Using Entropy Leads to a Better Understanding of Biological Systems
Abstract
:1. Introduction
2. A Comprehensive Perspective
2.1. The Core Aspect: Rules of Probability Theory, the Method of Maximum Entropy and Information Geometry
2.2. Comments
3. From Principles of Inference to Protein Folding Dynamics
3.1. Introduction
3.2. Theory
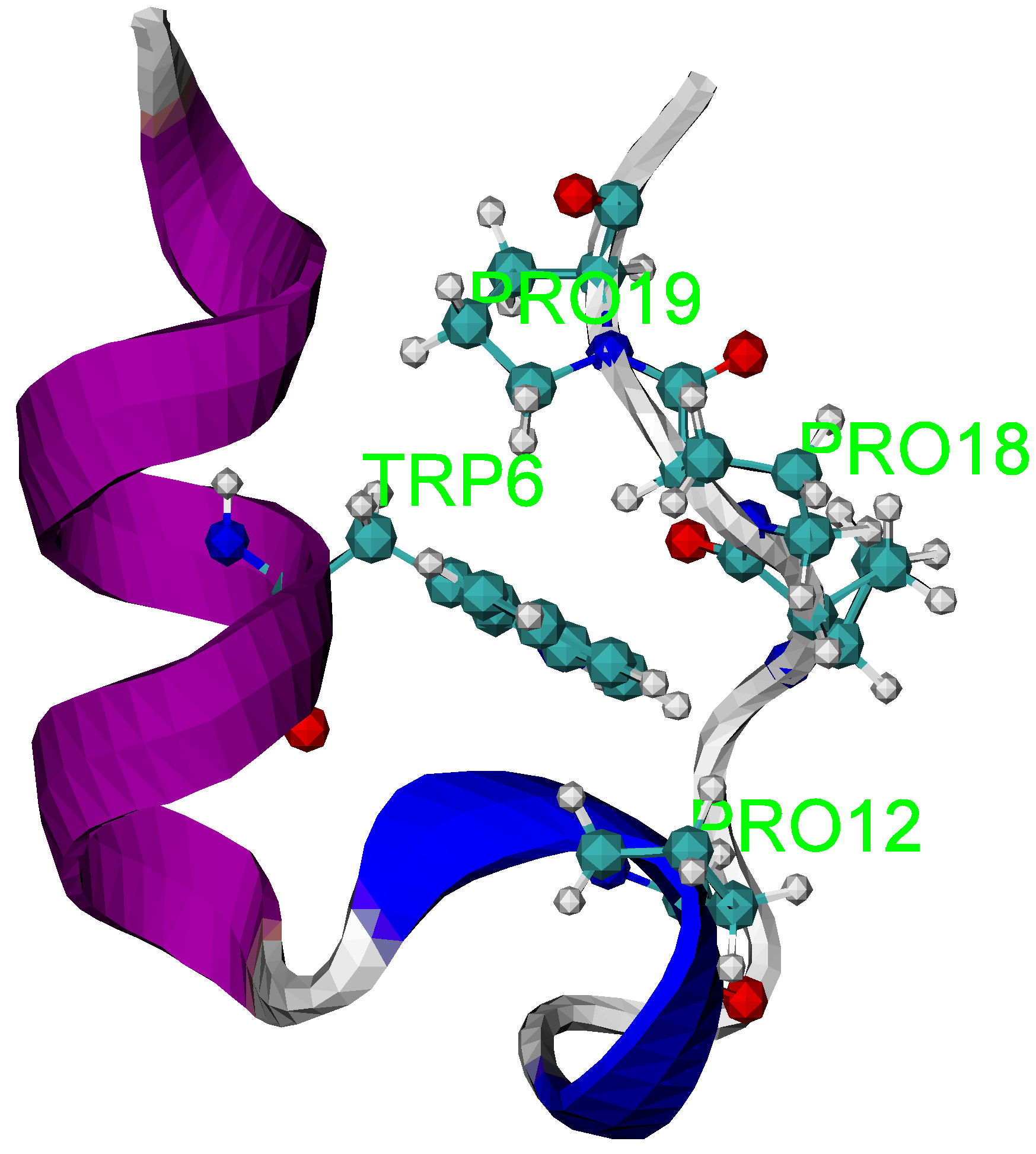
3.3. Folding Trajectories of Trp-Cage
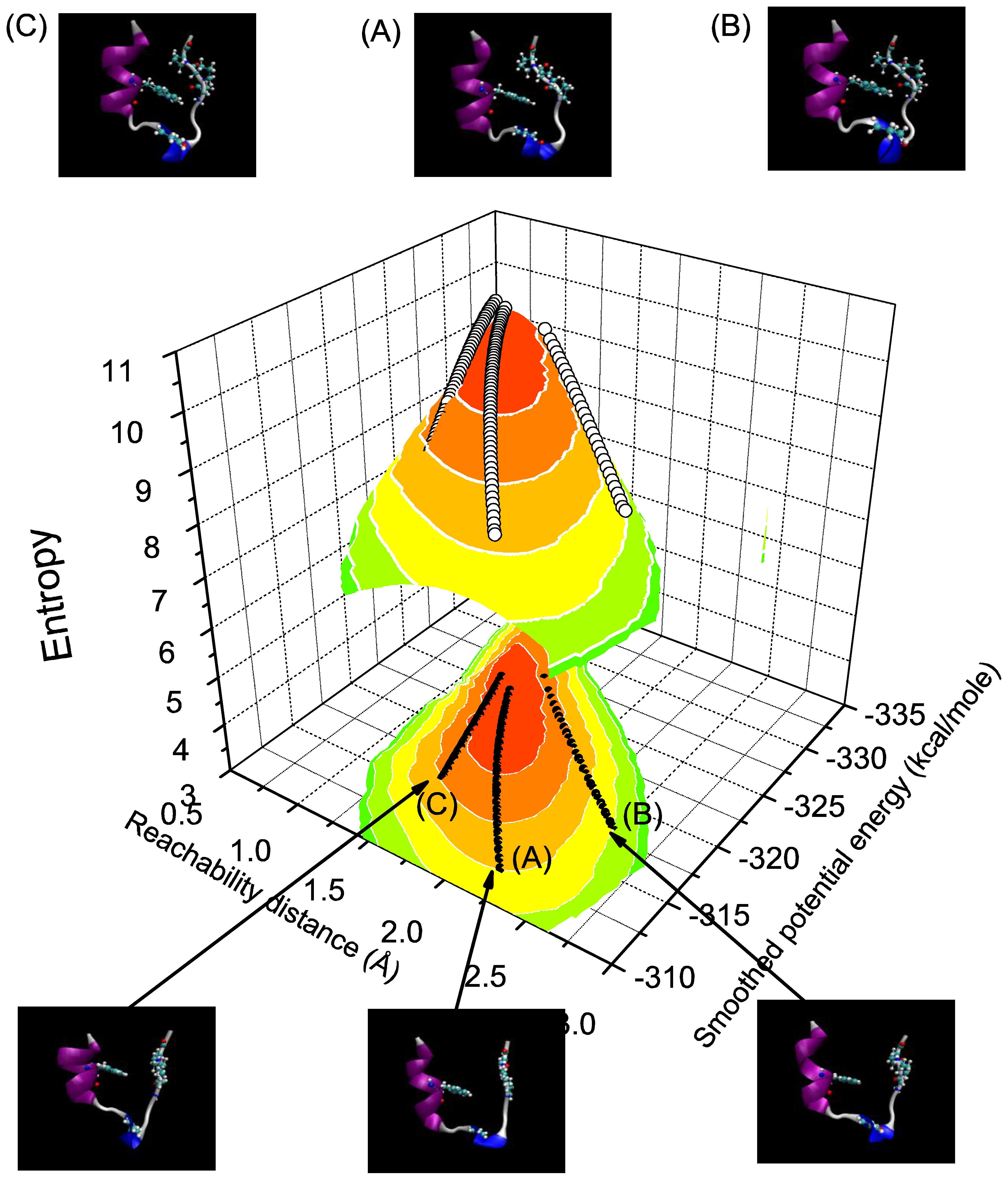
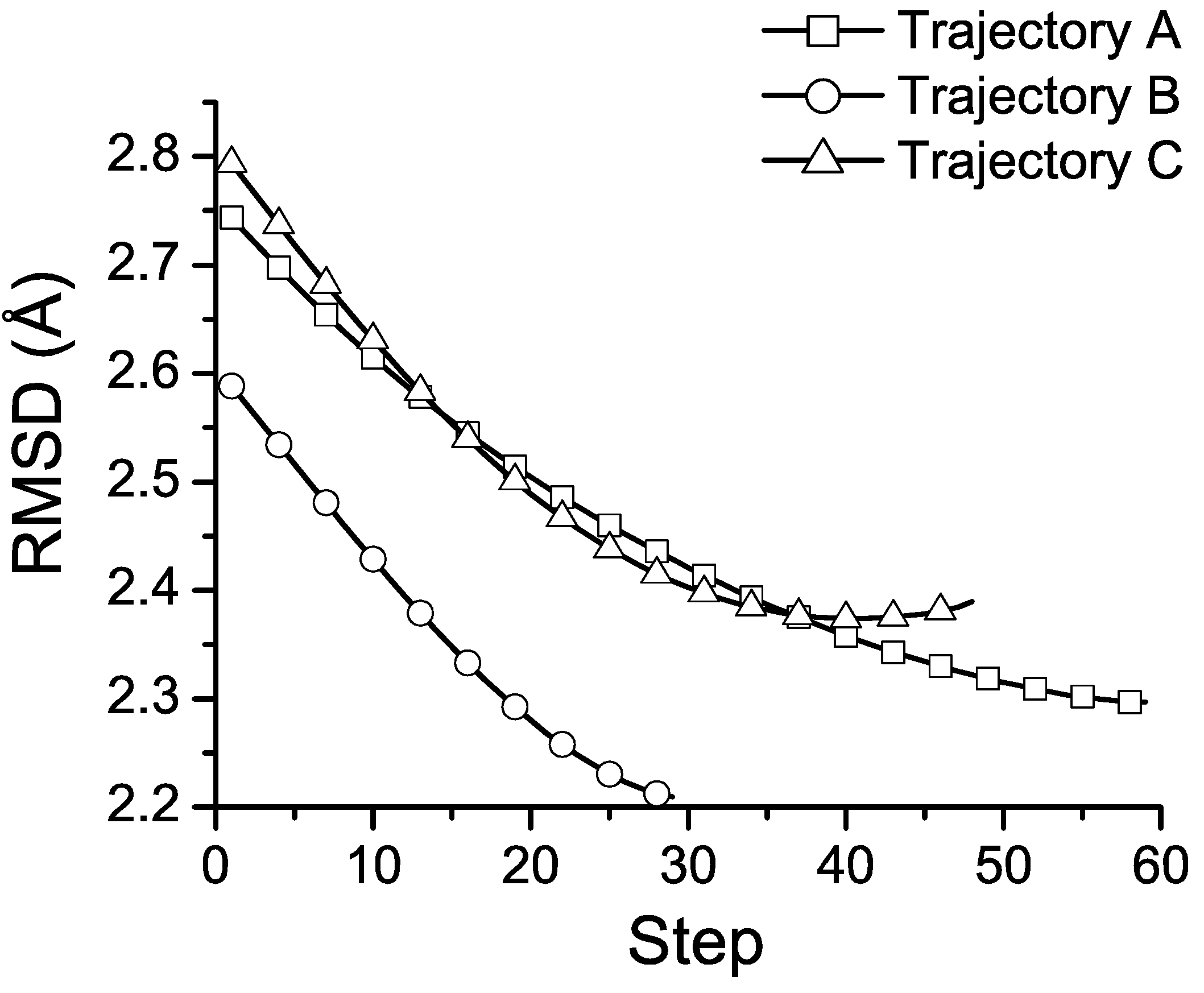
3.4. Discussion
4. The Relative Importance of Tubulin Isotypes Unveiled by the Maximum Entropy Approach
4.1. Introduction
| Cell Line | Origin | Media |
|---|---|---|
| A549 | Human lung carcinoma | RPMI with 10% fetal bovine serum (FBS) |
| MCF-7 | Human mammary gland adenocarcinoma | RPMI with 10% FBS |
| CEM | Human T-lymphoblastoid from ALL | RPMI with 10% FBS |
| HeLa | Human cervical carcinoma | DMEM with 10% FBS |
| M006X | Human glioma cells | DMEM-F12 with 10% FBS and 1% Glutamine |
| M010B | Human glioma cells | DMEM-F12 with 10% FBS and 1% Glutamine |
4.2. Method
4.3. Results and Discussion

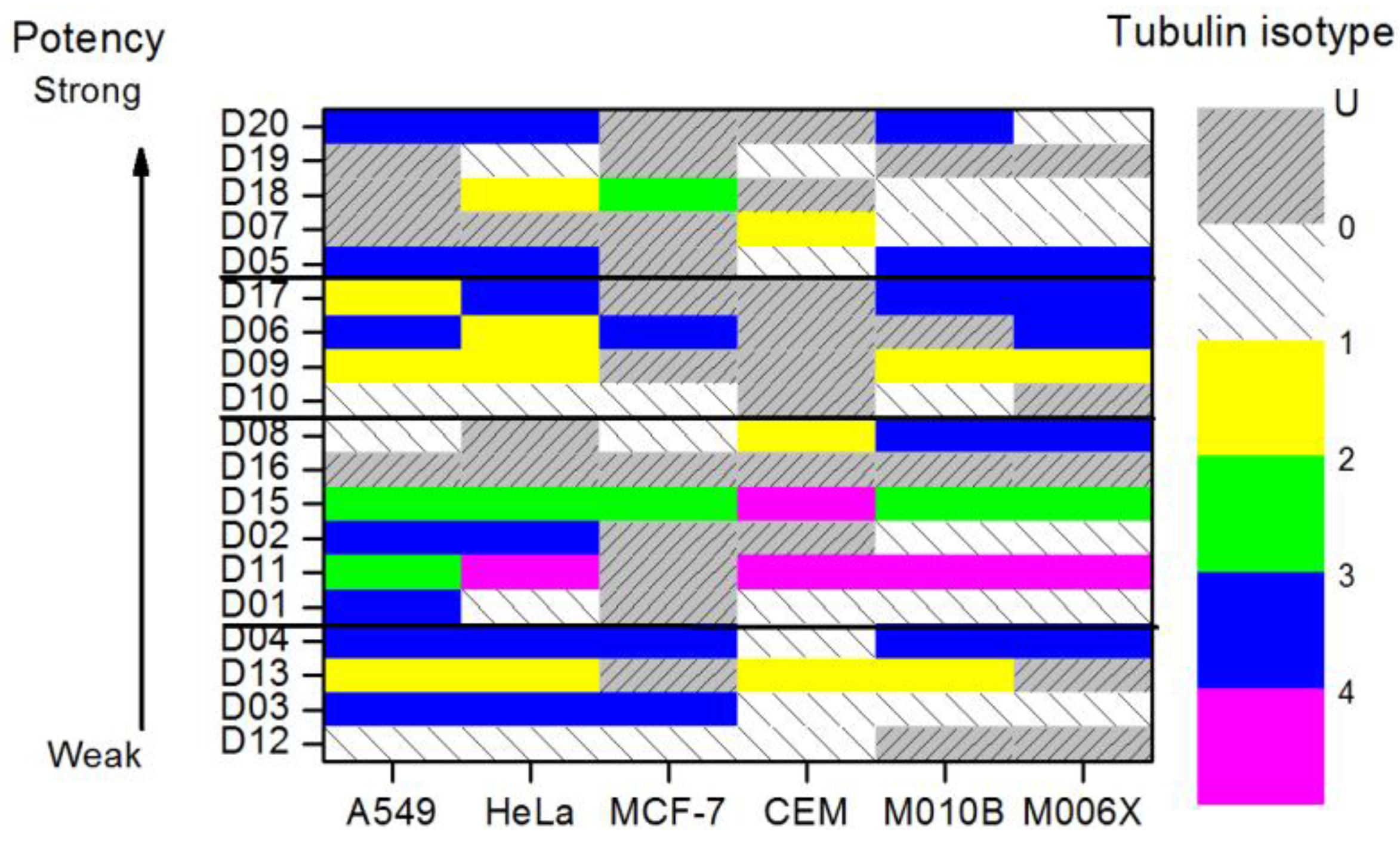
5. Entropic Fragment Based Aptamer Design
5.1. Introduction
5.2. Theory
5.3. Results and Discussions
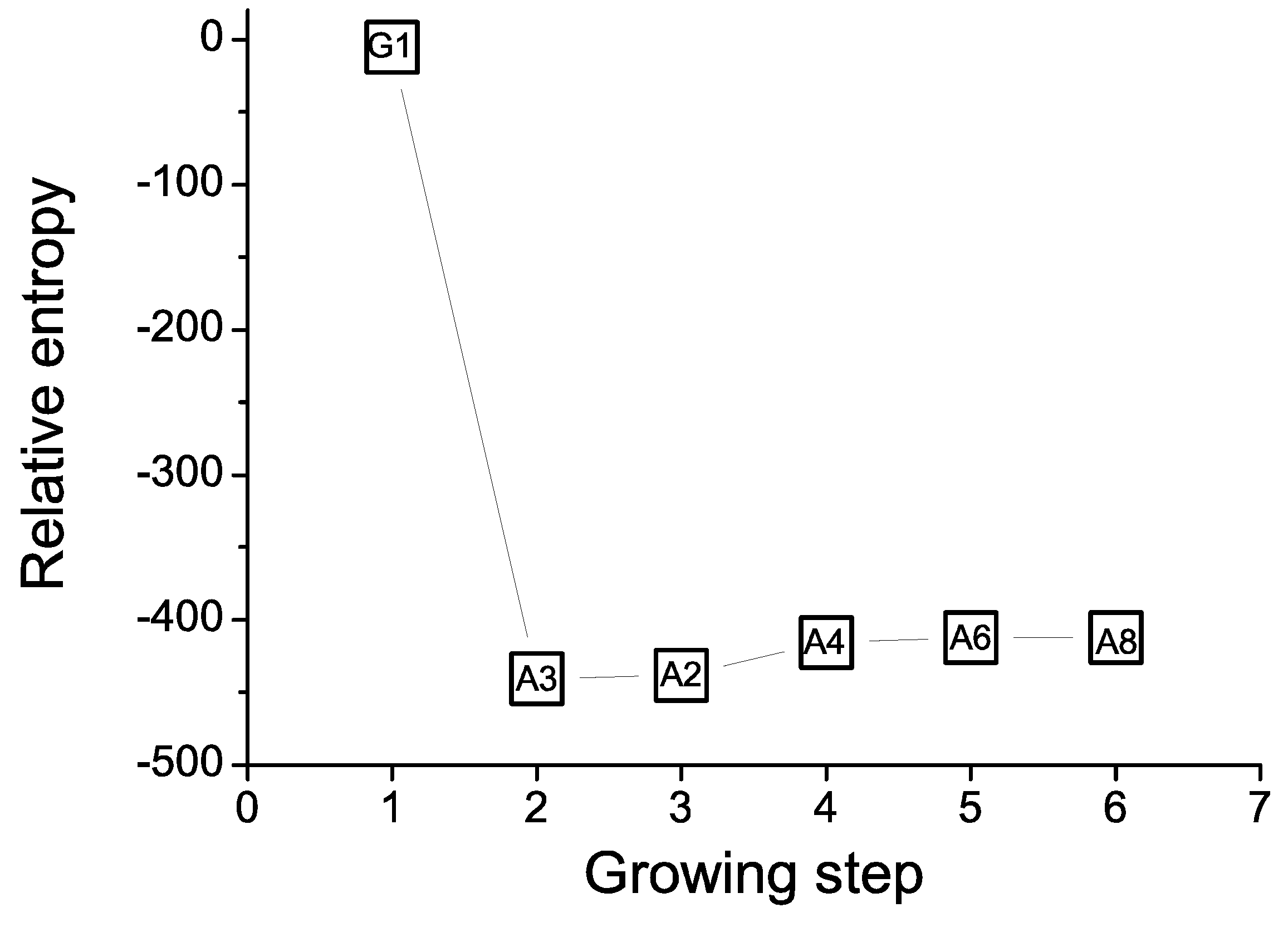
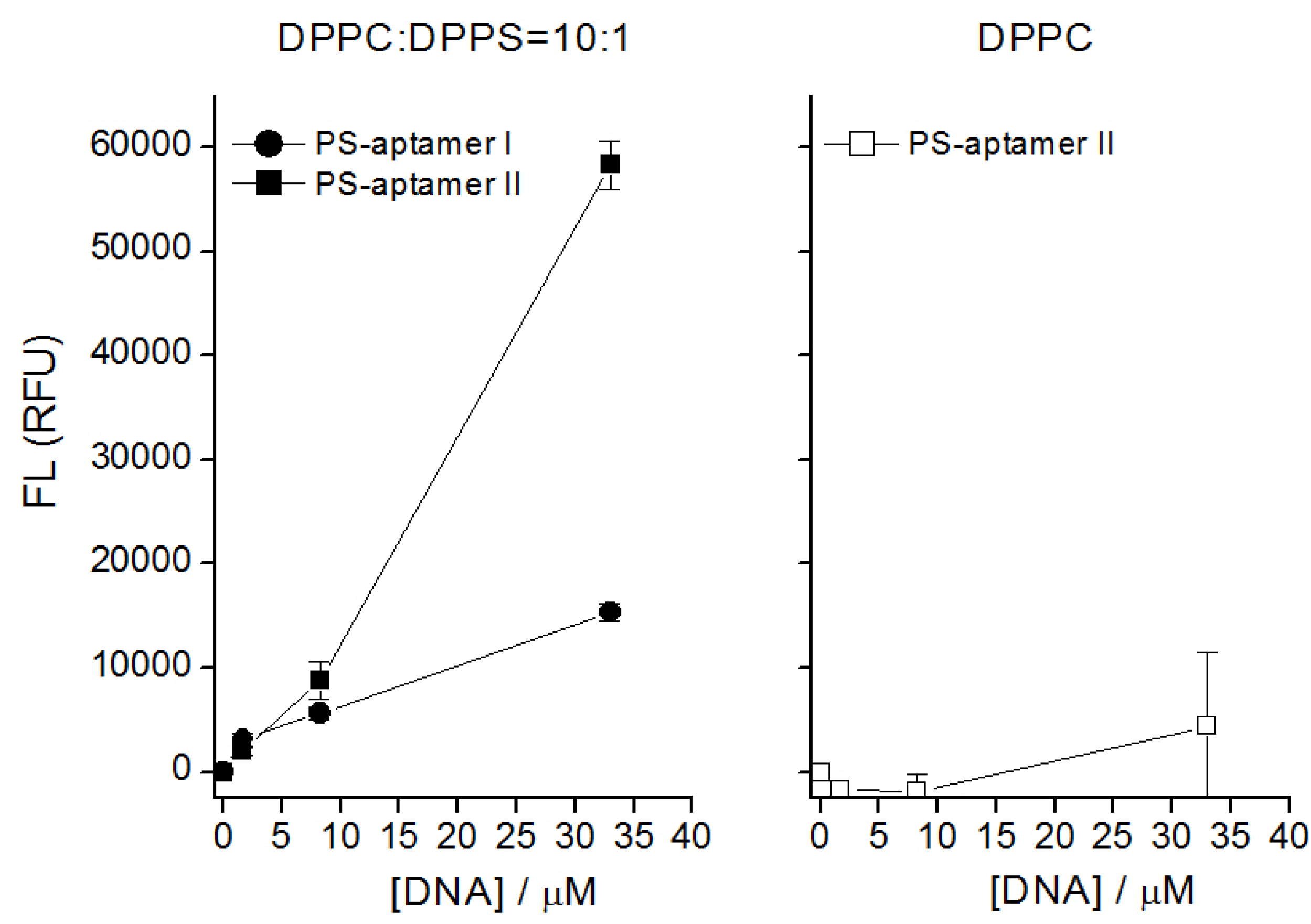
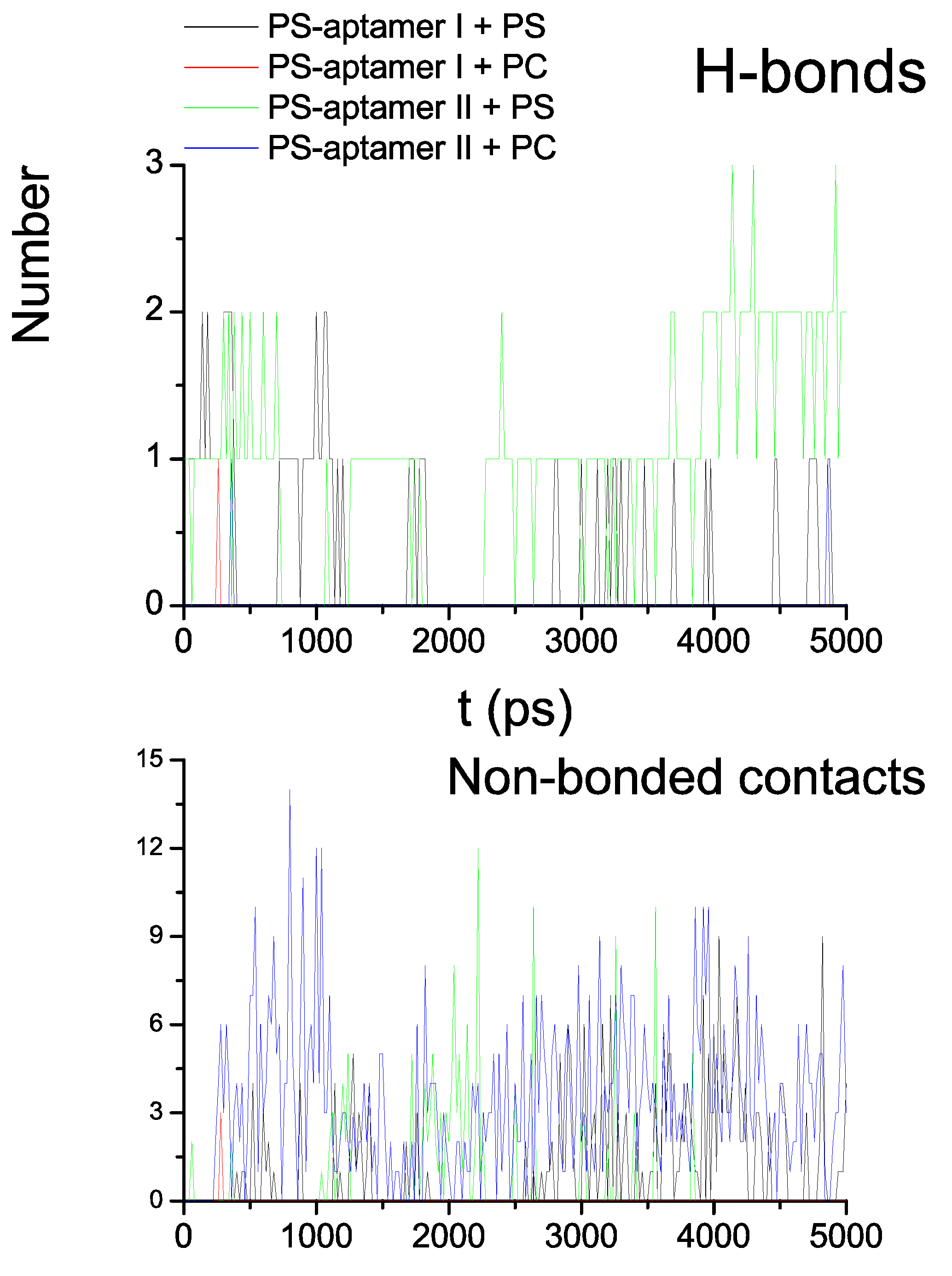
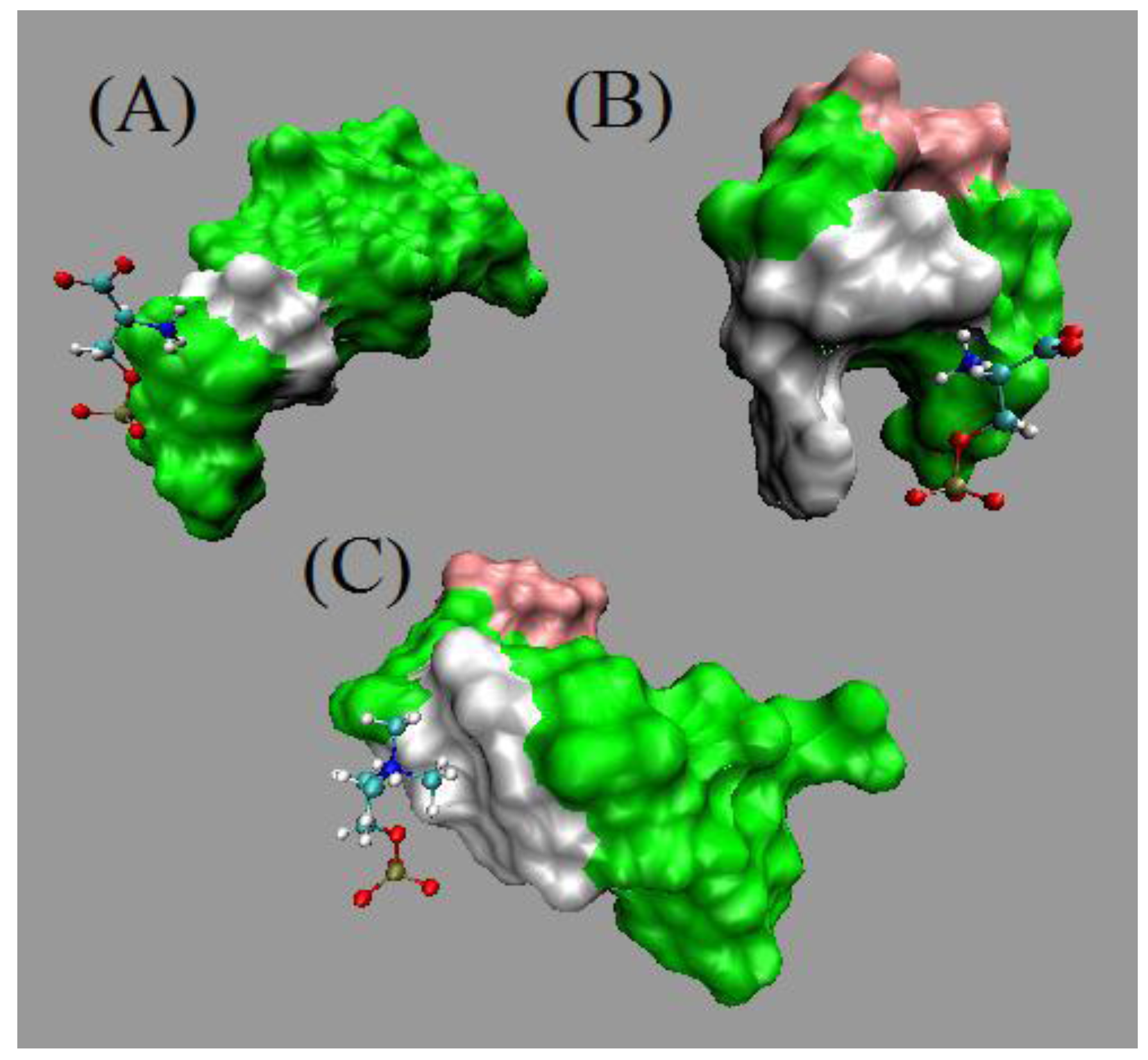
5.4. Comments
6. Conclusions
Acknowledgements
References
- Tseng, C.-Y.; Caticha, A. Using relative entropy to find optimal approximations: An application to simple fluids. Physica A 2008, 387, 6759–6770. [Google Scholar] [CrossRef]
- Dill, K.A. Theory for the folding and stability of globular proteins. Biochemistry 1985, 24, 1501–1509. [Google Scholar] [CrossRef] [PubMed]
- Shih, C.T.; Su, Z.Y.; Gwan, J.F.; Hao, B.L.; Hsieh, C.H.; Lee, H.C. Mean-field HP model, designability and alpha-helices in protein structures. Phys. Rev. Lett. 2000, 84, 386–389. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, J. Information, physics, quantum: The search for links. In Complexity, Entropy and the Physics of Information; Zurek, W.H., Ed.; Addison-Wesley: Redwood City, CA, USA, 1990; p. 1. [Google Scholar]
- Jaynes, E.T. Information theory and statistical mechanics. Phys. Rev. 1957, 106, 620–630. [Google Scholar] [CrossRef]
- Caticha, A. Entropic priors. In Bayesian Inference and Maximum Entropy Methods in Science and Engineering; Erickson, G., Zhai, Y., Eds.; AIP: Melville, NY, USA, 2004; AIP Conf. Proc. 707; p. 75. [Google Scholar]
- Shore, J.E.; Johnson, R.W. Axiomatic derivation of the principle of maximum entropy and the principle of minimum cross entropy. IEEE Trans. Inf. Theory 1980, IT-26, 26–37. [Google Scholar] [CrossRef]
- Shore, J.E.; Johnson, R.W. Properties of cross-entropy minimization. IEEE Trans. Inf. Theory 1981, IT-27, 472–482. [Google Scholar] [CrossRef]
- Giffin, A.; Caticha, A. Updating probabilities with data and moments. In Bayesian Inference and Maximum Entropy Methods in Science and Engineering; Knuth, K.H., Caticha, A., Center, J.L., Giffin, A., Rodríguez, C.C., Eds.; AIP: Melville, NY, USA, 2007; AIP Conf. Proc. 954; p. 74. [Google Scholar]
- Caticha, A. Maximum entropy, fluctuations and priors. In Maximum Entropy and Bayesian Methods in Science and Engineering; Mohammad-Djafari, A., Ed.; AIP: Melville. NY, USA, 2001; AIP Conf. Proc. 568; p. 72. [Google Scholar]
- Caticha, A. Entropic dynamics. In Maximum Entropy and Bayesian Methods in Science and Engineering; Fry, R.L., Ed.; AIP: Melville, NY, USA, 2002; AIP Conf Proc 617; p. 302. [Google Scholar]
- Caticha, A.; Cafaro, C. From information geometry to Newtonian dynamics. In Maximum Entropy and Bayesian Methods in Science and Engineering; Knuth, K.H., Caticha, A., Center, J.L., Giffin, A., Rodríguez, C.C., Eds.; AIP: Melville, NY, USA, 2007; AIP Conf. Proc. 954; p. 165. [Google Scholar]
- Caticha, A. From inference to physics. In Maximum Entropy and Bayesian Methods in Science and Engineering; de Souza Lauretto, M., de Bragança Pereira, C.A., Stern, J.M., Eds.; AIP: Melville, NY, USA, 2008; AIP Conf. Proc. 1073; p. 23. [Google Scholar]
- Tseng, C.-Y.; Yu, C.-P.; Lee, H.C. From laws of inference to protein folding dynamics. Phys. Rev. E 2010, 82, 0219141–0219149. [Google Scholar] [CrossRef]
- Tseng, C.-Y.; Mane, J.Y.; Winter, P.; Johnson, L.; Huzil, T.; Izbicka, E.; Luduena, R.F.; Tuszynski, J.A. Quantitative analysis of the effect of tubulin isotype expression on sensitivity of cancer cell lines to a set of novel colchicine derivatives. Mol. Canc. 2010, 9, 131. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.-Y.; Ashrafuzzaman, M.D.; Mane, J.Y.; Kapty, J.; Mercer, J.R.; Tuszynski, J.A. Entropic fragment based approach for aptamer design. Chem. Biol. Drug Des. 2010. submitted for publication. [Google Scholar] [CrossRef]
- Cox, R.T. Probability, frequency and reasonable expectation. Am. J. Phys. 1946, 14, 1–13. [Google Scholar] [CrossRef]
- Cox, R.T. The Algebra of Probable Inference; The Johns Hopkins Press: Baltimore, MD, US, 1961. [Google Scholar]
- Jaynes, E.T. Probability Theory: The Logic of Science; Cambridge University Press: Cambridge, UK, 2003. [Google Scholar]
- Giffin, A. Maximum Entropy: The Universal Method for Inference. Ph.D. Thesis, The State University of New York at Albany, NY, USA, 2008. [Google Scholar]
- Amari, S.; Nagaoka, H. Methods of Information Geometry; American Mathematical Society: Providence, RI, USA, 2000. [Google Scholar]
- Jaynes, E.T. Information theory and statistical mechanics II. Phys. Rev. 1957, 108, 171–190. [Google Scholar] [CrossRef]
- Fisher, R.A. Theory of statistical estimation. Proc. Camb. Phil. Soc. 1925, 22, 700–725. [Google Scholar] [CrossRef]
- Rao, C.R. Information and accuracy attainable in the estimation of statistical parameters. Bull. Calcutta Math. Soc. 1945, 37, 81–91. [Google Scholar]
- Amari, S. Differential-Geometrical Methods in Statistics; Springer-Verlag: New York, NY, USA, 1985. [Google Scholar]
- Rhee, Y.M.; Pande, V.S. Multiplexed-Replica Exchange Molecular Dynamics Method for Protein Folding Simulation. Biophys. J. 2003, 84, 775–786. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD-Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Han, J.; Kamber, M. Data Mining: Concept and Techniques, 2nd ed.; Morgan Kaufmann: San Francisco, CA, USA, 2006. [Google Scholar]
- Jaynes, E.T. Where do we stand on maximum entropy? In The Maximum Entropy Formalism; Levine, R.D., Tribus, M., Eds.; MIT Press: Cambridge, MA, USA, 1979; p. 15. [Google Scholar]
- Juraszek, J.; Bolhuis, P.G. Sampling the multiple folding mechanisms of Trp-cage in explicit solvent. Proc. Nat. Acad. Soc. U. S. A. 2006, 103, 15859–15864. [Google Scholar] [CrossRef] [PubMed]
- Dellago, C.; Bolhuis, P.G.; Csajka, F.S.; Chandler, D. Transition path sampling and the calculation of rate constants. J. Chem. Phys. 1998, 108, 1964–1977. [Google Scholar] [CrossRef]
- Bolhuis, P.G.; Chandler, D.; Dellago, C.; Geissler, P.I. Transition path sampling: Throwing ropes over rough mountain passes in the Dark. Annu. Rev. Phys. Chem. 2002, 53, 291–318. [Google Scholar] [CrossRef] [PubMed]
- Dellago, C.; Bolhuis, P.G.; Geissler, P.I. Transition Path Sampling. Adv. Chem. Phys. 2002, 123, 1–78. [Google Scholar]
- Owellen, R.J.; Owens, A.H.J.; Donigian, D.W. The binding of vincristine, vinblastine and colchicine to tubulin. Biochem. Biophys. Res. Commun. 1972, 47, 685–691. [Google Scholar] [CrossRef]
- Derry, W.B.; Wilson, L.; Khan, I.A.; Luduena, R.F.; Jordan, M.A. Taxol differentially modulates the dynamics of microtubules assembled from unfractionated and purified beta-tubulin isotypes. Biochemistry 1997, 36, 3554–3562. [Google Scholar] [CrossRef] [PubMed]
- Tembe, B.; McCammon, J. Ligand-receptor interactions. Comput. Chem. 1984, 8, 281–283. [Google Scholar] [CrossRef]
- Nimjee, S.M.; Rusconi, C.P.; Sullenger, B.A. Aptamers: An emerging class of therapeutics. Annu. Rev. Med. 2005, 56, 555–583. [Google Scholar] [CrossRef] [PubMed]
- Hamula, C.L.A.; Guthrie, J.W.; Zhang, H.; Li, X.F.; Chris, L.X. Selection and analytical applications of aptamers. Trends Anal. Chem. 2006, 25, 681–691. [Google Scholar] [CrossRef]
- James, W. Aptamer. In Encyclopedia of Analytical Chemistry; Wiley& Sons Inc.: Hoboken, NJ, USA, 2000; pp. 4848–4871. [Google Scholar]
- James, W. Aptamers in the virologists’ toolkit. J. Gen. Virol. 2007, 88, 351–364. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Gan, H.H.; Schlick, T. A computational proposal for designing structured RNA pools for in vitro selection of RNAs. RNA 2007, 13, 478–492. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Shin, J.S.; Elmetwaly, S.; Gan, H.H.; Schlick, T. RagPools: RNA-As Graph-Pools-a web server for assisting the design of structured RNA pools for in vitro selection. Bioinformatics 2007, 23, 2959–2960. [Google Scholar] [CrossRef] [PubMed]
- Chushak, Y.; Stone, M.O. In silico selection of RNA aptamers. Nucl. Acids Res. 2009, 37, e87. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.-Y. Entropic Criterion for Model Selection. Physica A 2006, 370, 530–538. [Google Scholar] [CrossRef]
- Chen, C.-C.; Tseng, C.-Y.; Dong, J.-J. New entropy-based method for variables selection and its application to the debris-flow hazard assessment. Eng. Geo. 2007, 94, 19–24. [Google Scholar] [CrossRef]
- Montaville, P.; Neumann, J.M.; Russo_Marie, F.; Ochsenbein, F. A new consensus sequence for phosphatidylserine recognition by annexins. J. Biol. Chem. 2002, 277, 24684–24693. [Google Scholar] [CrossRef] [PubMed]
© 2010 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Tseng, C.-Y.; Tuszynski, J.A. Using Entropy Leads to a Better Understanding of Biological Systems. Entropy 2010, 12, 2450-2469. https://doi.org/10.3390/e12122450
Tseng C-Y, Tuszynski JA. Using Entropy Leads to a Better Understanding of Biological Systems. Entropy. 2010; 12(12):2450-2469. https://doi.org/10.3390/e12122450
Chicago/Turabian StyleTseng, Chih-Yuan, and Jack A. Tuszynski. 2010. "Using Entropy Leads to a Better Understanding of Biological Systems" Entropy 12, no. 12: 2450-2469. https://doi.org/10.3390/e12122450





