Exposure to a PFOA, PFOS and PFHxS Mixture during Gestation and Lactation Alters the Liver Proteome in Offspring of CD-1 Mice
Abstract
:1. Introduction
2. Materials and Methods
- (1)
- Standard rodent diet (SD) + vehicle (VEH, 0.5% Tween 20, 10 mL/kg), n = 10
- (2)
- SD + PFOA (1 mg/kg), n = 10
- (3)
- SD + PFOS (1 mg/kg), n = 8
- (4)
- SD + PFHxS (1 mg/kg) n = 8
- (5)
- SD + PFAS mix (1 mg/kg PFOA, PFOS, and PFHxS), n = 6
- (6)
- 60% kCal high fat diet chow (HFD) + VEH, n = 10
- (7)
- HFD + PFOA (1 mg/kg), n = 12
- (8)
- HFD + PFOS (1 mg/kg), n = 6
- (9)
- HFD + PFHxS (1 mg/kg), n = 10
- (10)
- HFD + PFAS mix (1 mg/kg PFOA, PFOS, and PFHxS), n = 8
3. Results
3.1. Visualization of Global Proteome
3.2. Effect of PFAS Treatment on the Neonatal Liver Proteome in Dams Fed a Standard Chow Diet
3.2.1. Effect of Individual PFAS Treatment (PFOA, PFOS, PFHxS) on the Neonatal Liver Proteome in Mice Fed a Standard Diet
3.2.2. Common DEPs between PFOA SD and PFAS Mix SD Treatments Highlight Concordance between Treatments
3.2.3. DEPs Shared among Maternal PFOA SD, PFHxS SD, and PFAS Mix SD Exposures in the PND21 Neonatal Liver Proteome
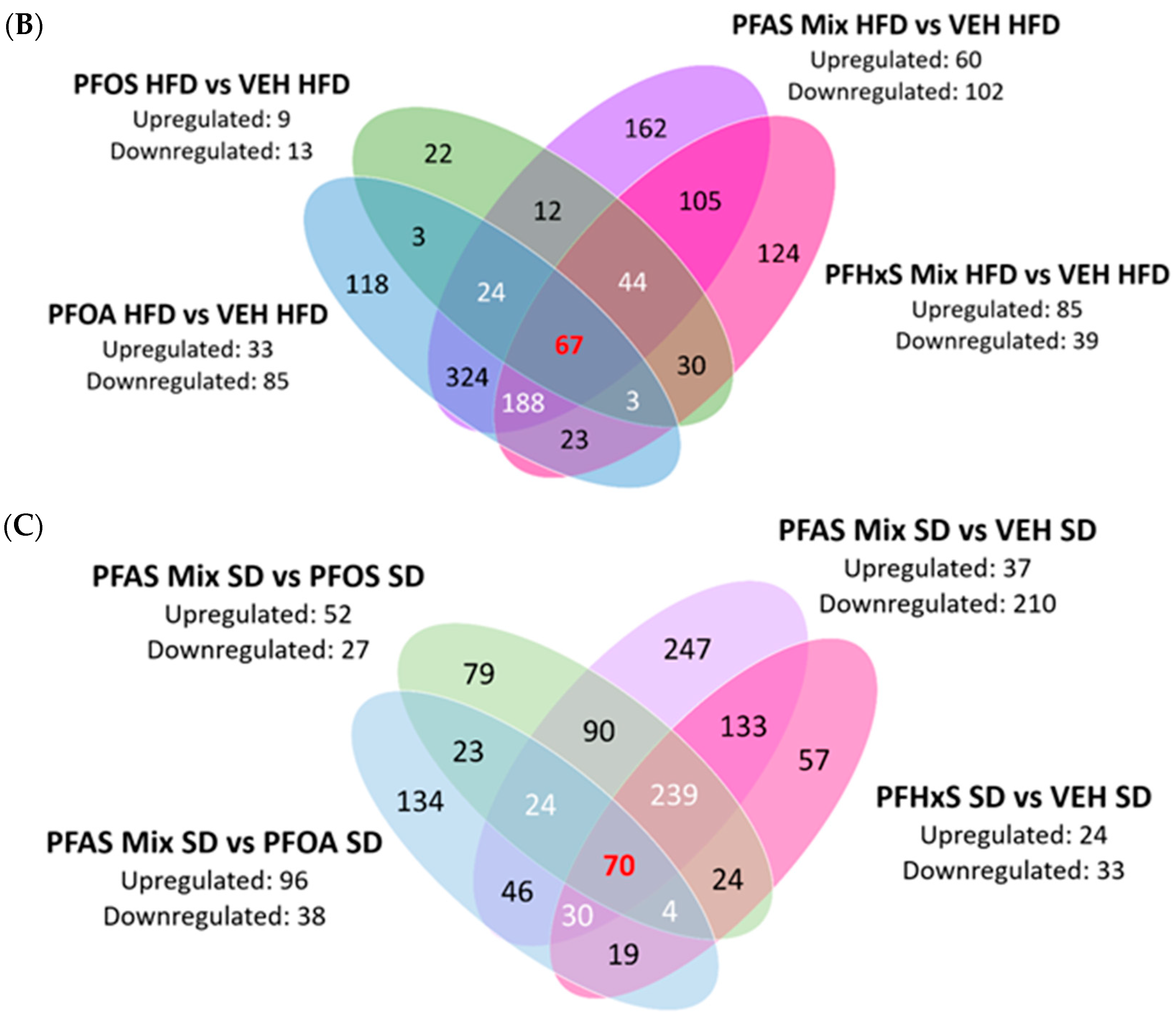
3.3. Combinatorial Effect of the PFAS Treatment and HFD on the Neonatal Liver Proteome
3.3.1. DEPs Shared among Maternal HFD and PFOA SD; PFHxS SD, and PFAS Mix SD Exposures in the PND21 Neonatal Liver Proteome
3.3.2. Shared DEPs at the Intersection of Maternal PFOA HFD, PFHxS HFD; and PFAS Mix HFD Treatments in the Neonatal Liver Proteome
3.4. Contributions of Individual PFAS within the PFAS Mixture
3.5. Pathway Modulation among Diet and Mixture-Specific Comparisons
3.5.1. Lipid, Xenobiotic and Inflammation Pathway Modulation within SD Comparisons
3.5.2. Lipid, Xenobiotic and Inflammation Pathway Modulation within HFD Comparisons
3.5.3. Lipid, Xenobiotic and Inflammation Pathway Modulation within the PFAS Mixture
3.5.4. Synthesis and Oxidation of Lipid Pathway Modulation within the PFAS Mixture
3.5.5. Proteins Modulated in Pathways Related to Liver Damage within the PFAS Mixture
3.5.6. Proteins Modulated in Pathways Related to Fatty Acid Metabolism, Oxidation and Transport within the PFAS Mixture
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sunderland, E.M.; Hu, X.C.; Dassuncao, C.; Tokranov, A.K.; Wagner, C.C.; Allen, J.G. A Review of the Pathways of Human Exposure to Poly- and Perfluoroalkyl Substances (PFASs) and Present Understanding of Health Effects. J. Exposure Sci. Environ. Epidemiol. 2019, 29, 131–147. [Google Scholar] [CrossRef] [PubMed]
- Olsen, G.W.; Burris, J.M.; Ehresman, D.J.; Froehlich, J.W.; Seacat, A.M.; Butenhoff, J.L.; Zobel, L.R. Half-Life of Serum Elimination of Perfluorooctanesulfonate, Perfluorohexanesulfonate, and Perfluorooctanoate in Retired Fluorochemical Production Workers. Environ. Health Perspect. 2007, 115, 1298–1305. [Google Scholar] [CrossRef] [PubMed]
- Dourson, M.; Gadagbui, B. The Dilemma of perfluorooctanoate (PFOA) human half-life. Regul. Toxicol. Pharmacol. 2021, 126, 105025. [Google Scholar] [CrossRef]
- Campbell, J.; Clewell, H.; Cox, T.; Dourson, M.; Ethridge, S.; Forsberg, N.; Gadagbui, B.; Hamade, A.; Naidu, R.; Pechacek, N.; et al. The Conundrum of the PFOA human half-life, an international collaboration. Regul. Toxicol. Pharmacol. 2022, 132, 105185. [Google Scholar] [CrossRef]
- Göckener, B.; Weber, T.; Rüdel, H.; Bücking, M.; Kolossa-Gehring, M. Human biomonitoring of per- and polyfluoroalkyl substances in German blood plasma samples from 1982 to 2019. Environ. Int. 2020, 145, 106123. [Google Scholar] [CrossRef] [PubMed]
- Pérez, F.; Nadal, M.; Navarro-Ortega, A.; Fàbrega, F.; Domingo, J.L.; Barceló, D.; Farré, M. Accumulation of perfluoroalkyl substances in human tissues. Environ. Int. 2013, 59, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Behr, A.-C.; Plinsch, C.; Braeuning, A.; Buhrke, T. Activation of human nuclear receptors by perfluoroalkylated substances (PFAS). Toxicol. Vitr. 2020, 62, 104700. [Google Scholar] [CrossRef] [PubMed]
- Ojo, A.F.; Peng, C.; Ng, J.C. Assessing the human health risks of per- and polyfluoroalkyl substances: A need for greater focus on their interactions as mixtures. J. Hazard. Mater. 2021, 407, 124863. [Google Scholar] [CrossRef] [PubMed]
- Das, K.P.; Wood, C.R.; Lin, M.T.; Starkov, A.A.; Lau, C.; Wallace, K.B.; Corton, J.C.; Abbott, B.D. Perfluoroalkyl acids-induced liver steatosis: Effects on genes controlling lipid homeostasis. Toxicology 2017, 378, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Fenton, S.E.; Ducatman, A.; Boobis, A.; DeWitt, J.C.; Lau, C.; Ng, C.; Smith, J.S.; Roberts, S.M. Per- and Polyfluoroalkyl Substance Toxicity and Human Health Review: Current State of Knowledge and Strategies for Informing Future Research. Environ. Toxicol. Chem. 2021, 40, 606–630. [Google Scholar] [CrossRef]
- Butenhoff, J. Toxicity of Ammonium Perfluorooctanoate in Male Cynomolgus Monkeys after Oral Dosing for 6 Months. Toxicol. Sci. 2002, 69, 244–257. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.T.; Zhao, Y.G.; Leung, P.Y.; Wong, C.K.C. Perinatal Exposure to Perfluorooctane Sulfonate Affects Glucose Metabolism in Adult Offspring. PLoS ONE 2014, 9, e87137. [Google Scholar] [CrossRef] [PubMed]
- Bassler, J.; Ducatman, A.; Elliott, M.; Wen, S.; Wahlang, B.; Barnett, J.; Cave, M.C. Environmental perfluoroalkyl acid exposures are associated with liver disease characterized by apoptosis and altered serum adipocytokines. Environ. Pollut. 2019, 247, 1055–1063. [Google Scholar] [CrossRef] [PubMed]
- Costello, E.; Rock, S.; Stratakis, N.; Eckel, S.P.; Walker, D.I.; Valvi, D.; Cserbik, D.; Jenkins, T.; Xanthakos, S.A.; Kohli, R.; et al. Exposure to per- and Polyfluoroalkyl Substances and Markers of Liver Injury: A Systematic Review and Meta-Analysis. Environ. Health Perspect. 2022, 130, 46001. [Google Scholar] [CrossRef] [PubMed]
- Rosen, M.B.; Das, K.P.; Rooney, J.; Abbott, B.; Lau, C.; Corton, J.C. PPARα-independent transcriptional targets of perfluoroalkyl acids revealed by transcript profiling. Toxicology 2017, 387, 95–107. [Google Scholar] [CrossRef]
- Gallo, V.; Leonardi, G.; Genser, B.; Lopez-Espinosa, M.-J.; Frisbee, S.J.; Karlsson, L.; Ducatman, A.M.; Fletcher, T. Serum Perfluorooctanoate (PFOA) and Perfluorooctane Sulfonate (PFOS) Concentrations and Liver Function Biomarkers in a Population with Elevated PFOA Exposure. Environ. Health Perspect. 2012, 120, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Salihovic, S.; Stubleski, J.; Kärrman, A.; Larsson, A.; Fall, T.; Lind, L.; Lind, P.M. Changes in markers of liver function in relation to changes in perfluoroalkyl substances—A longitudinal study. Environ. Int. 2018, 117, 196–203. [Google Scholar] [CrossRef]
- Jin, R.; McConnell, R.; Catherine, C.; Xu, S.; Walker, D.I.; Stratakis, N.; Jones, D.P.; Miller, G.W.; Peng, C.; Conti, D.V.; et al. Perfluoroalkyl Substances and Severity of Nonalcoholic Fatty Liver in Children: An Untargeted Metabolomics Approach. Environ. Int. 2020, 134, 105220. [Google Scholar] [CrossRef]
- Bjork, J.; Butenhoff, J.; Wallace, K.B. Multiplicity of nuclear receptor activation by PFOA and PFOS in primary human and rodent hepatocytes. Toxicology 2011, 288, 8–17. [Google Scholar] [CrossRef]
- Marques, E.; Pfohl, M.; Auclair, A.; Jamwal, R.; Barlock, B.J.; Sammoura, F.M.; Goedken, M.; Akhlaghi, F.; Slitt, A.L. Perfluorooctanesulfonic acid (PFOS) administration shifts the hepatic proteome and augments dietary outcomes related to hepatic steatosis in mice. Toxicol. Appl. Pharmacol. 2020, 408, 115250. [Google Scholar] [CrossRef]
- Roth, K.; Yang, Z.; Agarwal, M.; Liu, W.; Peng, Z.; Long, Z.; Birbeck, J.; Westrick, J.; Liu, W.; Petriello, M.C. Exposure to a mixture of legacy, alternative, and replacement per- and polyfluoroalkyl substances (PFAS) results in sex-dependent modulation of cholesterol metabolism and liver injury. Environ. Int. 2021, 157, 106843. [Google Scholar] [CrossRef]
- Pfohl, M.; Ingram, L.; Marques, E.; Auclair, A.; Barlock, B.; Jamwal, R.; Anderson, D.; Cummings, B.S.; Slitt, A.L. Perfluorooctanesulfonic Acid and Perfluorohexanesulfonic Acid Alter the Blood Lipidome and the Hepatic Proteome in a Murine Model of Diet-Induced Obesity. Toxicol. Sci. 2020, 178, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa, P.D.; Buss, C.; Entringer, S.; Swanson, J.M. Developmental Origins of Health and Disease: Brief History of the Approach and Current Focus on Epigenetic Mechanisms. Semin. Reprod. Med. 2009, 27, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Mora, A.M.; Fleisch, A.F.; Rifas-Shiman, S.L.; Baidal, J.A.W.; Pardo, L.; Webster, T.F.; Calafat, A.M.; Ye, X.; Oken, E.; Sagiv, S.K. Early life exposure to per- and polyfluoroalkyl substances and mid-childhood lipid and alanine aminotransferase levels. Environ. Int. 2018, 111, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mamsen, L.S.; Björvang, R.D.; Mucs, D.; Vinnars, M.-T.; Papadogiannakis, N.; Lindh, C.H.; Andersen, C.Y.; Damdimopoulou, P. Concentrations of perfluoroalkyl substances (PFASs) in human embryonic and fetal organs from first, second, and third trimester pregnancies. Environ. Int. 2019, 124, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Blake, B.E.; Fenton, S.E. Early life exposure to per- and polyfluoroalkyl substances (PFAS) and latent health outcomes: A review including the placenta as a target tissue and possible driver of peri- and postnatal effects. Toxicology 2020, 443, 152565. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Schreder, E.; Dempsey, J.C.; Uding, N.; Chu, V.; Andres, G.; Sathyanarayana, S.; Salamova, A. Per- and Polyfluoroalkyl Substances (PFAS) in Breast Milk: Concerning Trends for Current-Use PFAS. Environ. Sci. Technol. 2021, 55, 7510–7520. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Gao, Y.; Zhang, Y.; Qin, K.; Liew, Z.; Tian, Y. Associations of paternal and maternal per- and polyfluoroalkyl substances exposure with cord serum reproductive hormones, placental steroidogenic enzyme and birth weight. Chemosphere 2021, 285, 131521. [Google Scholar] [CrossRef] [PubMed]
- Filgo, A.J.; Quist, E.M.; Hoenerhoff, M.J.; Brix, A.E.; Kissling, G.E.; Fenton, S.E. Perfluorooctanoic acid (PFOA)-induced liver lesions in two strains of mice following developmental exposures: PPARα is not required. Toxicol. Pathol. 2015, 43, 558–568. [Google Scholar] [CrossRef]
- Chang, S.; Butenhoff, J.L.; Parker, G.A.; Coder, P.S.; Zitzow, J.D.; Krisko, R.M.; Bjork, J.A.; Wallace, K.B.; Seed, J.G. Reproductive and developmental toxicity of potassium perfluorohexanesulfonate in CD-1 mice. Reprod. Toxicol. 2018, 78, 150–168. [Google Scholar] [CrossRef]
- Marques, E.S.; Agudelo, J.; Kaye, E.M.; Modaresi, S.M.S.; Pfohl, M.; Bečanová, J.; Wei, W.; Polunas, M.; Goedken, M.; Slitt, A.L. The role of maternal high fat diet on mouse pup metabolic endpoints following perinatal PFAS and PFAS mixture exposure. Toxicology 2021, 462, 152921. [Google Scholar] [CrossRef]
- Lau, C.; Thibodeaux, J.R.; Hanson, R.G.; Narotsky, M.G.; Rogers, J.M.; Lindstrom, A.B.; Strynar, M.J. Effects of Perfluorooctanoic Acid Exposure during Pregnancy in the Mouse. Toxicol. Sci. 2006, 90, 510–518. [Google Scholar] [CrossRef]
- Jamwal, R.; Barlock, B.J.; Adusumalli, S.; Ogasawara, K.; Simons, B.L.; Akhlaghi, F. Multiplex and Label-Free Relative Quantification Approach for Studying Protein Abundance of Drug Metabolizing Enzymes in Human Liver Microsomes Using SWATH-MS. J. Proteome Res. 2017, 16, 4134–4143. [Google Scholar] [CrossRef]
- Jamwal, R.; de la Monte, S.M.; Ogasawara, K.; Adusumalli, S.; Barlock, B.B.; Akhlaghi, F. Nonalcoholic Fatty Liver Disease and Diabetes Are Associated with Decreased CYP3A4 Protein Expression and Activity in Human Liver. Mol. Pharm. 2018, 15, 2621–2632. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Hein, M.Y.; Cox, J.; Mann, M. A ‘Proteomic Ruler’ for Protein Copy Number and Concentration Estimation without Spike-in Standards. Mol. Cell. Proteom. 2014, 13, 3497–3506. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]
- Pfohl, M.; Marques, E.; Auclair, A.; Barlock, B.; Jamwal, R.; Goedken, M.; Akhlaghi, F.; Slitt, A.L. An ’Omics Approach to Unraveling the Paradoxical Effect of Diet on Perfluorooctanesulfonic Acid (PFOS) and Perfluorononanoic Acid (PFNA)-Induced Hepatic Steatosis. Toxicol. Sci. 2021, 180, 277–294. [Google Scholar] [CrossRef]
- Modaresi, S.M.S.; Wei, W.; Emily, M.; DaSilva, N.A.; Slitt, A.L. Per- and polyfluoroalkyl substances (PFAS) augment adipogenesis and shift the proteome in murine 3T3-L1 adipocytes. Toxicology 2022, 465, 153044. [Google Scholar] [CrossRef]
- Stratakis, N.; Conti, D.V.; Jin, R.; Margetaki, K.; Valvi, D.; Siskos, A.P.; Maitre, L.; Garcia, E.; Varo, N.; Zhao, Y.; et al. Prenatal Exposure to Perfluoroalkyl Substances Associated with Increased Susceptibility to Liver Injury in Children. Hepatology 2020, 72, 1758–1770. [Google Scholar] [CrossRef]
- Khusial, R.D.; Cioffi, C.E.; Caltharp, S.A.; Krasinskas, A.M.; Alazraki, A.; Knight-Scott, J.; Cleeton, R.; Castillo-Leon, E.; Jones, D.P.; Pierpont, B.; et al. Development of a Plasma Screening Panel for Pediatric Nonalcoholic Fatty Liver Disease Using Metabolomics. Hepatol. Commun. 2019, 3, 1311–1321. [Google Scholar] [CrossRef]









| Treatment | Log2FC | p-Value | Abbreviation | Protein Name | Function |
|---|---|---|---|---|---|
| PFOA SD | 1.75 | 0.00479 | Ctse | Cathepsin E | Role in activation-induced lymphocyte depletion |
| 1.40 | 6.90 × 10−7 | Gm4952 | Glycine N-acyltransferase-like protein | Enables glycine N-acyltransferase activity | |
| PFOS SD | 0.64 | 0.04073 | Ugt1a6 | UDP-glucuronosyltransferase 1–6 | Role in xenobiotic glucuronidation |
| 0.64 | 0.03844 | Fhit | Bis(5′-adenosyl)-triphosphatase | Involved in the diadenosine triphosphate catabolic process | |
| PFHxS SD | 0.89 | 0.04754 | Adsl | Adenylosuccinate lyase | Involved in the AMP biosynthetic process |
| 0.92 | 0.04504 | Cyp2a5 | Cytochrome p450 2a5 | Involved in arachidonic acid epoxygenase activity | |
| PFOA HFD | 0.83 | 0.03687 | Hsd3b5 | NADPH-dependent 3-keto-steroid reductase | Involved in the formation of steroids |
| 0.53 | 0.00257 | Acot13 | Acyl-coenzyme A thioesterase 13 | Involved in the lipid metabolic process | |
| PFOS HFD | 0.65 | 0.01482 | Mt-nd2 | NADH-ubiquinone oxidoreductase chain 2 | Responsible for electron transfer during oxidative phosphorylation |
| 0.57 | 0.02997 | Dpyd | Dihydropyrimidine dehydrogenase | Involved in pyrimidine nucleoside monophosphate catabolic process | |
| PFHxS HFD | 4.66 | 0.03114 | Slenbp2 | Selenium-binding protein 2 | Involved in detecting reactive xenobiotics in the cytoplasm |
| 1.98 | 4.51 × 10−4 | Cyp3a41 | Cytochrome p450 3a41 | Involved in lipid hydroxylation |
| Treatment | Log2FC | p-Value | Abbreviation | Protein Name | Function |
|---|---|---|---|---|---|
| PFOA SD | −1.10 | 1.65 × 10−6 | Gpt2 | Alanine aminotransferase 2 | Key intermediate protein in amino acid metabolism |
| −0.90 | 1.05 × 10−4 | Lpin1 | Phosphatidate phosphatase lpin1 | Involved in the cellular response to insulin stimulation | |
| PFOS SD | −0.68 | 0.01353 | Thnsl2 | Threonine synthase-like 2 | Involved in the serine family amino acid catabolic process |
| −0.68 | 0.01542 | Ldhb | L-lactate dehydrogenase B chain | Lactate metabolic process | |
| PFHxS SD | −1.05 | 0.01015 | Cyp2a12 | Cytochrome p450 2a12 | Arachidonic acid epoxygenase activity |
| −0.97 | 0.03609 | Atp5pf | ATP synthase-coupling factor 6, mitochondrial | Negative regulation of arachidonic acid secretion | |
| PFOA HFD | −1.62 | 0.01037 | Cyp2d9 | Cytochrome p450 2d9 | Role in the arachidonic acid metabolic process |
| −1.18 | 0.04546 | Cyp3a13 | Cytochrome p450 3a13 | Aids in removing methyl groups via oxidation | |
| PFOS HFD | −1.25 | 0.00885 | Ec:1.1.1.263 | 1,5-anhydro-D-fructose reductase | Responsible for the catalysis of redox reactions |
| −1.03 | 0.04074 | Apoa2 | Apolipoprotein A-II | Role in lipid binding | |
| PFHxS HFD | −0.77 | 0.00175 | Rcn2 | Reticulocalbin-2 | Involved in hepatic growth factor signaling |
| −0.70 | 0.01179 | Apoe | Apolipoprotein E | Regulate plasma lipoprotein metabolism |
| Pathway | Abbreviation | Protein Name | Function |
|---|---|---|---|
| Lipid Transport, Storage and Synthesis | Acaca | Acetyl-CoA carboxylase 1 | Enzyme that catalyzes the rate-limiting step in fatty acid synthesis |
| Apoe | Apolipoprotein E | Lipoprotein-mediated lipid transport | |
| Cd36 | Platelet glycoprotein 4 | Involved in long chain fatty acid uptake | |
| Fabp1 | Fatty acid-binding protein 1 | Role in fatty acid uptake, transport, and metabolism | |
| Fabp4 | Fatty acid-binding protein 4 | Role in fatty acid uptake, transport, and metabolism | |
| Fasn | Fatty acid synthase | Catalyzes long-chain saturated fatty acids from acetyl-coa and malonyl-coa | |
| Hmgcs1 | Hydroxymethylglutaryl-CoA synthase | Catalyzes the formation of HMG-coa | |
| Scd1 | Acyl-CoA desaturase 1 | Involved in fatty acid biosynthesis | |
| Slc27a2 | Very long-chain acyl-CoA synthetase | Catalyzing the formation of fatty acyl-coa | |
| Xenobiotic Metabolism and Transport | Aldh3a2 | Aldehyde dehydrogenase family 3 member a2 | Catalyzes the oxidation of medium and long-chain aliphatic aldehydes to fatty acids |
| Ces1 | Liver carboxylesterase 1 | Detoxification of xenobiotics | |
| Ces1d | Carboxylesterase 1d | Metabolism of xenobiotics and of natural substrates | |
| Ces2c | Acylcarnitine hydrolase | Prostaglandin metabolic process | |
| Cyp2b10 | Cytochrome p450 2b10 | Oxidizes steroids, fatty acids, and xenobiotics | |
| Cyp3a11 | Cytochrome p450 3a11 | Steroid metabolic process | |
| Ntcp | Sodium/bile acid cotransporter | Transporter of conjugated bile salts from plasma into the hepatocyte | |
| Oatp1a1 | Organic anion transporting polypeptide 1a1 | Mediates the Na+-independent transport of organic anions | |
| Oatp1b2 | Organic anion transporting polypeptide 1b2 | Mediates the Na+-independent uptake of organic anions | |
| Oatp2b1 | Organic anion transporting polypeptide 2b1 | Mediates the Na+-independent transport of organic anions | |
| Por | Cytochrome p450 oxidoreductase | Donate electrons directly from NADPH to all microsomal p450 enzymes | |
| Ugt1a1 | UDP-glucuronosyltransferase 1a1 | Catalyzes phase II biotransformation reactions in which lipophilic substrates are conjugated with glucuronic acid | |
| Inflammation | Ephx1 | Epoxide hydrolase 1 | Catalyzes the hydrolysis of arene and aliphatic epoxides |
| Gclc | Glutamate-Cysteine Ligase Catalytic Subunit | The first rate-limiting enzyme of glutathione synthesis | |
| Gstm3 | GSTM3 glutathione S-transferase mu 3 | Mediates uptake and detoxification of both endogenous compounds and xenobiotics | |
| Gstm5 | Glutathione S-transferase Mu 5 | Conjugation of reduced glutathione to exogenous/endogenous hydrophobic electrophiles | |
| Sod1 | Superoxide Dismutase 1 | Eliminates radicals that are toxic to biological systems | |
| Lipid Catabolism | Acot2 | Acyl-CoA Thioesterase 2 | Regulation of lipid metabolism/intracellular levels of free fatty acids |
| Acox1 | Peroxisomal acyl-coenzyme A oxidase 1 | Catalyzes the desaturation of acyl-coas to 2-trans-enoyl-coas | |
| Cpt1b | Carnitine palmitoyltransferase 1b | Rate-controlling enzyme of fatty acid beta-oxidation | |
| Cyp4a10 | Cytochrome p450 4a10 | Arachidonic acid metabolic process | |
| Cyp4a12a | Cytochrome p450 4a12a | Metabolism of fatty acids and oxylipins | |
| Cyp4a14 | Cytochrome p450 4a14 | Oxidation of medium chain fatty acids | |
| Ehhadh | Enoyl-CoA Hydratase And 3-Hydroxyacyl CoA Dehydrogenase | Enzyme in fatty acid beta-oxidation pathway |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaye, E.; Marques, E.; Agudelo Areiza, J.; Modaresi, S.M.S.; Slitt, A. Exposure to a PFOA, PFOS and PFHxS Mixture during Gestation and Lactation Alters the Liver Proteome in Offspring of CD-1 Mice. Toxics 2024, 12, 348. https://doi.org/10.3390/toxics12050348
Kaye E, Marques E, Agudelo Areiza J, Modaresi SMS, Slitt A. Exposure to a PFOA, PFOS and PFHxS Mixture during Gestation and Lactation Alters the Liver Proteome in Offspring of CD-1 Mice. Toxics. 2024; 12(5):348. https://doi.org/10.3390/toxics12050348
Chicago/Turabian StyleKaye, Emily, Emily Marques, Juliana Agudelo Areiza, Seyed Mohamad Sadegh Modaresi, and Angela Slitt. 2024. "Exposure to a PFOA, PFOS and PFHxS Mixture during Gestation and Lactation Alters the Liver Proteome in Offspring of CD-1 Mice" Toxics 12, no. 5: 348. https://doi.org/10.3390/toxics12050348





