]
- Beylerli, O.; Khasanov, D.; Gareev, I.; Valitov, E.; Sokhatskii, A.; Wang, C.; Pavlov, V.; Khasanova, G.; Ahmad, A. Differential non-coding RNAs expression profiles of invasive and non-invasive pituitary adenomas. Non-Coding RNA Res. 2021, 6, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Gui, F.; Peng, H.; Liu, Y. Elevated circulating lnc-ANRIL/miR-125a axis level predicts higher risk, more severe disease condition, and worse prognosis of sepsis. J. Clin. Lab. Anal. 2019, 33, e22917. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Meng, F.; Ma, G.; Lei, H.; Liu, J. An increase in a long noncoding RNA ANRIL in peripheral plasma is an indicator of stable angina. Clinics 2023, 78, 100289. [Google Scholar] [CrossRef]
- Ge, J.; Geng, S.; Jiang, H. Long noncoding RNA s antisense noncoding RNA in the INK 4 locus (ANRIL) correlates with lower acute exacerbation risk, decreased inflammatory cytokines, and mild GOLD stage in patients with chronic obstructive pulmonary disease. J. Clin. Lab. Anal. 2019, 33, e22678. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Guo, J.; Ai, F. Circulating long noncoding RNA ANRIL downregulation correlates with increased risk, higher disease severity and elevated pro-inflammatory cytokines in patients with acute ischemic stroke. J. Clin. Lab. Anal. 2019, 33, e22629. [Google Scholar] [CrossRef]
- Zheng, M.; Zheng, Y.; Gao, M.; Ma, H.; Zhang, X.; Li, Y.; Wang, F.; Huang, H. Expression and clinical value of lncRNA MALAT1 and lncRNA ANRIL in glaucoma patients. Exp. Ther. Med. 2019, 19, 1329–1335. [Google Scholar] [CrossRef] [PubMed]
- Fenoglio, C.; Oldoni, E.; Serpente, M.; De Riz, M.A.; Arcaro, M.; D’Anca, M.; Pietroboni, A.M.; Calvi, A.; Lecchi, E.; Goris, A.; et al. LncRNAs expression profile in peripheral blood mononuclear cells from multiple sclerosis patients. J. Neuroimmunol. 2018, 324, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Li, S.; He, Y.; Ren, Q.; Qin, S. Long non-coding RNA ANRIL serves as a potential marker of disease risk, inflammation, and disease activity of pediatric inflammatory bowel disease. Clin. Res. Hepatol. Gastroenterol. 2022, 46, 101895. [Google Scholar] [CrossRef]
- Ayoub, S.E.; Shaker, O.G.; Aboshama, R.A.; Etman, M.K.; Khalefa, A.A.; Elguaad, M.M.K.A.; Zaki, O.M.; Ali, D.Y.; Hemeda, N.F.; Amin, A.; et al. Expression profile of LncRNA ANRIL, miR-186, miR-181a, and MTMR-3 in patients with preeclampsia. Non-Coding RNA Res. 2023, 8, 481–486. [Google Scholar] [CrossRef]
- Huang, T.; Wang, J.; Zhou, Y.; Zhao, Y.; Hang, D.; Cao, Y. LncRNA CASC2 is up-regulated in osteoarthritis and participates in the regulation of IL-17 expression and chondrocyte proliferation and apoptosis. Biosci. Rep. 2019, 39, BSR20182454. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, N.; Wu, C. LncRNA CASC 2 is upregulated in aphthous stomatitis and predicts the recurrence. BMC Oral Health 2020, 20, 12. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; e Kan, Q.; Su, Y.; Man, H. Long Non-Coding RNA CASC2 Improves Diabetic Nephropathy by Inhibiting JNK Pathway. Exp. Clin. Endocrinol. Diabetes 2019, 127, 533–537. [Google Scholar] [CrossRef] [PubMed]
- Refai, N.S.; Louka, M.L.; Halim, H.Y.; Montasser, I. Long non-coding RNAs (CASC2 and TUG1) in hepatocellular carcinoma: Clinical significance. J. Gene Med. 2019, 21, e3112. [Google Scholar] [CrossRef]
- Yang, Y.; Sun, Z.; Ren, T.; Lei, W. Differential Expression of lncRNA CASC2 in the Serum of Childhood Asthma and Its Role in Airway Smooth Muscle Cells Proliferation and Migration. J. Asthma Allergy 2022, 15, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Su, N.; Zhang, Y.; Wang, G. Clinical Significance of Serum lncRNA Cancer Susceptibility Candidate 2 (CASC2) for Chronic Renal Failure in Patients with Type 2 Diabetes. Med. Sci. Monit. 2018, 24, 6079–6084. [Google Scholar] [CrossRef]
- Dong, Y.; Wu, W. Downregulation of lncRNA CASC2 promotes the postoperative local recurrence of early oral squamous cell carcinoma. Eur. Arch. Oto-Rhino-Laryngol. 2019, 276, 605–610. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Wei, L.; Yin, X.; Li, H.; Qin, G.; Li, S.; Peng, T.; Liu, B.; Zhao, S.; Zhuo, Q. Long non-coding RNA cancer susceptibility candidate 2 regulates the function of human fibroblast-like synoviocytes via the microRNA-18a-5p/B-cell translocation gene 3 signaling axis in rheumatoid arthritis. Bioengineered 2022, 13, 3240–3250. [Google Scholar] [CrossRef]
- Liu, C.; Guo, X.; Bai, S.; Zeng, G.; Wang, H. LncRNA CASC2 downregulation participates in rheumatoid arthritis, and CASC2 overexpression promotes the apoptosis of fibroblast like synoviocytes by downregulating IL 17. Mol. Med. Rep. 2020, 21, 2131–2137. [Google Scholar] [CrossRef]
- Wang, M.; Wei, J.; Shang, F.; Zang, K.; Ji, T. Long non coding RNA CASC2 ameliorates sepsis induced acute kidney injury by regulating the miR 155 and NF κB pathway. Int. J. Mol. Med. 2020, 45, 1554–1562. [Google Scholar] [CrossRef]
- Wang, R.; Zhao, J.; Wei, Q.; Wang, H.; Zhao, C.; Hu, C.; Han, Y.; Hui, Z.; Yang, L.; Dai, Q.; et al. Potential of circulating lncRNA CASC2 as a biomarker in reflecting the inflammatory cytokines, multi-organ dysfunction, disease severity, and mortality in sepsis patients. J. Clin. Lab. Anal. 2022, 36, e24569. [Google Scholar] [CrossRef] [PubMed]
- Hola, M.A.M.; Ali, M.A.M.; ElNahass, Y.; Salem, T.A.E.; Mohamed, M.R. Expression and prognostic relevance of long noncoding RNAs CRNDE and AOX2P in adult acute myeloid leukemia. Int. J. Lab. Hematol. 2021, 43, 732–742. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Shi, L.; Song, L.; Long, Y.; Yuan, K.; Ding, W.; Deng, L. PLncRNA CRNDE and lncRNA SNHG7 are Promising Biomarkers for Prognosis in Synchronous Colorectal Liver Metastasis Following Hepatectomy/p. Cancer Manag. Res. 2020, 12, 1681–1692. [Google Scholar] [CrossRef] [PubMed]
- Graham, L.D.; Pedersen, S.K.; Brown, G.S.; Ho, T.; Kassir, Z.; Moynihan, A.T.; Vizgoft, E.K.; Dunne, R.; Pimlott, L.; Young, G.P.; et al. Colorectal Neoplasia Differentially Expressed (CRNDE), a Novel Gene with Elevated Expression in Colorectal Adenomas and Adenocarcinomas. Genes Cancer 2012, 2, 829–840. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zhu, H.; Xiao, M.; Zhou, S. Serum exosomal long noncoding RNA CRNDE as a prognostic biomarker for hepatocellular carcinoma. J. Clin. Lab. Anal. 2021, 35, e23959. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, C. Serum Colorectal Neoplasia Differentially Expressed Level and Prognostic Factors in Patients with RLNM of NPC. Altern. Ther. Health Med. 2023; ahead of print. [Google Scholar]
- Yuan, R.; Dai, C.; Chen, P.; Lv, M.; Shu, Y.; Wang, Z.; Xu, Y.; Li, J. Circulating TP73-AS1 and CRNDE serve as diagnostic and prognostic biomarkers for non-small cell lung cancer. Cancer Med. 2023, 12, 1655–1672. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, W.; Xu, M.; Yu, L. Long non-coding RNA CRNDE and toll-like receptor 3 correlate with disease severity, inflammation, and mortality in sepsis. J. Clin. Lab. Anal. 2020, 34, 9. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Ma, L. Clinical and prognostic significance of long non-coding RNA CRNDE expression in severe pneumonia and its correlation with inflammatory factor levels. Am. J. Transl. Res. 2023, 15, 1798–1806. [Google Scholar] [PubMed]
- Shehata, A.M.F.; Gohar, S.F.; Muharram, N.M.; Soliman, S.S.; Shalaby, H.M.; Eldin, S.M.K.; El-Bassal, F.I. LncRNA CRNDE is downregulated and associated with poor prognostic markers in chronic lymphocytic leukemia. Int. J. Lab. Hematol. 2024, 46, 107–112. [Google Scholar] [CrossRef]
- Yue, C.; He, M.; Teng, Y.; Bian, X. Long non-coding RNA metastasis-related lung adenocarcinoma transcript 1 (MALAT1) forms a negative feedback loop with long non-coding RNA colorectal neoplasia differentially expressed (CRNDE) in sepsis to regulate lung cell apoptosis. Bioengineered 2022, 13, 8201–8207. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, G.; Zou, C.; Gong, Z.; Wang, S.; Liu, J.; Ma, G.; Liu, X.; Zhang, W.; Jiang, P. Long noncoding RNA DGCR5 suppresses gastric cancer progression by acting as a competing endogenous RNA of PTEN and BTG1. J. Cell. Physiol. 2019, 234, 11999–12010. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Wang, X.; Zhang, W.; Zhangyuan, G.; Jin, K.; Yu, W.; Xie, Y.; Xu, X.; Wang, H.; Sun, B. Down-Regulation of LncRNA DGCR5 Correlates with Poor Prognosis in Hepatocellular Carcinoma. Cell. Physiol. Biochem. 2016, 40, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Gu, Y. LncRNA DLEU1 promotes angiogenesis in diabetic foot ulcer wound healing by regulating miR-96-5p. Ir. J. Med. Sci. 2024, 193, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Shan, L.; Zhao, T.; Wang, Y. Upregulation of Serum lncRNA DLEU1 Predicts Progression of Premalignant Endometrial Lesion and Unfavorable Clinical Outcome of Endometrial Cancer. Technol. Cancer Res. Treat. 2020, 19, 1533033820965589. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lu, X.; Yang, M.; Shangguan, J.; Yin, Y. GAS5 knockdown suppresses inflammation and oxidative stress induced by oxidized low-density lipoprotein in macrophages by sponging miR-135a. Mol. Cell. Biochem. 2021, 476, 949–957. [Google Scholar] [CrossRef] [PubMed]
- Weber, D.G.; Casjens, S.; Brik, A.; Raiko, I.; Lehnert, M.; Taeger, D.; Gleichenhagen, J.; Kollmeier, J.; Bauer, T.T.; et al.; The MoMar Study Group Circulating long non-coding RNA GAS5 (growth arrest-specific transcript 5) as a complement marker for the detection of malignant mesothelioma using liquid biopsies. Biomark. Res. 2020, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Senousy, M.A.; Shaker, O.G.; Sayed, N.H.; Fathy, N.; Kortam, M.A. LncRNA GAS5 and miR-137 Polymorphisms and Expression are Associated with Multiple Sclerosis Risk: Mechanistic Insights and Potential Clinical Impact. ACS Chem. Neurosci. 2020, 11, 1651–1660. [Google Scholar] [CrossRef] [PubMed]
- Fantini, S.; Rontauroli, S.; Sartini, S.; Mirabile, M.; Bianchi, E.; Badii, F.; Maccaferri, M.; Guglielmelli, P.; Ottone, T.; Palmieri, R.; et al. Increased Plasma Levels of lncRNAs LINC01268, GAS5 and MALAT1 Correlate with Negative Prognostic Factors in Myelofibrosis. Cancers 2021, 13, 4744. [Google Scholar] [CrossRef] [PubMed]
- Cong, C.; Tian, J.; Gao, T.; Zhou, C.; Wang, Y.; Cui, X.; Zhu, L. PlncRNA GAS5 Is Upregulated in Osteoporosis and Downregulates miR-21 to Promote Apoptosis of Osteoclasts/p. Clin. Interv. Aging 2020, 15, 1163–1169. [Google Scholar] [CrossRef]
- Visconti, V.V.; Fittipaldi, S.; Ciuffi, S.; Marini, F.; Isaia, G.; D’amelio, P.; Migliaccio, S.; Marcocci, C.; Minisola, S.; Nuti, R.; et al. Circulating Long Non-Coding RNA GAS5 Is Overexpressed in Serum from Osteoporotic Patients and Is Associated with Increased Risk of Bone Fragility. Int. J. Mol. Sci. 2020, 21, 6930. [Google Scholar] [CrossRef]
- Wang, C.; Yue, S.; Jiang, Y.; Mao, Y.; Zhao, Z.; Liu, X.; Zhang, X.; Pei, D.; Li, Y. LncRNA GAS5 is upregulated in polycystic ovary syndrome and regulates cell apoptosis and the expression of IL-6. J. Ovarian Res. 2020, 13, 145. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Hodges, T.R.; Song, R.; Gong, Y.; Calin, G.A.; Heimberger, A.B.; Zhao, H. Serum HOTAIR and GAS5 levels as predictors of survival in patients with glioblastoma. Mol. Carcinog. 2018, 57, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Toraih, E.A.; El-Wazir, A.; Al Ageeli, E.; Hussein, M.H.; Eltoukhy, M.M.; Killackey, M.T.; Kandil, E.; Fawzy, M.S. Unleash multifunctional role of long noncoding RNAs biomarker panel in breast cancer: A predictor classification model. Epigenomics 2020, 12, 1215–1237. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.; Shaker, O.G.; Khalifa, A.A.; Ezzat, E.M.; Elghobary, H.A.; Mawla, T.S.A.; Elkhateeb, A.F.; Elebiary, A.M.A.; Elamir, A.M. LncRNAs NEAT1, HOTAIR, and GAS5 expression in hypertensive and non-hypertensive associated cerebrovascular stroke patients, and its link to clinical characteristics and severity score of the disease. Non-Coding RNA Res. 2023, 8, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.; Wu, A.; Liu, M. Plasma Long Non-Coding RNA (lncRNA) GAS5 is a New Biomarker for Coronary Artery Disease. Med. Sci. Monit. 2017, 23, 6042–6048. [Google Scholar] [CrossRef] [PubMed]
- Ayeldeen, G.; Shaker, O.G.; Amer, E.; Zaafan, M.A.; Herzalla, M.R.; Keshk, M.A.; Abdelhamid, A.M. The Impact of lncRNA-GAS5/miRNA-200/ACE2 Molecular Pathway on the Severity of COVID-19. Curr. Med. Chem. 2024, 31, 1142–1151. [Google Scholar] [CrossRef]
- Sun, H.; Chen, T.; Li, X.; Zhu, Y.; Zhang, S.; He, P.; Peng, Y.; Fan, Q. The relevance of the non-invasive biomarkers lncRNA GAS5/miR-21 ceRNA regulatory network in the early identification of diabetes and diabetic nephropathy. Diabetol. Metab. Syndr. 2023, 15, 197. [Google Scholar] [CrossRef]
- Wang, Y.; Jing, W.; Ma, W.; Liang, C.; Chai, H.; Tu, J. Down-regulation of long non-coding RNA GAS5-AS1 and its prognostic and diagnostic significance in hepatocellular carcinoma. Cancer Biomark. 2018, 22, 227–236. [Google Scholar] [CrossRef]
- Guo, Y.; Li, C.; Zhang, R.; Zhan, Y.; Yu, J.; Tu, J.; Zheng, J. Epigenetically-regulated serum GAS5 as a potential biomarker for patients with chronic hepatitis B virus infection. Cancer Biomark. 2021, 32, 137–146. [Google Scholar] [CrossRef]
- Li, Y.; Sun, L.; Liu, J.; Xu, G.; Hu, Y.; Qin, A. Down-regulation of GAS5 has diagnostic value for tuberculosis and regulates the inflammatory response in mycobacterium tuberculosis infected THP-1 cells. Tuberculosis 2022, 132, 102141. [Google Scholar] [CrossRef]
- Li, C.; Lv, Y.; Shao, C.; Chen, C.; Zhang, T.; Wei, Y.; Fan, H.; Lv, T.; Liu, H.; Song, Y. Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J. Cell. Physiol. 2019, 234, 20721–20727. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Lv, T.; Shi, X.; Liu, H.; Zhu, Q.; Zeng, J.; Yang, W.; Yin, J.; Song, Y. Circulating long noncoding RNA GAS5 is a novel biomarker for the diagnosis of nonsmall cell lung cancer. Medicine 2016, 95, e4608. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Li, Q.; Liu, Y.-F.; Li, Y. LncRNA GAS5 regulates angiogenesis by targeting miR 10a 3p/VEGFA in osteoporosis. Mol. Med. Rep. 2021, 24, 4. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Xing, W.; Li, Y.; Xie, Y.; Tang, X.; Zhang, Q. Downregulation of serum long noncoding RNA GAS5 may contribute to insulin resistance in PCOS patients. Gynecol. Endocrinol. 2018, 34, 784–788. [Google Scholar] [CrossRef] [PubMed]
- Tofigh, R.; Hosseinpourfeizi, M.; Safaralizadeh, R.; Ghoddusifar, S.; Baradaran, B. Serum Levels of Long Non-coding RNAs NEAT1, GAS5, and GAPLINC Altered in Rheumatoid Arthritis. Curr. Rheumatol. Rev. 2024, 20, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Wang, W.; Li, P. LncRNA GAS5 overexpression downregulates IL-18 and induces the apoptosis of fibroblast-like synoviocytes. Clin. Rheumatol. 2019, 38, 3275–3280. [Google Scholar] [CrossRef]
- Zeng, Z.; Lan, Y.; Chen, Y.; Zuo, F.; Gong, Y.; Luo, G.; Peng, Y.; Yuan, Z. LncRNA GAS5 suppresses inflammatory responses by inhibiting HMGB1 release via miR-155-5p/SIRT1 axis in sepsis. Eur. J. Pharmacol. 2023, 942, 175520. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.-C.; Li, J.; Leng, R.-X.; Li, X.-P.; Li, X.-M.; Wang, D.-G.; Pan, H.-F.; Ye, D.-Q. Identification of long non-coding RNAs GAS5, linc0597 and lnc-DC in plasma as novel biomarkers for systemic lupus erythematosus. Oncotarget 2017, 8, 23650–23663. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Liu, L.; Chen, J.; Zhi, J.; Li, J.; Li, L.; Jiang, Z. LncRNA HOTAIR in exercise-induced neuro-protective function in Alzheimer’s disease. Folia Neuropathol. 2022, 60, 414–420. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, K.; Luo, Z.; Liu, L.; Wu, L.; Liu, J. Circulating long non-coding HOX transcript antisense intergenic ribonucleic acid in plasma as a potential biomarker for diagnosis of breast cancer. Thorac. Cancer 2016, 7, 627–632. [Google Scholar] [CrossRef]
- Zhang, L.; Song, X.; Wang, X.; Xie, Y.; Wang, Z.; Xu, Y.; You, X.; Liang, Z.; Cao, H. Circulating DNA of HOTAIR in serum is a novel biomarker for breast cancer. Breast Cancer Res. Treat. 2015, 152, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Song, M.; Zhang, J.; Kuerban, M.; Wang, H. Combined identification of long non-coding RNA CCAT1 and HOTAIR in serum as an effective screening for colorectal carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 14131–14140. [Google Scholar] [PubMed]
- Jiang, Y.; Mo, H.; Luo, J.; Zhao, S.; Liang, S.; Zhang, M.; Yuan, J. HOTAIR Is a Potential Novel Biomarker in Patients with Congenital Heart Diseases. BioMed Res. Int. 2018, 2018, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Avazpour, N.; Hajjari, M.; Yazdankhah, S.; Sahni, A.; Foroughmand, A.M. Circulating HOTAIR LncRNA Is Potentially Up-regulated in Coronary Artery Disease. Genom. Inform. 2018, 16, e25. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xia, Y.; Zhang, Y. Diagnostic significance of serum lncRNA HOTAIR and its predictive value for the development of chronic complications in patients with type 2 diabetes mellitus. Diabetol. Metab. Syndr. 2021, 13, 97. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; He, X.; Zheng, Z.; Ma, X.; Hu, X.; Wu, D.; Wang, M. Serum HOTAIR as a novel diagnostic biomarker for esophageal squamous cell carcinoma. Mol. Cancer 2017, 16, 75. [Google Scholar] [CrossRef] [PubMed]
- Elsayed, E.T.; Salem, P.E.; Darwish, A.M.; Fayed, H.M. Plasma long non-coding RNA HOTAIR as a potential biomarker for gastric cancer. Int. J. Biol. Markers 2018, 33, 528–533. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Liu, Z.; Xiao, H.; Yang, X.; Li, T.; Huang, W.; Zhou, H. Effect of tumor exosome-derived Lnc RNA HOTAIR on the growth and metastasis of gastric cancer. Clin. Transl. Oncol. 2023, 25, 3447–3459. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Wu, X.; Ke, F. Long Non-Coding RNA HOTAIR Expression and Clinical Significance in Patients with Gestational Diabetes. Int. J. Gen. Med. 2021, 14, 9945–9950. [Google Scholar] [CrossRef]
- Wang, X.; Yu, X.; Xu, H.; Wei, K.; Wang, S.; Wang, Y.; Han, J. Serum-derived extracellular vesicles facilitate temozolomide resistance in glioblastoma through a HOTAIR-dependent mechanism. Cell Death Dis. 2022, 13, 4. [Google Scholar] [CrossRef]
- Lou, Z.-H.; Xu, K.-Y.; Qiao, L.; Su, X.-Q.; Ou-Yang, Y.; Miao, L.-B.; Liu, F.; Wang, Y.; Fu, A.; Ren, X.-H.; et al. Diagnostic Potential of the Serum lncRNAs HOTAIR, BRM and ICR for Hepatocellular Carcinoma. Front. Biosci.-Landmark 2022, 27, 264. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, Y.; Lu, J.; Sun, Y.; Xiao, H.; Liu, M.; Tian, L. Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Med. Oncol. 2014, 31, 9. [Google Scholar] [CrossRef] [PubMed]
- Shehata, A.M.F.; Eldin, S.M.K.; Osman, N.F.; Helwa, M.A. Deregulated Expression of Long Non-coding RNA HOX Transcript Antisense RNA (HOTAIR) in Egyptian Patients with Multiple Myeloma. Indian J. Hematol. Blood Transfus. 2020, 36, 271–276. [Google Scholar] [CrossRef]
- Li, N.; Wang, Y.; Liu, X.; Luo, P.; Jing, W.; Zhu, M.; Tu, J. Identification of Circulating Long Noncoding RNA HOTAIR as a Novel Biomarker for Diagnosis and Monitoring of Non–Small Cell Lung Cancer. Technol. Cancer Res. Treat. 2017, 16, 1060–1066. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Liu, J.; Luo, T.; Chen, Q.; Lu, M.; Meng, D. LncRNA PACER is down-regulated in osteoarthritis and regulates chondrocyte apoptosis and lncRNA HOTAIR expression. Biosci. Rep. 2019, 39, 6. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, J.; Li, Z.; Qiu, S.; Cao, J.; Zhao, Y.; Huang, Z.; He, J.; Luo, F.; Yang, K. Diagnostic Value of Serum lncRNA HOTAIR Combined with Galectin-3 in Benign and Papillary Thyroid Carcinoma. Cancer Manag. Res. 2021, 13, 6517–6525. [Google Scholar] [CrossRef]
- Tan, J.; Dan, J.; Liu, Y. Clinical Efficacy of Methotrexate Combined with Iguratimod on Patients with Rheumatoid Arthritis and Its Influence on the Expression Levels of HOTAIR in Serum. BioMed Res. Int. 2021, 2021, 2486617. [Google Scholar] [CrossRef]
- Mahmoud, R.H.; Fouad, N.A.; Hefzy, E.M.; Shaker, O.G.; Ahmed, T.I.; Hussein, H.A.; Nasr, M.H.; Zaki, O.M.; Abdelghaffar, N.K.; Abdelaleem, O.O. The potential role of serum expression profile of long non coding RNAs, Cox2 and HOTAIR as novel diagnostic biomarkers in systemic lupus erythematosus. PLoS ONE 2022, 17, e0268176. [Google Scholar] [CrossRef]
- Chen, H.; Li, X.; Chen, W.; Wu, T.; Liu, S. LncRNA HOTAIR Inhibits miR-19a-3p to Alleviate Foam Cell Formation and Inflammatory Response in Atherosclerosis. Int. J. Med. Sci. 2024, 21, 521–529. [Google Scholar] [CrossRef]
- Hameed, N.A.A.; Shaker, O.G.; Hasona, N.A. LINC00641/miR-378a and Their Cross-Talk with TNF-α/IFN-γ as Potential Biomarkers in Ulcerative Colitis and Crohn’s Diseases. J. Interferon Cytokine Res. 2023, 43, 531–537. [Google Scholar] [CrossRef]
- Lu, H.; Wang, G.; Zhao, J.; Jiang, H. Knockdown of lncRNA MALAT1 ameliorates acute kidney injury by mediating the miR-204/APOL1 pathway. J. Clin. Lab. Anal. 2021, 35, e23881. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Niu, Z.; Zhang, R.; Peng, Z.; Wang, L.; Liu, Z.; Gao, Y.; Pei, H.; Pan, L. MALAT1 shuttled by extracellular vesicles promotes M1 polarization of macrophages to induce acute pancreatitis via miR-181a-5p/HMGB1 axis. J. Cell. Mol. Med. 2021, 25, 9241–9254. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Jin, J.; Liu, E.; Zhang, J. A novel circulating biomarker lnc-MALAT1 for acute myocardial infarction: Its relationship with disease risk, features, cytokines, and major adverse cardiovascular events. J. Clin. Lab. Anal. 2022, 36, e24771. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Fan, R.; Chen, L.; Qian, H. Clinical Significance of Long Non-coding RNA MALAT1 Expression in Tissue and Serum of Breast Cancer. Ann. Clin. Lab. Sci. 2016, 46, 418–424. [Google Scholar] [PubMed]
- Xia, H.; Chen, Q.; Chen, Y.; Ge, X.; Leng, W.; Tang, Q.; Ren, M.; Chen, L.; Yuan, D.; Zhang, Y.; et al. The lncRNA MALAT1 is a novel biomarker for gastric cancer metastasis. Oncotarget 2016, 7, 56209–56218. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, H.; Wang, F.; Ye, M.; Zhu, H.; Bu, S. Long non-coding RNA MALAT 1 expression in patients with gestational diabetes mellitus. Int. J. Gynecol. Obstet. 2018, 140, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Wang, Y.; Hu, P.; Wu, J. Long Non-Coding RNA Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT1) Promotes Hypertension by Modulating the Hsa-miR-124-3p/Nuclear Receptor Subfamily 3, Group C, Member 2 (NR3C2) and Hsa-miR-135a-5p/NR3C2 Axis. Med. Sci. Monit. 2020, 26, e920478-1–e920478-10. [Google Scholar] [CrossRef] [PubMed]
- Shaker, O.G.; Mahmoud, R.H.; Abdelaleem, O.O.; Ibrahem, E.G.; Mohamed, A.A.; Zaki, O.M.; Abdelghaffar, N.K.; Ahmed, T.I.; Hemeda, N.F.; Ahmed, N.A.; et al. LncRNAs, MALAT1 and lnc-DC as potential biomarkers for multiple sclerosis diagnosis. Biosci. Rep. 2019, 39, 1. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Zeng, J.; Chao, W.; Chen, X.; Huang, Y.; Deng, K.; Huang, Z.; Li, J.; Dai, M.; Chen, S.; et al. Serum long non-coding RNAs MALAT1, AFAP1-AS1 and AL359062 as diagnostic and prognostic biomarkers for nasopharyngeal carcinoma. Oncotarget 2017, 8, 41166–41177. [Google Scholar] [CrossRef]
- Fernandes, M.; Marques, H.; Teixeira, A.L.; Medeiros, R. CeRNA Network of lncRNA/miRNA as Circulating Prognostic Biomarkers in Non-Hodgkin Lymphomas: Bioinformatic Analysis and Assessment of Their Prognostic Value in an NHL Cohort. Int. J. Mol. Sci. 2022, 23, 201. [Google Scholar] [CrossRef]
- Zhang, R.; Xia, Y.; Wang, Z.; Zheng, J.; Chen, Y.; Li, X.; Wang, Y.; Ming, H. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2017, 490, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Huo, Y.; Li, Q.; Wang, X.; Jiao, X.; Zheng, J.; Li, Z.; Pan, X. MALAT1 predicts poor survival in osteosarcoma patients and promotes cell metastasis through associating with EZH2. Oncotarget 2017, 8, 46993–47006. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Fu, Z.; Dai, C.; Cao, J.; Liu, X.; Xu, J.; Lv, M.; Gu, Y.; Zhang, J.; Hua, X.; et al. LncRNAs expression profiling in normal ovary, benign ovarian cyst and malignant epithelial ovarian cancer. Sci. Rep. 2016, 6, 38983. [Google Scholar] [CrossRef] [PubMed]
- Yang, H. LncRNA MALAT1 potentiates inflammation disorder in Parkinson’s disease. Int. J. Immunogenet. 2021, 48, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Pan, Z.; Wang, P. The Value of Combined Detection of Serum PSA, MALAT1 and TMPRSS2-ETV1 in Evaluating the Progress and Prognosis of Prostate Cancer. Arch. Españoles De Urol. 2023, 76, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Bhattcharjee, D.; Misra, S.; Saha, A.; Bhattacharyya, N.P.; Ghosh, A. Increase in MEG3, MALAT1, NEAT1 significantly predicts the clinical parameters in patients with rheumatoid arthritis. Pers. Med. 2020, 17, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; He, Y.; Zhou, L.; Deng, Y.; Si, L. Long non coding RNA MALAT1 serves as an independent predictive biomarker for the diagnosis, severity and prognosis of patients with sepsis. Mol. Med. Rep. 2020, 21, 1365–1373. [Google Scholar] [CrossRef]
- Yan, L.-P.; Liu, Z.-B.; Wu, M.; Ge, Y.-P.; Zhang, Q. Effect of lncRNA MALAT1 expression on survival status of elderly patients with severe pneumonia. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 3959–3964. [Google Scholar] [PubMed]
- Ye, D.; Deng, Y.; Shen, Z. The Role and Mechanism of MALAT1 Long Non-Coding RNA in the Diagnosis and Treatment of Head and Neck Squamous Cell Carcinoma. OncoTargets Ther. 2021, 14, 4127–4136. [Google Scholar] [CrossRef]
- Zhu, M.; Xie, J. LncRNA MALAT1 Promotes Ulcerative Colitis by Upregulating lncRNA ANRIL. Dig. Dis. Sci. 2020, 65, 3191–3196. [Google Scholar] [CrossRef]
- Tello-Flores, V.A.; Valladares-Salgado, A.; Ramírez-Vargas, M.A.; Cruz, M.; Del-Moral-Hernández, O.; Cahua-Pablo, J.; Ramírez, M.; Hernández-Sotelo, D.; Armenta-Solis, A.; Flores-Alfaro, E. Altered levels of MALAT1 and H19 derived from serum or serum exosomes associated with type-2 diabetes. Non-Coding RNA Res. 2020, 5, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.-X.; Gao, H.-X.; Xu, X.-Y.; Ding, F.-K. Effects of lncRNA MALAT1 and lncRNA NKILA on proliferation, invasion and apoptosis of retinoblastoma. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 8296–8307. [Google Scholar] [CrossRef] [PubMed]
- Min, W.; Dai, D.; Wang, J.; Zhang, D.; Zhang, Y.; Han, G.; Zhang, L.; Chen, C.; Li, X.; Li, Y.; et al. Long Noncoding RNA miR210HG as a Potential Biomarker for the Diagnosis of Glioma. PLoS ONE 2016, 11, e0160451. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zheng, Y.; Li, L. LncRNA RPSAP52 regulates miR-423-5p/GSTM1 axis to suppress hypoxia-induced renal proximal tubular epithelial cell apoptosis. Arch. Physiol. Biochem. 2022, 128, 1066–1070. [Google Scholar] [CrossRef] [PubMed]
- Niu, T.; An, Y.; Lv, T.; Liu, D. Long non coding RNA RPSAP52 upregulates Timp3 by serving as the endogenous sponge of microRNA 365 in diabetic retinopathy. Exp. Ther. Med. 2020, 20, 1. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Wang, X.; Liu, S.; Chen, C.; Jiang, F.; Mao, K.; Zeng, F. LncRNA SAMMSON overexpression distinguished glioblastoma patients from patients with diffuse neurosarcoidosis. NeuroReport 2019, 30, 817–821. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Tian, X.; Zhang, Q.; Shi, P.; Li, S. Long non-coding RNA SAMMSON as a novel potential diagnostic and prognostic biomarker for oral squamous cell carcinoma. J. Dent. Sci. 2020, 15, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Sun, W.; Wang, Z.; Dong, W.; Qin, Y. Long noncoding RNA SAMMSON promotes papillary thyroid carcinoma progression through p300/Sp1 axis and serves as a novel diagnostic and prognostic biomarker. IUBMB Life 2020, 72, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Yang, Y.; Wang, L.; Shi, Q.; Ma, H.; He, S.; Feng, L.; Fang, J. SOX2-OT Binds with ILF3 to Promote Head and Neck Cancer Progression by Modulating Crosstalk between STAT3 and TGF-β Signaling. Cancers 2023, 15, 5766. [Google Scholar] [CrossRef]
- Teng, Y.; Kang, H.; Chu, Y. Identification of an Exosomal Long Noncoding RNA SOX2-OT in Plasma as a Promising Biomarker for Lung Squamous Cell Carcinoma. Genet. Test. Mol. Biomark. 2019, 23, 235–240. [Google Scholar] [CrossRef]
- Lai, Y.; Dong, L.; Jin, H.; Li, H.; Sun, M.; Li, J. Exosome long non-coding RNA SOX2-OT contributes to ovarian cancer malignant progression by miR-181b-5p/SCD1 signaling. Aging 2021, 13, 23726–23738. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Hei, B.; Hao, W.; Lin, S.; Wang, Y.; Liu, X.; Meng, X.; Guan, Z. Clinical value of lncRNA SOX2-OT in pulmonary arterial hypertension and its role in pulmonary artery smooth muscle cell proliferation, migration, apoptosis, and inflammatory. Heart Lung 2022, 55, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-G.; Zhou, M.-W.; Bai, L.; Han, R.-Y.; Lv, K.; Wang, Z. Extracellular vesicles promote esophageal cancer progression by delivering IncZEB1-AS1 between cells. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2667–2670. [Google Scholar]
- Kato, T.; Kawakami, K.; Mizutani, K.; Ando, T.; Sakai, Y.; Sakurai, K.; Toyota, S.; Ehara, H.; Ito, H.; Ito, M. H19 in Serum Extracellular Vesicles Reflects Resistance to AR Axis-targeted Therapy among CRPC Patients. Cancer Genom.-Proteom. 2023, 20, 456–468. [Google Scholar] [CrossRef] [PubMed]
- Rey, F.; Maghraby, E.; Messa, L.; Esposito, L.; Barzaghini, B.; Pandini, C.; Bordoni, M.; Gagliardi, S.; Diamanti, L.; Raimondi, M.T.; et al. Identification of a novel pathway in sporadic Amyotrophic Lateral Sclerosis mediated by the long non-coding RNA ZEB1-AS1. Neurobiol. Dis. 2023, 178, 106030. [Google Scholar] [CrossRef]
- Gu, L.; Sun, H.; Yan, Z. LncRNA ZEB1 AS1 is downregulated in diabetic lung and regulates lung cell apoptosis. Exp. Ther. Med. 2020, 20, 225. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, A.A.; Ashoori, H.; Vahidian, F.; Mosleh, I.S.; Kamian, S. Long non-coding RNA panel as a molecular biomarker in glioma. J. Egypt. Natl. Cancer Inst. 2021, 33, 1. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yang, Z.; Deng, Z.; Zhou, Y.; Gong, Q.; Zhao, R.; Chen, T. Upregulated lncRNA ADAMTS9-AS2 suppresses progression of lung cancer through inhibition of miR-223-3p and promotion of TGFBR3. IUBMB Life 2018, 70, 536–546. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Askari, A.; Hussen, B.M.; Baniahmad, A.; Taheri, M.; Mokhtari, M. A review on the role of ADAMTS9-AS2 in different disorders. Pathol.-Res. Pract. 2023, 243, 154346. [Google Scholar] [CrossRef]
- Jin, D.; Song, Y.; Chen, Y.; Zhang, P. Identification of Three lncRNAs as Potential Predictive Biomarkers of Lung Adenocarcinoma. BioMed Res. Int. 2020, 2020, 7573689. [Google Scholar] [CrossRef]
- Liu, D.; Wu, K.; Yang, Y.; Zhu, D.; Zhang, C.; Zhao, S. Long noncoding RNA ADAMTS9-AS2 suppresses the progression of esophageal cancer by mediating CDH3 promoter methylation. Mol. Carcinog. 2020, 59, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Chen, X.; Chen, S.; Yang, J.; Liu, Q.; Cheng, Y. Screening of tumor grade-related mRNAs and lncRNAs for Esophagus Squamous Cell Carcinoma. J. Clin. Lab. Anal. 2021, 35, e23797. [Google Scholar] [CrossRef] [PubMed]
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016, 7, 12429. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.; Lhuillier, J.; Grosjean, G.; Ayadi, L.; Maenner, S. The Long Non-Coding RNA ANRIL in Cancers. Cancers 2023, 15, 4160. [Google Scholar] [CrossRef] [PubMed]
- Lou, N.; Liu, G.; Pan, Y. Long noncoding RNA ANRIL as a novel biomarker in human cancer. Future Oncol. 2020, 16, 2981–2995. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.C.; Awasthee, N.; Rai, V.; Chava, S.; Gunda, V.; Challagundla, K.B. Long non-coding RNAs and nuclear factor-κB crosstalk in cancer and other human diseases. Biochim. Et Biophys. Acta (BBA)-Rev. Cancer 2020, 1873, 188316. [Google Scholar] [CrossRef] [PubMed]
- Wufuer, A.; Luohemanjiang, X.; Du, L.; Lei, J.; Shabier, M.; Han, D.F.; Ma, J. ANRIL overexpression globally induces expression and alternative splicing of genes involved in inflammation in HUVECs. Mol. Med. Rep. 2022, 27, 27. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zheng, H.; Tse, G.; Zhang, L.; Wu, W.K.K. CASC 2: An emerging tumour-suppressing long noncoding RNA in human cancers and melanoma. Cell Prolif. 2018, 51, e12506. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Chen, J.-Y.; Meng, J.; Chen, Z. Inflammation and DNA methylation coregulate the CtBP-PCAF-c-MYC transcriptional complex to activate the expression of a long non-coding RNA CASC2 in acute pancreatitis. Int. J. Biol. Sci. 2020, 16, 2116–2130. [Google Scholar] [CrossRef]
- Jiang, C.; Shen, F.; Du, J.; Fang, X.; Li, X.; Su, J.; Wang, X.; Huang, X.; Liu, Z. Upregulation of CASC2 sensitized glioma to temozolomide cytotoxicity through autophagy inhibition by sponging miR-193a-5p and regulating mTOR expression. Biomed. Pharmacother. 2018, 97, 844–850. [Google Scholar] [CrossRef]
- Wang, R.; Li, Y.; Zhu, G.; Tian, B.; Zeng, W.; Yang, Y.; Li, Z. Long noncoding RNA CASC2 predicts the prognosis of glioma patients and functions as a suppressor for gliomas by suppressing Wnt/β-catenin signaling pathway. Neuropsychiatr. Dis. Treat. 2017, 13, 1805–1813. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Shen, L.; Zhao, H.; Liu, Q.; Fu, J.; Guo, Y.; Peng, R.; Cheng, L. LncRNA CASC2 Interacts With miR-181a to Modulate Glioma Growth and Resistance to TMZ Through PTEN Pathway. J. Cell. Biochem. 2017, 118, 1889–1899. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yin, M.; Peng, G.; Zhao, Y. CRNDE: An important oncogenic long non-coding RNA in human cancers. Cell Prolif. 2018, 51, e12440. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wang, L.; Li, Z.; Zheng, Y.; Shi, Z.; Wang, G. Long noncoding RNA CRNDE functions as a diagnostic and prognostic biomarker in osteosarcoma, as well as promotes its progression via inhibition of miR-335-3p. J. Biochem. Mol. Toxicol. 2021, 35, e22734. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Sha, H.; Sun, X.; Zhang, Y.; Wu, Y.; Zhang, J.; Zhang, H.; Wu, J.; Feng, J. CRNDE: An oncogenic long non-coding RNA in cancers. Cancer Cell Int. 2020, 20, 162. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Ma, B.; Gao, Q.; Zhan, H.; Liu, Y.; Chen, Z.; Ye, S.; Li, J.; Yao, L.; Huang, W. Long non-coding RNA CRNDE in cancer prognosis: Review and meta-analysis. Clin. Chim. Acta 2018, 485, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Zottel, A.; Šamec, N.; Paska, A.V.; Jovčevska, I. Coding of Glioblastoma Progression and Therapy Resistance through Long Noncoding RNAs. Cancers 2020, 12, 1842. [Google Scholar] [CrossRef]
- Kiang, K.M.-Y.; Zhang, X.-Q.; Zhang, G.P.; Li, N.; Cheng, S.Y.; Poon, M.-W.; Pu, J.K.-S.; Lui, W.-M.; Leung, G.K.-K. CRNDE Expression Positively Correlates with EGFR Activation and Modulates Glioma Cell Growth. Target. Oncol. 2017, 12, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Guan, G.; Li, X.; Wei, C.; Wu, J.; Cheng, P.; Wu, A.; Cheng, W. Profiling pro-neural to mesenchymal transition identifies a lncRNA signature in glioma. J. Transl. Med. 2020, 18, 378. [Google Scholar] [CrossRef]
- Xue, C.; Chen, C.; Gu, X.; Li, L. Progress and assessment of lncRNA DGCR5 in malignant phenotype and immune infiltration of human cancers. Am. J. Cancer Res. 2021, 11, 1–13. [Google Scholar]
- Johnson, R. Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol. Dis. 2012, 46, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Yang, X.; Chen, J.; Fu, J.; Chen, C.; Wen, J.; Mo, Q. LncRNA DGCR5 inhibits the proliferation of colorectal cancer cells by downregulating miR 21. Oncol. Lett. 2019, 18, 3331–3336. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chang, Y.; Lu, S.; Xiang, Y. Downregulation of long noncoding RNA DGCR5 contributes to the proliferation, migration, and invasion of cervical cancer by activating Wnt signaling pathway. J. Cell. Physiol. 2019, 234, 11662–11669. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; He, W.; Xu, T.; Dai, J.; Xu, L.; Sun, F. Upregulation of lncRNA DGCR5 correlates with better prognosis and inhibits bladder cancer progression via transcriptionally facilitating P21 expression. J. Cell. Physiol. 2019, 234, 6254–6262. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Shi, M.; Xiang, T.; Bu, Y. Long noncoding RNA DGCR5 represses hepatocellular carcinoma progression by inactivating Wnt signaling pathway. J. Cell. Biochem. 2019, 120, 275–282. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, J.; Shi, M.; Chen, F. LncRNA DGCR5 represses the development of hepatocellular carcinoma by targeting the miR-346/KLF14 axis. J. Cell. Physiol. 2019, 234, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Wang, R.; Jin, X.; Zeng, J.; Pan, J. LncRNA DGCR5 promotes lung adenocarcinoma (LUAD) progression via inhibiting hsa-mir-22-3p. J. Cell. Physiol. 2018, 233, 4126–4136. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Dong, H.; Zeng, J.; Pan, J.; Jin, X. LncRNA DGCR5 contributes to CSC-like properties via modulating miR-330-5p/CD44 in NSCLC. J. Cell. Physiol. 2018, 233, 7447–7456. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Chu, B.; Cai, C.; Wu, X.; Yao, W.; Wu, Z.; Yang, Z.; Li, F.; Liu, Y.; Dong, P.; et al. DGCR5 Promotes Gallbladder Cancer by Sponging MiR-3619-5p via MEK/ERK1/2 and JNK/p38 MAPK Pathways. J. Cancer 2020, 11, 5466–5477. [Google Scholar] [CrossRef]
- Wu, X.; Hou, P.; Qiu, Y.; Wang, Q.; Lu, X. Large-Scale Analysis Reveals the Specific Clinical and Immune Features of DGCR5 in Glioma/p. OncoTargets Ther. 2020, 13, 7531–7543. [Google Scholar] [CrossRef]
- Zhang, B.; Cheng, Y.; Li, R.; Lian, M.; Guo, S.; Liang, C. Development of a novel angiogenesis-related lncRNA signature to predict the prognosis and immunotherapy of glioblastoma multiforme. Transl. Cancer Res. 2023, 12, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xie, Z.; Lei, X.; Gan, R. Long non coding RNA GAS5 in human cancer (Review). Oncol. Lett. 2020, 20, 2587–2594. [Google Scholar] [CrossRef]
- Yu, Y.; Hann, S.S. Novel Tumor Suppressor lncRNA Growth Arrest-Specific 5 (GAS5) In Human Cancer. OncoTargets Ther. 2019, 12, 8421–8436. [Google Scholar] [CrossRef]
- Huang, H.; Du, J.; Jin, B.; Pang, L.; Duan, N.; Huang, C.; Hou, J.; Yu, W.; Hao, H.; Li, H. Combination of Urine Exosomal mRNAs and lncRNAs as Novel Diagnostic Biomarkers for Bladder Cancer. Front. Oncol. 2021, 11, 667212. [Google Scholar] [CrossRef]
- Wu, S.; Ren, K.; Zhao, J.; Li, J.; Jia, B.; Wu, X.; Dou, Y.; Fei, X.; Huan, Y.; He, X.; et al. LncRNA GAS5 represses stemness and malignancy of gliomas via elevating the SPACA6-miR-125a/let-7e Axis. Front. Oncol. 2022, 12, 803652. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Luo, Q.; Xiao, Y.; Zhu, J.; Zhang, Y.; Ding, J.; Li, J. LINC00467: An oncogenic long noncoding RNA. Cancer Cell Int. 2022, 22, 303. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Li, J.; Bo, H.; He, D.; Xiao, M.; Xiang, L.; Gong, L.; Hu, Y.; Zhang, Y.; Cheng, Y.; et al. LINC00467 is up-regulated by TDG-mediated acetylation in non-small cell lung cancer and promotes tumor progression. Oncogene 2020, 39, 6071–6084. [Google Scholar] [CrossRef]
- Yang, J.; Liu, Y.; Mai, X.; Lu, S.; Jin, L.; Tai, X. STAT1-induced upregulation of LINC00467 promotes the proliferation migration of lung adenocarcinoma cells by epigenetically silencing DKK1 to activate Wnt/β-catenin signaling pathway. Biochem. Biophys. Res. Commun. 2019, 514, 118–126. [Google Scholar] [CrossRef]
- Ding, H.; Luo, Y.; Hu, K.; Liu, P.; Xiong, M. PLinc00467 promotes lung adenocarcinoma proliferation via sponging miR-20b-5p to activate CCND1 expression/p. OncoTargets Ther. 2019, 12, 6733–6743. [Google Scholar] [CrossRef]
- Deng, L.-H.; Zhao, H.; Bai, L.-P.; Xie, J.; Liu, K.; Yan, F. Linc00467 promotion of gastric cancer development by directly regulating miR-7-5p expression and downstream epidermal growth factor receptor. Bioengineered 2021, 12, 9484–9495. [Google Scholar] [CrossRef]
- Changizian, M.; Nourisanami, F.; Hajpoor, V.; Parvaresh, M.; Bahri, Z.; Motovali-Bashi, M. LINC00467: A key oncogenic long non-coding RNA. Clin. Chim. Acta 2022, 536, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Bo, H.; Liang, Y.; Li, G. LINC00467 Is Upregulated by DNA Copy Number Amplification and Hypomethylation and Shows ceRNA Potential in Lung Adenocarcinoma. Front. Endocrinol. 2022, 12, 802463. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Liu, C.; Ye, Z.; Wu, C.; Ding, Y.; Huang, J. Overexpressed LINC00467 promotes the viability and proliferation yet inhibits apoptosis of gastric cancer cells via raising ITGB3 level. Tissue and Cell 2021, 73, 101644. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Nie, P.; Xu, S. Long noncoding RNA linc00467 plays an oncogenic role in hepatocellular carcinoma by regulating the miR-18a-5p/NEDD9 axis. J. Cell. Biochem. 2020, 121, 3135–3144. [Google Scholar] [CrossRef]
- Ge, Q.; Jia, D.; Cen, D.; Qi, Y.; Shi, C.; Li, J.; Sang, L.; Yang, L.-J.; He, J.; Lin, A.; et al. Micropeptide ASAP encoded by LINC00467 promotes colorectal cancer progression by directly modulating ATP synthase activity. J. Clin. Investig. 2021, 131, 22. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Deng, W.; Zhu, K.; Zeng, Z.; Hu, B.; Zhou, Z.; Xie, A.; Zhang, C.; Fu, B.; Zhou, X.; et al. LINC00467 Promotes Prostate Cancer Progression via M2 Macrophage Polarization and the miR-494-3p/STAT3 Axis. Front. Oncol. 2021, 11, 661431. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.; Tang, Y. LINC00467 knockdown repressed cell proliferation but stimulated cell apoptosis in glioblastoma via miR-339-3p/IP6K2 axis. Cancer Biomark. 2020, 28, 169–180. [Google Scholar] [CrossRef]
- Han, X.; Zhang, S. Role of Long Non-Coding RNA LINC00641 in Cancer. Front. Oncol. 2022, 11, 829137. [Google Scholar] [CrossRef]
- Tabatabaiefar, M.A.; Sajjadi, R.S.; Modarressi, M.H. JPX and LINC00641 ncRNAs expression in prostate tissue: A case-control study. Res. Pharm. Sci. 2021, 16, 5. [Google Scholar] [CrossRef]
- Li, Z.; Hong, S.; Liu, Z. LncRNA LINC00641 predicts prognosis and inhibits bladder cancer progression through miR-197-3p/KLF10/PTEN/PI3K/AKT cascade. Biochem. Biophys. Res. Commun. 2018, 503, 1825–1829. [Google Scholar] [CrossRef]
- Mao, Q.; Lv, M.; Li, L.; Sun, Y.; Liu, S.; Shen, Y.; Liu, Z.; Luo, S. Long intergenic noncoding RNA 00641 inhibits breast cancer cell proliferation, migration, and invasion by sponging miR-194-5p. J. Cell. Physiol. 2020, 235, 2668–2675. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Zuo, C.; Fang, P.; Liu, G.; Qiu, Y.; Huang, Y.; Tang, R. Targeting Glioblastoma Stem Cells: A Review on Biomarkers, Signal Pathways and Targeted Therapy. Front. Oncol. 2021, 11, 701291. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Su, Y.; Lei, X.; Zhao, H.; Wang, L.; Xu, T.; Guo, J.; Yang, W.; Zhang, X. LINC00641/miR-582-5p mediate oxaliplatin resistance by activating autophagy in gastric adenocarcinoma. Sci. Rep. 2020, 10, 14981. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, H.; Long, H.; Li, D.; Zheng, X. LINC00641 regulates autophagy and intervertebral disc degeneration by acting as a competitive endogenous RNA of miR-153-3p under nutrition deprivation stress. J. Cell. Physiol. 2019, 234, 7115–7127. [Google Scholar] [CrossRef]
- Chen, Q.; Yan, J.; Xie, W.; Xie, W.; Li, M.; Ye, Y. LncRNA LINC00641 Sponges miR-497-5p to Ameliorate Neural Injury Induced by Anesthesia via Up-Regulating BDNF. Front. Mol. Neurosci. 2020, 13, 95. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, H.; Xu, P.; Tan, Y.; Xu, Y.; Wang, L.; Liu, B.; Chen, Q.; Tian, D. Identification and validation of a five-lncRNA prognostic signature related to Glioma using bioinformatics analysis. BMC Cancer 2021, 21, 251. [Google Scholar] [CrossRef]
- Yang, J.; Yu, D.; Liu, X.; Changyong, E.; Yu, S. LINC00641/miR-4262/NRGN axis confines cell proliferation in glioma. Cancer Biol. Ther. 2020, 21, 758–766. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.; Zhi, Y.; Zheng, G.; Zhang, B.; Zhu, H.; Wang, M. Analysis of long non-coding RNAs in glioblastoma for prognosis prediction using weighted gene co-expression network analysis, Cox regression, and L1-LASSO penalization. OncoTargets Ther. 2019, 12, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Zhu, D.; Zhang, X.; Wang, J.; Cao, H.; Li, L. The crucial role of LncRNA MIR210HG involved in the regulation of human cancer and other disease. Clin. Transl. Oncol. 2023, 25, 137–150. [Google Scholar] [CrossRef]
- Lei, D.; Fang, C.; Deng, N.; Yao, B.; Fan, C. Long noncoding RNA expression profiling identifies MIR210HG as a novel molecule in severe preeclampsia. Life Sci. 2021, 270, 119121. [Google Scholar] [CrossRef]
- Li, D.; Qian, X.; Xu, P.; Wang, X.; Li, Z.; Qian, J.; Yao, J. Identification of lncRNAs and Their Functional Network Associated with Chemoresistance in SW1990/GZ Pancreatic Cancer Cells by RNA Sequencing. DNA Cell Biol. 2018, 37, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, W.; Chen, X.; Li, Y.; Wen, P.; Xu, F. MIR210HG predicts poor prognosis and functions as an oncogenic lncRNA in hepatocellular carcinoma. Biomed. Pharmacother. 2019, 111, 1297–1301. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Li, G.; Wang, C.; Gong, G.; Wang, L.; Li, C.; Chen, Y.; Wang, X. MIR210HG regulates glycolysis, cell proliferation, and metastasis of pancreatic cancer cells through miR-125b-5p/HK2/PKM2 axis. RNA Biol. 2021, 18, 2513–2530. [Google Scholar] [CrossRef] [PubMed]
- Ata-Abadi, N.S.; Mowla, S.J.; Aboutalebi, F.; Dormiani, K.; Kiani-Esfahani, A.; Tavalaee, M.; Nasr-Esfahani, M.H. Hypoxia-related long noncoding RNAs are associated with varicocele-related male infertility. PLoS ONE 2020, 15, e0232357. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.-H.; Wei, S.-T.; Liu, J.-J.; Chang, Y.-J.; Lin, Y.-F.; Yu, C.-S.; Chang, S.L.-Y. Recognition of a Novel Gene Signature for Human Glioblastoma. Int. J. Mol. Sci. 2022, 23, 4157. [Google Scholar] [CrossRef] [PubMed]
- Zuo, X.-L.; Cai, J.; Chen, Z.-Q.; Zhang, Y.; Liang, L.-H.; Wang, J.-F.; Wang, J.-G.; Wu, J.; Mao, J.-D. The utility of long non-coding RNA ZEB1-AS1 as a prognostic biomarker in human solid tumors: A meta-analysis. Clin. Chim. Acta 2018, 485, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Guo, Y.; Wang, C.; Wang, Q.; Yan, G. Long Noncoding RNA ZEB1-AS1 Downregulates miR-23a, Promotes Tumor Progression, and Predicts the Survival of Oral Squamous Cell Carcinoma Patients. OncoTargets Ther. 2021, 14, 2699–2710. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Pan, J.; Li, H.; Long, J.; Fang, F.; Chen, J.; Zhu, X.; Xiang, X.; Zhang, D. LncRNA ZEB1-AS1 Was Suppressed by p53 for Renal Fibrosis in Diabetic Nephropathy. Mol. Ther.-Nucleic Acids 2018, 12, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Cheng, B.; Yang, L.; Li, G.; Yuan, Y.; Luo, G.; Shu, Z.; Jiang, H. LncRNA ZEB1-AS1 knockdown alleviates oxidative low-density lipoprotein-induced endothelial cell injury via the miR-590-5p/ HDAC9 axis. Cent. Eur. J. Immunol. 2021, 46, 325–335. [Google Scholar] [CrossRef]
- Lv, Q.-L.; Hu, L.; Chen, S.-H.; Sun, B.; Fu, M.-L.; Qin, C.-Z.; Qu, Q.; Wang, G.-H.; He, C.-J.; Zhou, H.-H. A Long Noncoding RNA ZEB1-AS1 Promotes Tumorigenesis and Predicts Poor Prognosis in Glioma. Int. J. Mol. Sci. 2016, 17, 1431. [Google Scholar] [CrossRef]





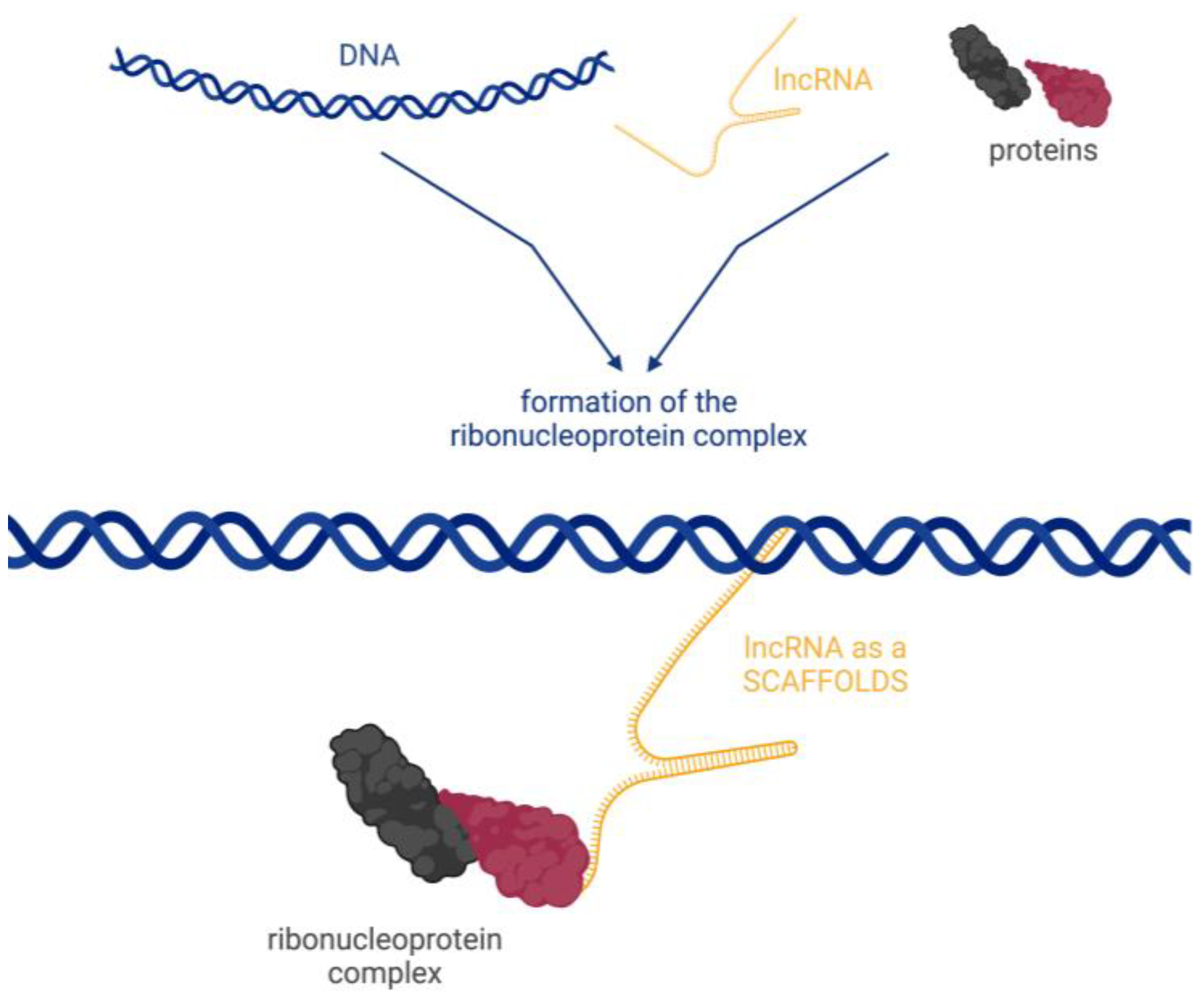

| lncRNA Name /Alternative Transcript Name/ | Gene Location Class Ensembl Gene ID | Role in GBM | Expression | Function in GBM | Data | Ref. |
|---|---|---|---|---|---|---|
| AC016405.3 /RP11-44N11.2/ /lnc-DERL1-3/ | 8q24.13 bidirectional ENSG00000272384 | suppressor | ↓ | suppressing proliferation and invasion | clinical association; GBM primary tissue; GBM cell line | [33] |
| ADAMTs9-AS2 /NONHSAT090261/ | 03p14.1 antisense ENSG00000241684 | oncogenic | ↑ | TMZ resistance | clinical association; GBM cell line | [49] |
| AGAP2-AS1 /HSALNG0091650/ | 02q14.1 antisense ENSG00000255737 | oncogenic | ↑ | proliferation, viability | GBM primary tissue; GBM cell line | [52] |
| AHIF /lnc-TMEM30B-9/ /HIFiA-AS2/ | 14q23.2 antisense ENSG00000258777 | protumour | ↑ | invasion, viability, GSC, radiation resistance | GBM cell line; GSC mesenchymal line | [53] |
| ANRIL /CDKN2B-AS1/ | 09p21.3 antisense ENSG00000240498 | oncogenic | ↑ | cell proliferation | GBM cell line; GBM tissue; GBM patient serum | [13,54] |
| lncRNA-ATB | 14q11.2 intronic - | protumour | ↑ | invasion of cell | GBM cell line | [40] |
| CASC2 | 10q26.11 antisense ENSG00000177640 | suppressor | ↓ | inhibitor of proliferation | GBM cell line; GBM tissue; xenograft | [55] |
| CASC7 /lnc-AGO2-1/ | 8q24.3 intronic ENSG00000259758 | suppressor | ↓ | inhibitor proliferation, regulation of cell cycle | GBM primary tissue; GBM cell line | [43] |
| CASC9 /LINC00981/ /RP11-697M17.1-003/ | 8q21.13 intronic ENSG00000249395 | oncogenic | ↑ | tumourigenesis | GBM cell line | [56] |
| CCND2-AS1 | 12p13.32 antisense ENSG00000255920 | protumour | ↑ | proliferation and growth | GBM cell line; GBM patient tissue | [57] |
| CRNDE /lnc-IRX3-80/ | 16q12.2 antisense ENSG00000245694 | oncogenic | ↑ | proliferation, invasion, migration, inhibition of apoptosis | GBM cell line; GBM patient tissue | [58] |
| DCST1-AS1 | 01q21.3 antisense ENSG00000232093 | protumour | ↑ | proliferation | clinical related; GBM primary tissue; primary cultivation | [59] |
| DGCR5 | 22q11.21 antisense ENSG00000237517 | suppressor | ↓ | proliferation, migration, invasion, apoptosis | GBM cell line; GBM tissue | [60] |
| DLEU1-AS1 | 13q14.3 intronic ENSG00000186047 | biomarker | ↑ | proliferation, cell cycle, autophagy; correlation with prognosis | GBM cell line; GBM tissue | [61] |
| ECONEXIN /LINC00461/ | 05q14.3 intronic ENSG00000245526 | protumour | ↑ | proliferation | GBM cell line; GBM tissue; TCGA data | [62] |
| FAM66C | 12p13.31 antisense ENSG00000226711 | - | ↑↓ | tumour microenvironment | GBM cell line; GBM tissue; TCGA data | [44] |
| GAS5 | 01q25.1 antisense ENSG00000234741 | suppressor | ↓ | inhibition of proliferation, invasion and viability | GBM cell line; GBM tissue; GBM patient serum | [63] |
| H19 /D11S813E/ /ASM1/ | 11p15.5 intronic ENSG00000130600 | protumour | ↑ | proliferation, invasion, angiogenesis | GBM cell line | [64] |
| HMMR-AS1 | 05q34 antisense ENSG00000251018 | protumour | ↑ | tumourigenesis, proliferation, invasion, radiation resistance | GBM cell line | [50] |
| HOTAIR | 12q13.13 antisense ENSG00000228630 | protumour | ↑ | proliferation, invasion, therapy resistance, chromatin remodelling | clinical association; GBM patient tissue/serum; cell line; xenoimplants | [65,66] |
| HOTAIRM1 /HOXA-AS1/ | 07p15.2 antisense ENSG00000233429 | oncogenic | ↑ | proliferation, invasion, viability | clinical association TCGA; GBM primary tissue; GBM cell line | [67] |
| HOXA-AS2 | 07p15.2 antisense ENSG00000253552 | protumour | ↑ | migration, invasion, viability | GBM tissue; GBM cell line | [68] |
| HOXB13-AS1 /lncHOXB13-1/ | 17q21.2 intronic ENSG00000159184 | protumour | ↑ | proliferation, progress | GBM tissue; GBM cell line | [69] |
| HOTTIP /HOXA-AS6/ | 07p15.2 antisense ENSG00000243766 | antitumour | ↓ | inhibition of cell cycle, induction of apoptosis | GBM tissue; GBM cell line | [70] |
| HULC /lnc-BMP6-106/ | 06p24.3 intronic ENSG00000285219 | protumour | ↑ | proliferation, angiogenesis, activity of MGMT | GBM cell line; GBM patient tissue | [29] |
| KTN1-AS1 | 14q22.3 antisense ENSG00000186615 | tumour suppressor | ↑ | viability and invasion of cell; correlation with prognosis | GBM tissue; GBM cell line; TCGA data | [71] |
| LINC00467 /NR_026761/ | 01q32.3 intronic ENSG00000153363 | protumour | ↑ | proliferation and invasion | GBM cell line | [72] |
| LINC00565 | 13q34 intronic ENSG00000260910 | unknown; biomarker | ↑ | correlation with prognosis | GBM patient serum | [73] |
| LINC00641 | 14q11.2 intronic ENSG00000258441 | unknown; biomarker | ↑ | correlation with prognosis | GBM patient serum | [73] |
| LINC01393 | 07q31.2 intronic ENSG00000225535 | unknown; biomarker | ↑ | tumour progress; correlation with prognosis | GBM tissue; GBM cell line; TCGA data | [74] |
| LINC01426 | 21q22.12 intronic ENSG00000234380 | oncogenic | ↑ | proliferation, invasion, viability | clinical association TCGA; GBM primary tissue; GBM cell line | [75] |
| LINC01446 | 07p12.1 intronic ENSG00000205628 | protumour | ↑ | tumourigenesis, progress | clinical association; GBM cell line; xenografts | [76] |
| LINC01494 | 02q35 intronic ENSG00000228135 | oncogenic | ↑ | proliferation, invasion | clinical association; GBM tissue; GBM cell line | [77] |
| LINC01503 | 9q34.11 intronic ENSG00000233901 | oncogenic; biomarker | ↑ | migration, invasion, apoptosis; correlation with malignancy grade and prognosis | GBM tissue; GBM cell line; TCGA data | [78] |
| LINC01711 | 20q13.32 intronic ENSG00000268941 | protumour | ↑ | proliferation, migration, invasion correlation with prognosis | GBM tissue; GBM cell line | [79] |
| LINC02283 | 04q12 intronic ENSG00000248184 | oncogenic | ↑ | correlation with expression of PDGFRA, malignancy | patient GSC lines; xenoimplants; GBM tissue | [80] |
| LINC-ROR /ROR/ | 18q21.31 intronic ENSG00000258609 | unknown | ↑↓ | GSC | GBM tissue; GBM cell line | [81] |
| lnc-TALC /LNCARSR/ /linc-GNAQ-7/ | 09q21.31 intronic ENSG00000233086 | protumour | ↑ | TMZ resistance, tumour relapse | TMZ-selected GBM cell lines | [48] |
| MAFG-DT /MAFG-AS1-001/ | 17q25.3 intronic ENSG00000265688 | protumour | ↑ | proliferation | GBM tissue; GBM cell lines | [82] |
| MALAT1 | 11q13.1 intronic ENSG00000251562 | unknown | ↑↓ | invasion, proliferation, migration, apoptosis, permeability of BBB, chemosensitivity | clinical association; GBM patient tissue and serum; GBM cell lines; xenografts | [38] |
| MATN1-AS1 | 01p35.2 intronic ENSG00000186056 | suppressor | ↓ | inhibition of proliferation and invasion | GBM primary tissue lines; GBM cell lines | [38] |
| MDC1-AS | 06p21.33 antisense ENSG00000224328 | suppressor | ↓ | inhibition of proliferation | GBM cell lines | [83] |
| MEG3 /lnc-DLK1-3/ | 14q32.2 intronic ENSG00000214548 | suppressor | ↓ | inhibition of proliferation | GBM tissue; GBM cell lines | [84] |
| MIAT | 22q12.1 intronic ENSG00000225783 | oncogenic | ↑ | proliferation, migration, metastasis | GBM tissue; GBM cell lines | [37] |
| MIR210HG | 11p15.5 intronic ENSG00000247095 | unknown; biomarker | ↑ | hypoxia, invasion, TMZ resistance, correlation with prognosis | GBM cell line; xenografts; TCGA data; GBM patient plasma | [45] |
| MNX1-AS1 /CCAT5/ /LOC645249/ | 07q36.3 intronic ENSG00000243479 | oncogenic | ↑ | proliferation, migration, invasion | GBM tissue; GBM cell lines | [85] |
| NCK1-AS1 /SLC35G2-AS1/ /NCK1-DT/ | 03q22.3 antisense ENSG00000239213 | protumour | ↑ | TMZ resistance | primary tissue; GBM cell lines | [86] |
| NEAT1 /LINC00084/ | 11q13.1 intronic ENSG00000245532 | protumour | ↑ | proliferation, glycolysis | GBM primary tissue; cell lines; xenografts | [87] |
| PART1 | 05q12.1 antisense ENSG00000152931 | tumour suppressor | ↓ | inhibition of progression and tumour growth | clinical association TCGA; GBM tissue; GBM cell lines | [88] |
| PARTICL /PARTICLE/ | 2p11.2 circulating ENSG00000286532 | regulation of tumour suppressors | - | tumour microenvironment, chromatin dynamics | GBM cell lines; GBM tissue | [32,89] |
| PCAT1 /PCA1/ | 08q24.21 intronic ENSG00000253438 | unknown | ↑↓ | viability, DNA repair | GBM cell lines | [42] |
| PVT1 /lncRNA1331/ | 08q24.2 intronic ENSG00000249859 | oncogenic | ↑ | tumourigenesis, progress | GBM tissue; GBM cell lines; xenoimplants | [90] |
| RBPMS-AS1 | 08p12 antisense ENSG00000254109 | antitumour | ↓ | radiosensitivity, apoptosis | GBM tissue; GBM cell line; xenoimplants | [91] |
| RPSAP52 | 12q14.3 antisense ENSG00000241749 | unknown; biomarker | ↑ | correlation with prognosis | clinical association; GBM primary tissue; GBM cell line | [92] |
| RUNX1-IT1 | 21q22.12 intronic ENSG00000159216 | protumour | ↑ | cell cycles, proliferation | GBM tissue; GBM cell line | [93] |
| SAMMSON /LINC01212/ | 03p13 intronic ENSG00000240405 | oncogenic; potential biomarker | ↑ | proliferation, viability, invasion, apoptosis | GBM tissue; GBM cell line; GBM patient serum | [94] |
| SOX2-OT | 3q26.3 overlapping ENSG00000242808 | unknown; biomarker | ↑ | migration and invasion; correlation with prognosis | GBM tissue; GBM cell lines | [87] |
| TALNEC2 /LINC01116/ | 02q31.1 intronic ENSG00000163364 | protumour | ↑ | tumourigenesis, radiation resistance | clinical association TCGA; GBM primary tissue; GBM cell lines | [95] |
| TP73-AS1 /lnc-LRRC47-78/ /KIAA0495/ | 01p36.32 antisense ENSG00000227372 | unknown; biomarker | ↑ | correlation with prognosis; resistance and metabolism, TMZ in GSC | clinical association TCGA; GSC lines | [96] |
| TSLC1-AS1 /lnc-NXPE2-1/ /RP11-713B9/ | 11q23.2 antisense ENST00000546273 | tumour suppressor | ↓ | inhibition of cell proliferation, migration and invasion | GBM tissue; GBM cell lines | [97] |
| TUSC7 /LINC00902/ | 03q13.31 antisense ENSG00000243197 | tumour suppressor | ↓ | inhibition, resistance to TMZ, tumour malignancy | GBM cell line; GBM patients resistant to TMZ tissue | [98] |
| TUG1 | 22q12.2 antisense ENSG00000253352 | unknown | ↑↓ | permeability of BBB | GBM tissue; GBM cell line | [46] |
| TUNAR | 14q32.2 intronic ENSG00000250366 | unknown | ↑ | regulation of tumour progress, cell cycles | GBM cell lines | [99] |
| UCA1 /UCAT1/ /oncolncRNA-36/ | 19p13.12 intronic ENSG00000214049 | protumour | ↑ | proliferation, invasion, migration; glycolysis | GBM tissue; GBM cell lines | [100,101] |
| XIST | Xq13.2 intronic ENSG00000229807 | protumour | ↑ | permeability of BBB, angiogenesis, proliferation of CSC, migration, invasion | GBM tissue; GBM cell line | [102] |
| ZEB1-AS1 | 10p11.22 antisense ENSG00000237036 | protumour | ↑ | cell proliferation, migration, invasion | GBM cell line | [103] |
| ZBED3-AS1 | 05q13.3 antisense ENSG00000250802 | unknown; biomarker | ↓ | TMZ resistance | TMZ-resistant GBM cell line a tissue | [104] |
| lncRNA | Expression | Disease | Expression Level Correlates | Fluids | Ref. |
|---|---|---|---|---|---|
| ADAMTs9-AS2 | ↓ | ischemic stroke | severity of disability | plasma | [111] |
| ↓ | non-small cell lung cancer | aggressive tumour behaviour | serum | [112] | |
| ANRIL | ↑ | breast cancer | metastasis | serum | [113] |
| ↑ | coronary artery disease | prognosis, degree of inflammation, severity of disability | plasma | [114] | |
| ↑ | COVID-19 | severity of disability | blood | [115] | |
| ↑ | Crohn’s disease | diagnosis | serum | [116] | |
| ↑ | diabetes mellitus | diagnosis | serum | [117] | |
| ↑ | glioma | tumour grade and prognosis | serum | [54] | |
| ↑ | intraductal papillary mucinous neoplasms of the pancreas | malignant prediction | plasma | [118] | |
| ↑ | ischemic stroke | severity of disability | serum | [119,120] | |
| ↑ | multiple myeloma | prognosis | plasma | [121] | |
| ↑ | neonatal sepsis | higher risk of mortality | plasma | [122] | |
| ↑ | non-small cell lung cancer | prognosis | serum, plasma | [123,124] | |
| ↑ | pituitary adenomas | prognosis | plasma | [125] | |
| ↑ | sepsis | severity of disability and prognosis | plasma | [126] | |
| ↑ | stable angina | level of troponin 1 | plasma | [127] | |
| ↑ | ulcerative colitis | diagnosis | serum | [116] | |
| ↓ | acute exacerbation of chronic obstructive pulmonary disease | levels of inflammatory cytokines | plasma | [128] | |
| ↓ | acute ischemic stroke | clinico-pathological symptoms | plasma | [129] | |
| ↓ | glaucoma | clinico-pathological symptoms | serum | [130] | |
| ↓ | multiple sclerosis | diagnosis | blood | [131] | |
| ↓ | paediatric inflammatory bowel disease | diagnosis | serum | [132] | |
| ↓ | preeclampsia | diagnosis | serum | [133] | |
| CASC2 | ↑ | osteoarthritis | level of IL-17 | plasma | [134] |
| ↑ | aphthous stomatitis | level of IL-6 and IL-18plasma | plasma | [135] | |
| ↓ | diabetic nephropathy | diagnosis | serum | [136] | |
| ↓ | hepatocellular carcinoma | tumour grade | serum | [137] | |
| ↓ | childhood asthma | diagnosis | serum | [138] | |
| ↓ | chronic renal failure | diagnosis | serum | [139] | |
| ↓ | oral squamous | prognosis | plasma | [140] | |
| ↓ | rheumatoid arthritis | diagnosis | serum, plasma | [141,142] | |
| ↓ | sepsis | clinico-pathological symptoms | serum, blood | [143,144] | |
| CRNDE | ↑ | acute myeloid leukaemia | clinico-pathological symptoms | blood | [145] |
| ↑ | colorectal carcinoma | aggressive tumour and liver metastasis | serum, plasma | [146,147] | |
| ↑ | hepatocellular carcinoma | tumour size and differentiation | serum | [148] | |
| ↑ | nasopharyngeal carcinoma | lymph node metastasis | serum | [149] | |
| ↑ | non-small cell lung cancer | diagnosis | plasma | [150] | |
| ↑ | sepsis | severity of disability | serum | [151] | |
| ↑ | severe pneumonia | prognosis | serum | [152] | |
| ↓ | chronic lymphocytic leukaemia | prognosis | serum | [153] | |
| ↓ | sepsis | increasing levels after treatment | plasma | [154] | |
| DGCR5 | ↓ | gastric cancer | clinico-pathological symptoms, metastasis | plasma | [155] |
| ↓ | hepatocellular carcinoma | diagnosis | serum | [156] | |
| DLEU-AS1 | ↑ | diabetic foot ulcer | diagnosis | serum | [157] |
| ↑ | endometrial cancer | clinico-pathological symptoms | serum | [158] | |
| GAS5 | ↑ | atherosclerosis | diagnosis | serum | [159] |
| ↑ | malignant mesothelioma | diagnosis | plasma | [160] | |
| ↑ | multiple sclerosis | clinico-pathological symptoms | serum | [161] | |
| ↑ | myelofibrosis | clinico-pathological symptoms | plasma | [162] | |
| ↑ | osteoporosis | diagnosis | plasma | [163] | |
| ↑ | osteoporosis with fractures | upregulated in the presence of a fracture | serum | [164] | |
| ↑ | polycystic ovary syndrome | diagnosis | plasma | [165] | |
| ↓ | glioblastoma multiforme | prognosis | serum | [166] | |
| ↓ | breast cancer | diagnosis | serum | [167] | |
| ↓ | cerebrovascular stroke | diagnosis | serum | [168] | |
| ↓ | coronary artery disease | diagnosis | plasma | [169] | |
| ↓ | COVID-19 | severity of disability | serum | [170] | |
| ↓ | diabetes mellitus 2 | diagnosis | serum | [171] | |
| ↓ | hepatocellular carcinoma | diagnosis | plasma | [172] | |
| ↓ | chronic hepatitis B virus infection | liver fibrosis | serum | [173] | |
| ↓ | mycobacterium tuberculosis | diagnosis | serum | [174] | |
| ↓ | non-small cell lung cancer | tumour size and metastasis | serum, plasma | [175,176] | |
| ↓ | osteoporosis | diagnosis | serum | [177] | |
| ↓ | polycystic ovary syndrome | biomarker of insulin resistance | serum | [178] | |
| ↓ | rheumatoid arthritis | diagnosis | serum, plasma | [179,180] | |
| ↓ | sepsis | diagnosis | serum | [181] | |
| ↓ | systemic lupus erythematosus | diagnosis | plasma | [182] | |
| HOTAIR | ↑ | Alzheimer’s disease | clinico-pathological symptoms, decreasing levels after treatment (exercises) | serum | [183] |
| ↑ | breast cancer | lymph node metastasis | plasma, serum | [184,185] | |
| ↑ | colorectal carcinoma | diagnosis | plasma | [186] | |
| ↑ | congenital heart diseases | diagnosis | plasma | [187] | |
| ↑ | coronary artery disease | diagnosis | blood | [188] | |
| ↑ | diabetes mellitus 2 | complications of diabetes | serum | [189] | |
| ↑ | oesophageal squamous cell carcinoma | tumour grade, decreasing levels after treatment | serum | [190] | |
| ↑ | gastric cancer | tumour grade and metastasis | plasma, serum | [191,192] | |
| ↑ | gestational diabetes | body mass index, fasting plasma glucose | plasma | [193] | |
| ↑ | glioblastoma multiforme | level of tissue expression, progression | serum | [66,166,194] | |
| ↑ | hepatocellular carcinoma | diagnosis | serum | [195] | |
| ↑ | laryngeal squamous cell carcinoma | lymph node metastasis | serum | [196] | |
| ↑ | multiple myeloma | disease stage | plasma | [197] | |
| ↑ | non-small cell lung cancer | histology subtype and tumour-node-metastasis stage | plasma | [198] | |
| ↑ | osteoarthritis | diagnosis | plasma | [199] | |
| ↑ | papillary thyroid carcinoma | tumour grade | serum | [200] | |
| ↑ | rheumatoid arthritis | decreasing levels after treatment | serum | [201] | |
| ↑ | systemic lupus erythematosus | level of IL-6 | serum | [202] | |
| ↓ | acute myocardial infarction | diagnosis | plasma | [203] | |
| ↓ | atherosclerosis | diagnosis | plasma | [203] | |
| LINC00467 | ↓ | acute myeloid leukaemia | increasing levels after treatment | serum | [101] |
| LINC00565 | ↑ | glioblastoma multiforme | survival pattern patients | serum | [73] |
| LINC00641 | ↑ | Crohn’s disease | diagnosis | serum | [204] |
| ↑ | glioblastoma multiforme | survival pattern patients | serum | [73] | |
| ↑ | ulcerative colitis | diagnosis | serum | [204] | |
| MALAT1 | ↑ | acute kidney injury | diagnosis | serum | [205] |
| ↑ | acute pancreatitis | diagnosis | serum | [206] | |
| ↑ | angina pectoris | severity of disability | serum | [207] | |
| ↑ | breast cancer | diagnosis | serum | [208] | |
| ↑ | gastric cancer | metastasis | plasma | [209] | |
| ↑ | gestational diabetes | diagnosis | serum | [210] | |
| ↑ | glioblastoma multiforme | TMZ chemoresistance | serum | [38] | |
| ↑ | hypertension | diagnosis | plasma | [211] | |
| ↑ | multiple sclerosis | diagnosis | serum | [212] | |
| ↑ | nasopharyngeal carcinoma | tumour stage, decreasing levels after treatment | serum | [213] | |
| ↑ | non-Hodgkin lymphoma | tumour stage | plasma | [214] | |
| ↑ | non-small cell lung cancer | tumour grade and metastasis | serum | [215] | |
| ↑ | osteosarcoma | survival pattern patients | serum | [216] | |
| ↑ | ovarian cancer | metastasis | serum | [217] | |
| ↑ | Parkinson’s disease | degree of inflammation | serum | [218] | |
| ↑ | prostate cancer | diagnosis | serum | [219] | |
| ↑ | rheumatoid arthritis | diagnosis | plasma | [220] | |
| ↑ | sepsis | clinico-pathological symptoms | plasma | [221] | |
| ↑ | severe pneumonia | prediction of survival of patients | serum | [222] | |
| ↑ | tongue squamous cell carcinoma | diagnosis | serum, plasma | [223] | |
| ↑ | ulcerative colitis | diagnosis | plasma | [224] | |
| ↓ | diabetes mellitus 2 | diagnosis | serum | [225] | |
| ↓ | retinoblastoma | diagnosis | serum | [226] | |
| MIR210HG | ↑ | glioma | diagnosis | serum | [227] |
| RPSAP52 | ↑ | renal failure | diagnosis | plasma | [228] |
| ↓ | diabetic retinopathy | diagnosis | plasma | [229] | |
| SAMMSON | ↑ | glioblastoma multiforme | diagnosis | plasma | [230] |
| ↑ | oral squamous cell carcinoma | levels of tissue expression, decreasing levels after treatment | serum | [231] | |
| ↑ | papillary thyroid carcinoma | diagnosis | plasma | [232] | |
| SOX2-OT | ↑ | head and neck squamous cell carcinoma | diagnosis | plasma | [233] |
| ↑ | lung squamous cell carcinoma | tumour size and lymph node metastasis, decreasing levels after treatment | plasma | [234] | |
| ↑ | ovarian cancer | diagnosis | plasma | [235] | |
| ↑ | pulmonary arterial hypertension | diagnosis | serum | [236] | |
| TP73-AS1 | ↑ | non-small cell lung cancer | prognosis | serum | [150] |
| ↑ | NK/T-cell lymphoma | diagnosis | blood | [96] | |
| ZEB1-AS1 | ↑ | oesophageal carcinoma | diagnosis | serum | [237] |
| ↑ | prostate cancer | diagnosis | serum | [238] | |
| ↓ | amyotrophic lateral sclerosis | diagnosis | blood | [239] | |
| ↓ | diabetes mellitus | complications of diabetes (diabetic lung) | plasma | [240] |
| Disease | Deregulated Levels of lncRNA Biomarkers in Peripheral Blood |
|---|---|
| breast cancer | ANRIL (↑), HOTAIR (↑), MALAT1 (↑), GAS5 (↓) |
| coronary artery disease | ANRIL (↑), HOTAIR (↑), GAS5 (↓) |
| diabetes mellitus 2 | ANRIL (↑), HOTAIR (↑), GAS5 (↓), MALAT1 (↓), ZEB1-AS1 (↓) |
| gastric cancer | HOTAIR (↑), MALAT1 (↑), DGCR5 (↓), |
| glioblastoma multiforme | ANRIL (↑), HOTAIR (↑), LINC00641 (↑), LINC00565 (↑), MALAT1 (↑), SAMMSON (↑), GAS5 (↓) |
| hepatocellular carcinoma | CRNDE (↑), HOTAIR (↑), CASC2 (↓), DGCR5 (↓), GAS5 (↓) |
| multiple sclerosis | GAS5 (↑), MALAT1 (↑), ANRIL (↓) |
| non-small cell lung cancer | ANRIL (↑), CRNDE (↑), GAS5 (↑), HOTAIR (↑), MALAT1 (↑), TP73-AS1 (↑), ADAMTs9-AS2 (↓) |
| rheumatoid arthritis | HOTAIR (↑), MALAT1 (↑), CASC2 (↓), GAS5 (↓) |
| sepsis | ANRIL (↑), CRNDE (↑↓), CASC2 (↓), GAS5 (↓) |
| ulcerative colitis | ANRIL (↑), LINC00641 (↑), MALAT (↑) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pokorná, M.; Černá, M.; Boussios, S.; Ovsepian, S.V.; O’Leary, V.B. lncRNA Biomarkers of Glioblastoma Multiforme. Biomedicines 2024, 12, 932. https://doi.org/10.3390/biomedicines12050932
Pokorná M, Černá M, Boussios S, Ovsepian SV, O’Leary VB. lncRNA Biomarkers of Glioblastoma Multiforme. Biomedicines. 2024; 12(5):932. https://doi.org/10.3390/biomedicines12050932
Chicago/Turabian StylePokorná, Markéta, Marie Černá, Stergios Boussios, Saak V. Ovsepian, and Valerie Bríd O’Leary. 2024. "lncRNA Biomarkers of Glioblastoma Multiforme" Biomedicines 12, no. 5: 932. https://doi.org/10.3390/biomedicines12050932







