Effect of Conformational Entropy on the Nanomechanics of Microcantilever-Based Single-Stranded DNA Sensors
Abstract
:1. Introduction
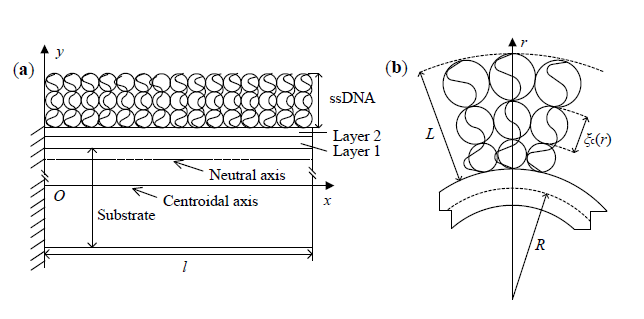
2. Theoretical Model
2.1. Conformational Free Energy
2.2. Mechanical Energy
3. Results and Discussion
3.1. Effect of Curvature
3.2. Effect of Chain Length and Packing Density of ssDNA
3.3. Effect of Thickness and Elastic Modulus of Substrate
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Huber, F.; Lang, H.P.; Backmann, N.; Rimoldi, D.; Gerber, C. Direct detection of a BRAF mutation in total RNA from melanoma cells using cantilever arrays. Nat. Nanotechnol 2013, 8, 125–129. [Google Scholar]
- Zhang, N.H.; Tan, Z.Q.; Li, J.J.; Meng, W.L.; Xu, L.W. Interactions of single-stranded DNA on microcantilevers. Curr. Opin. Colloid Interface Sci 2011, 16, 592–596. [Google Scholar]
- Arlett, J.L.; Myers, E.B.; Roukes, M.L. Comparative advantages of mechanical biosensors. Nat. Nanotechnol 2011, 6, 203–215. [Google Scholar]
- Fritz, J.; Baller, M.K.; Lang, H.P.; Rothuizen, H.; Vettiger, P.; Meyer, E.; Guntherodt, H.J.; Gerber, C.; Gimzewski, J.K. Translating biomolecular recognition into nanomechanics. Science 2000, 288, 316–318. [Google Scholar]
- Braun, T.; Ghatkesar, M.K.; Backmann, N.; Grange, W.; Boulanger, P.; Letellier, L.; Lang, H.P.; Bietsch, A.; Gerber, C.; Hegner, M. Quantitative time-resolved measurement of membrane protein-ligand interactions using microcantilever array sensors. Nat. Nanotechnol 2009, 4, 179–185. [Google Scholar]
- Wu, G.H.; Ji, H.F.; Hansen, K.; Thundat, T.; Datar, R.; Cote, R.; Hagan, M.F.; Chakraborty, A.K.; Majumdar, A. Origin of nanomechanical cantilever motion generated from biomolecular interactions. Proc. Natl. Acad. Sci. USA 2001, 98, 1560–1564. [Google Scholar]
- McKendry, R.; Zhang, J.Y.; Arntz, Y.; Strunz, T.; Hegner, M.; Lang, H.P.; Baller, M.K.; Certa, U.; Meyer, E.; Guntherodt, H.J.; Gerber, C. Multiple label-free biodetection and quantitative DNA-binding assays on a nanomechanical cantilever array. Proc. Natl. Acad. Sci. USA 2002, 99, 9783–9788. [Google Scholar]
- Mertens, J.; Rogero, C.; Calleja, M.; Ramos, D.; Martin-Gago, J.A.; Briones, C.; Tamayo, J. Label-free detection of DNA hybridization based on hydration-induced tension in nucleic acid films. Nat. Nanotechnol 2008, 3, 301–307. [Google Scholar]
- Sushko, M.L.; Harding, J.H.; Shluger, A.L.; McKendry, R.A.; Watari, M. Physics of nanomechanical biosensing on cantilever arrays. Adv. Mater 2008, 20, 3848–3853. [Google Scholar]
- Hagan, M.F.; Majumdar, A.; Chakraborty, A.K. Nanomechanical forces generated by surface grafted DNA. J. Phys. Chem. B 2002, 106, 10163–10173. [Google Scholar]
- Zhang, N.H.; Shan, J.Y. An energy model for nanomechanical deflection of cantilever-DNA chip. J. Mech. Phys. Solids 2008, 56, 2328–2337. [Google Scholar]
- Liu, F.; Zhang, Y.; Ou-Yang, Z.C. Flexoelectric origin of nanomechanic deflection in DNA-microcantilever system. Biosens. Bioelectron 2003, 18, 655–660. [Google Scholar]
- Begley, M.R.; Utz, M.; Komaragiri, U. Chemo-mechanical interactions between adsorbed molecules and thin elastic films. J. Mech. Phys. Solids 2005, 53, 2119–2140. [Google Scholar]
- Sushko, M.L. Nanomechanics of organic/inorganic interfaces: A theoretical insight. Faraday Discuss 2009, 143, 63–80. [Google Scholar]
- Zhang, N.H.; Shan, J.Y.; Xing, J.J. Piezoelectric properties of single-strand DNA molecular brush biolayers. Acta Mech. Solida Sin 2007, 20, 206–210. [Google Scholar]
- Utz, M.; Begley, M.R. Scaling theory of adsorption-induced stresses in polymer brushes grafted onto compliant structures. J. Mech. Phys. Solids 2008, 56, 801–814. [Google Scholar]
- Begley, M.R.; Utz, M. Multiscale modeling of adsorbed molecules on freestanding microfabricated structures. J. Appl. Mech 2008, 75. [Google Scholar] [CrossRef]
- Hariharan, R.; Biver, C.; Mays, J.; Russel, W.B. Ionic strength and curvature effects in flat and highly curved polyelectrolyte brushes. Macromolecules 1998, 31, 7506–7513. [Google Scholar]
- Rubinstein, M.; Colby, R.H. Polymer Physics; Oxford University Press: Oxford, UK, 2003. [Google Scholar]
- Zhang, N.H.; Xing, J.J. An alternative model for elastic bending deformation of multilayered beams. J. Appl. Phys 2006, 100. [Google Scholar] [CrossRef]
- Zhang, N.H. Thermoelastic stresses in multilayered beams. Thin Solid Films 2007, 515, 8402–8406. [Google Scholar]
- Zhang, N.H.; Chen, J.Z. An alternative two-variable model for bending problems of multilayered beams. J. Appl. Mech 2008, 75. [Google Scholar] [CrossRef]
- Zhang, N.H.; Chen, J.Z. Elastic bending analysis of bilayered beams by an alternative two-variable method. Eur. J. Mech. A 2009, 28, 284–288. [Google Scholar]
- Zhang, N.H.; Chen, J.Z. An alternative model for elastic thermal stresses in two materials joined by a graded layer. Compos. Part B 2010, 41, 375–379. [Google Scholar]
- Zhang, N.H.; Meng, W.L.; Aifantis, E.C. Elastic bending analysis of bilayered beams containing a gradient layer by an alternative two-variable method. Compos. Struct 2011, 93, 3130–3139. [Google Scholar]
- Herrera-May, A.L.; Aguilera-Cortes, L.A.; Plascencia-Mora, H.; Rodriguez-Morales, A.L.; Lu, J. Analytical modeling for the bending resonant frequency of multilayered microresonators with variable cross-section. Sensors 2011, 11, 8203–8226. [Google Scholar]
- Steel, A.B.; Levicky, R.L.; Herne, T.M.; Tarlov, M.J. Immobilization of nucleic acids at solid surfaces: Effect of oligonucleotide length on layer assembly. Biophys. J 2000, 79, 975–981. [Google Scholar]
- Alvarez, M.; Carrascosa, L.G.; Moreno, M.; Calle, A.; Zaballos, A.; Lechuga, L.M.; Martinez, A.C.; Tamayo, J. Nanomechanics of the formation of DNA self-assembled monolayers and hybridization on microcantilevers. Langmuir 2004, 20, 9663–9668. [Google Scholar]
- Stachowiak, J.C.; Yue, M.; Castelino, K.; Chakraborty, A.; Majumdar, A. Chemomechanics of surface stresses induced by DNA hybridization. Langmuir 2006, 22, 263–268. [Google Scholar]
- Yue, M.; Lin, H.; Dedrick, D.E.; Satyanarayana, S.; Majumdar, A.; Bedekar, A.S.; Jenkins, J.W.; Sundaram, S. A 2-D microcantilever array for multiplexed biomolecular analysis. J. Microelectromech. Syst 2004, 13, 290–299. [Google Scholar]
- Shi, M.X.; Liu, B.; Zhang, Z.Q.; Zhang, Y.W.; Gao, H.J. Direct influence of residual stress on the bending stiffness of cantilever beams. Proc. R. Soc. A 2012, 468, 2595–2613. [Google Scholar]
- Calleja, M.; Tamayo, J.; Nordstrom, M.; Boisen, A. Low-noise polymeric nanomechanical biosensors. Appl. Phys. Lett 2006, 88. [Google Scholar] [CrossRef]
- Zhang, N.H.; Meng, W.L.; Tan, Z.Q. A multi-scale model for the analysis of the inhomogeneity of elastic properties of DNA biofilm on microcantilevers. Biomaterials 2013, 34, 1833–1842. [Google Scholar]
- Zhang, N.H.; Chen, J.Z.; Li, J.J.; Tan, Z.Q. Mechanical properties of DNA biofilms adsorbed on microcantilevers in label-free biodetections. Biomaterials 2010, 31, 6659–6666. [Google Scholar]
- Tang, H.S.; Meng, W.L.; Zhang, N.H. Mechanical properties of double-stranded DNA biofilm with Gaussian distribution. Acta Mech. Sin 2014, 30, 15–19. [Google Scholar]
- Meng, W.L.; Tan, Z.Q.; Tang, H.S.; Zhang, N.H. Elastic properties of single-stranded DNA biofilm with strong interactions. Proceedings of Sixth International Conference on Nonlinear Mechanics (ICNM-6), Shanghai, China, 12–15 August 2013; Zhou, Z.W., Ed.; DES Tech Publications: Lancaster, PA, USA, 2013; pp. 154–157. [Google Scholar]






© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Tan, Z.-Q.; Zhang, N.-H. Effect of Conformational Entropy on the Nanomechanics of Microcantilever-Based Single-Stranded DNA Sensors. Entropy 2014, 16, 4923-4936. https://doi.org/10.3390/e16094923
Tan Z-Q, Zhang N-H. Effect of Conformational Entropy on the Nanomechanics of Microcantilever-Based Single-Stranded DNA Sensors. Entropy. 2014; 16(9):4923-4936. https://doi.org/10.3390/e16094923
Chicago/Turabian StyleTan, Zou-Qing, and Neng-Hui Zhang. 2014. "Effect of Conformational Entropy on the Nanomechanics of Microcantilever-Based Single-Stranded DNA Sensors" Entropy 16, no. 9: 4923-4936. https://doi.org/10.3390/e16094923