Single-Cell Reprogramming in Mouse Embryo Development through a Critical Transition State
Abstract
:1. Introduction
- (i)
- SOC control of overall expression represents the self-organization of coexisting critical states (distinct response expression domains) through a critical transition. Temporal variance expression (normalized root mean square fluctuation: nrmsf; see Methods) acts as an order parameter in self-organization. Distinct critical states can be observed in a transitional behavior of the ensemble expression profile (e.g., bimodality coefficient) or bifurcation of the ensemble expression state (e.g., probability density profile) according to nrmsf. Coherent behaviors emerge from stochastic expression within critical states (coherent-stochastic behaviors; see non-equilibrium statistical mechanism underpinning the spontaneous emergence of order out of disorder [7]) as the collective behaviors of groups with more than around 50 genes (mean-field approach) [6,8,9].
- (ii)
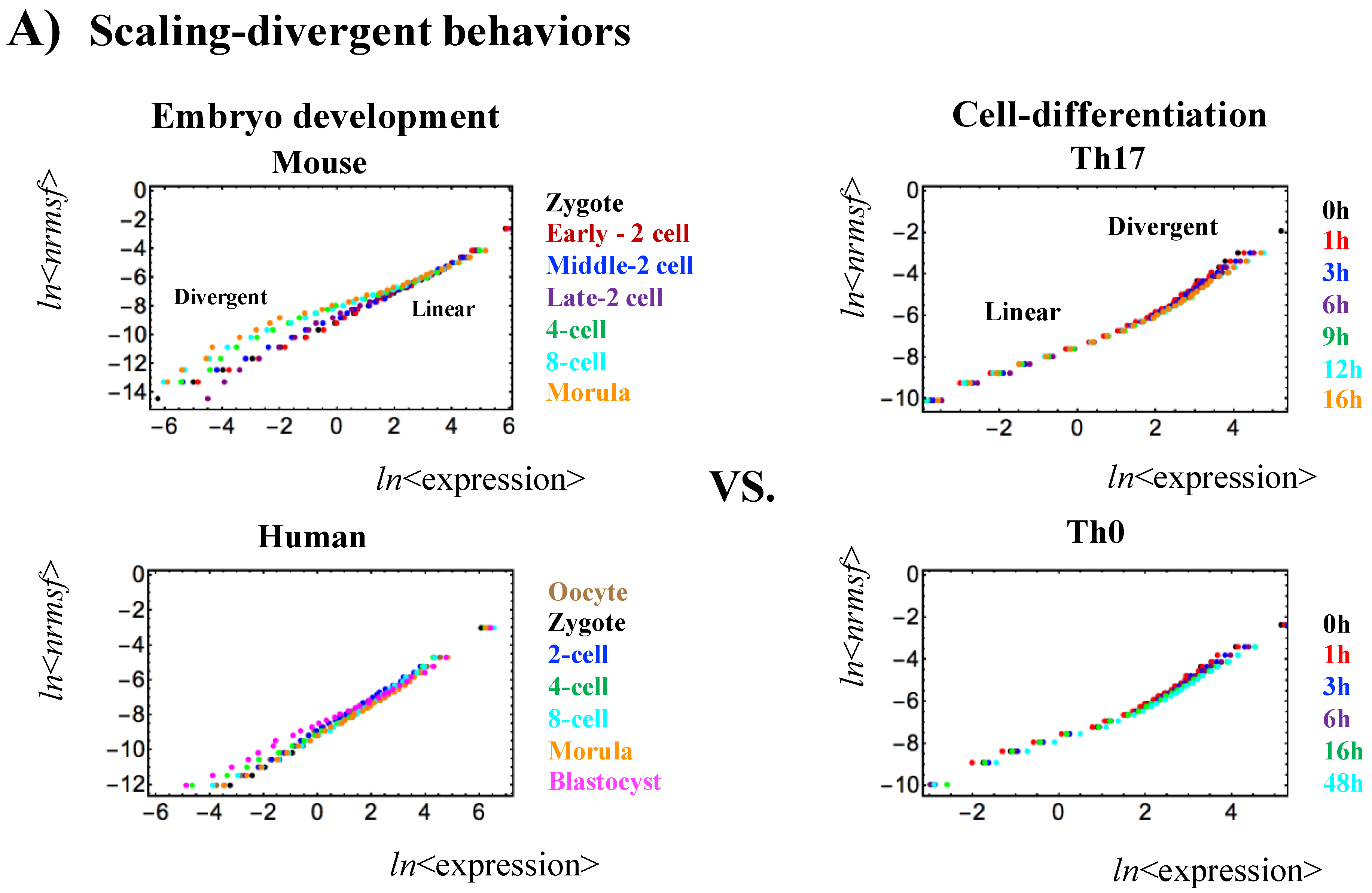
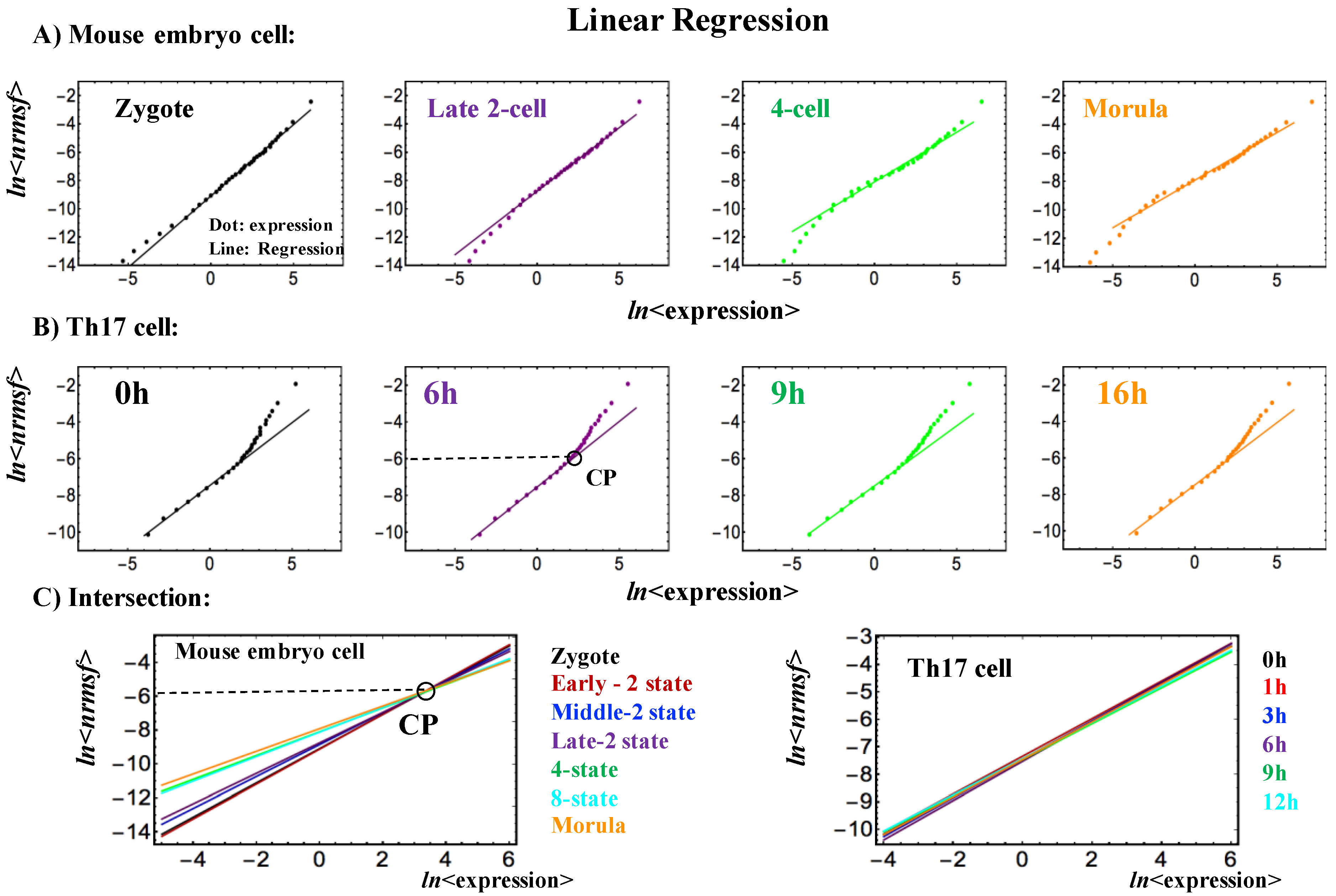
- Self-organization based on SOC occurs through distinctive critical behaviors: sandpile-type critical behavior (criticality) and scaling-divergent behavior. In sandpile-type criticality, the divergence of two different regulatory behaviors occurs at a critical point (summit of the sandpile) that neatly separates up- and down-regulation. Sandpile-type criticality emerges spontaneously by the grouping of different gene expressions according to their fold-change. Reprogramming or cell-fate change is observed as erasure of criticality of an initial-state (such as an early embryo state). Scaling-divergent behavior emerges upon averaging the behavior of gene-group expression according to nrmsf, producing both linear (scaling) and divergent domains in a log-log plot. This divergent behavior anticipates a transition [10]. Since nrmsf is an order parameter, the above separation of scaling domains (Figure 1) shows how self-organization occurs in overall expression and that it is not confined to a few “master genes”.
2. Results
2.1. Dynamic Interaction of Critical States
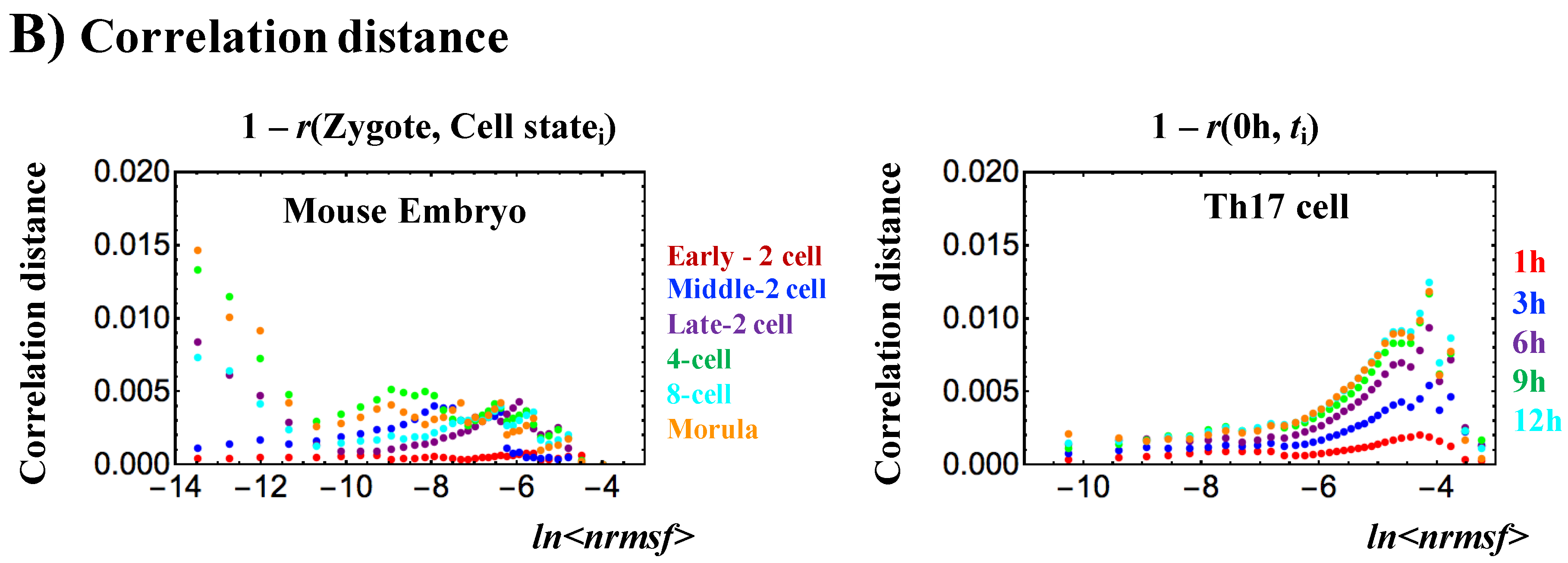
2.1.1. Sloppiness of Mouse RNA Expression Dynamics: Coherent-Stochastic Behaviors in Critical States
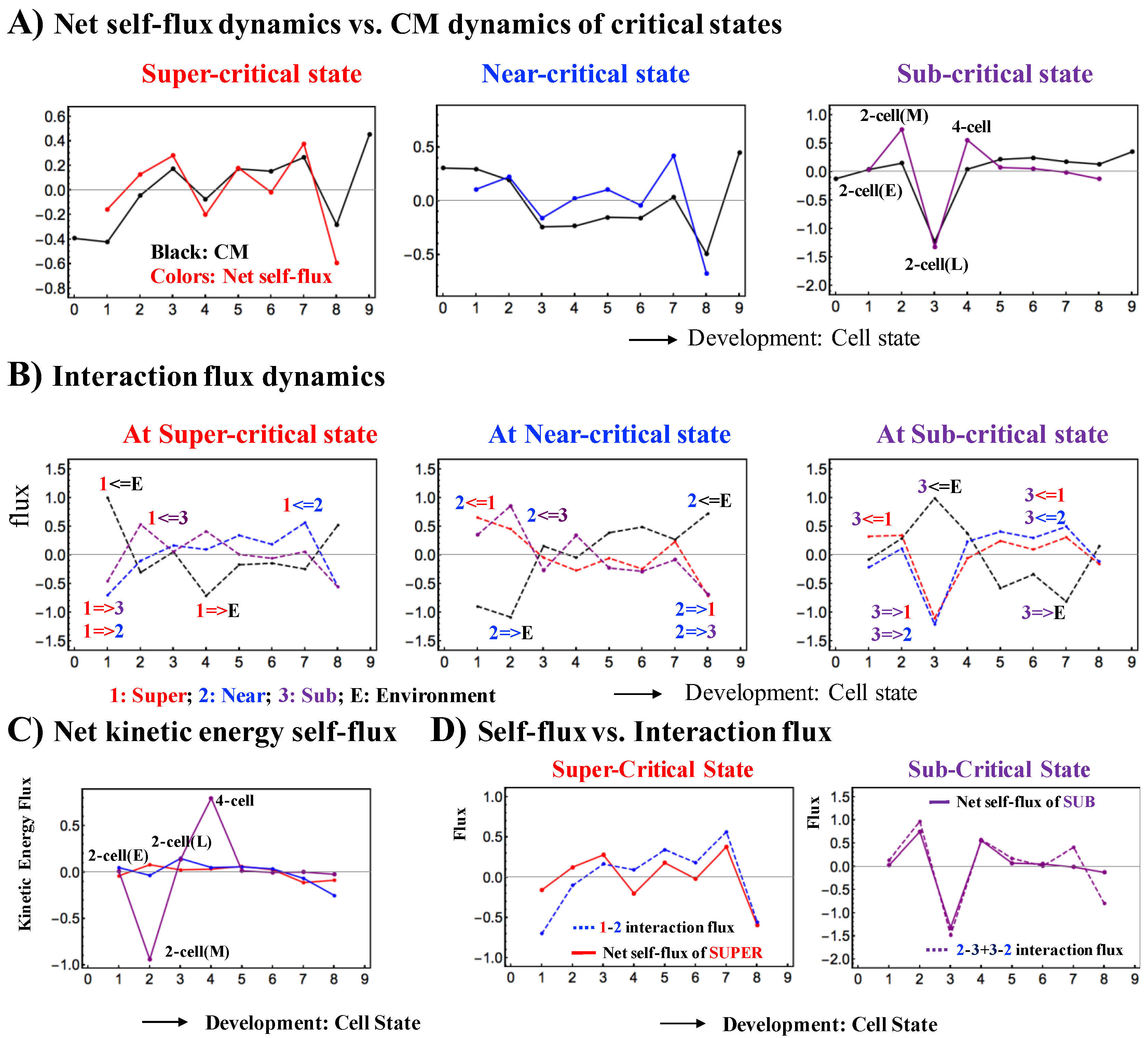
2.1.2. Expression Flux Dynamics Representing a Change in Genetic Activity
2.1.3. Sub-Critical State as a Generator of Perturbation in Genome-Wide Self-Organization
2.1.4. Change in Criticality: Global Impact on the Whole Genome-Expression System
2.2. SOC Control Mechanism of Genome Reprogramming through a Critical Transition State
- (i)
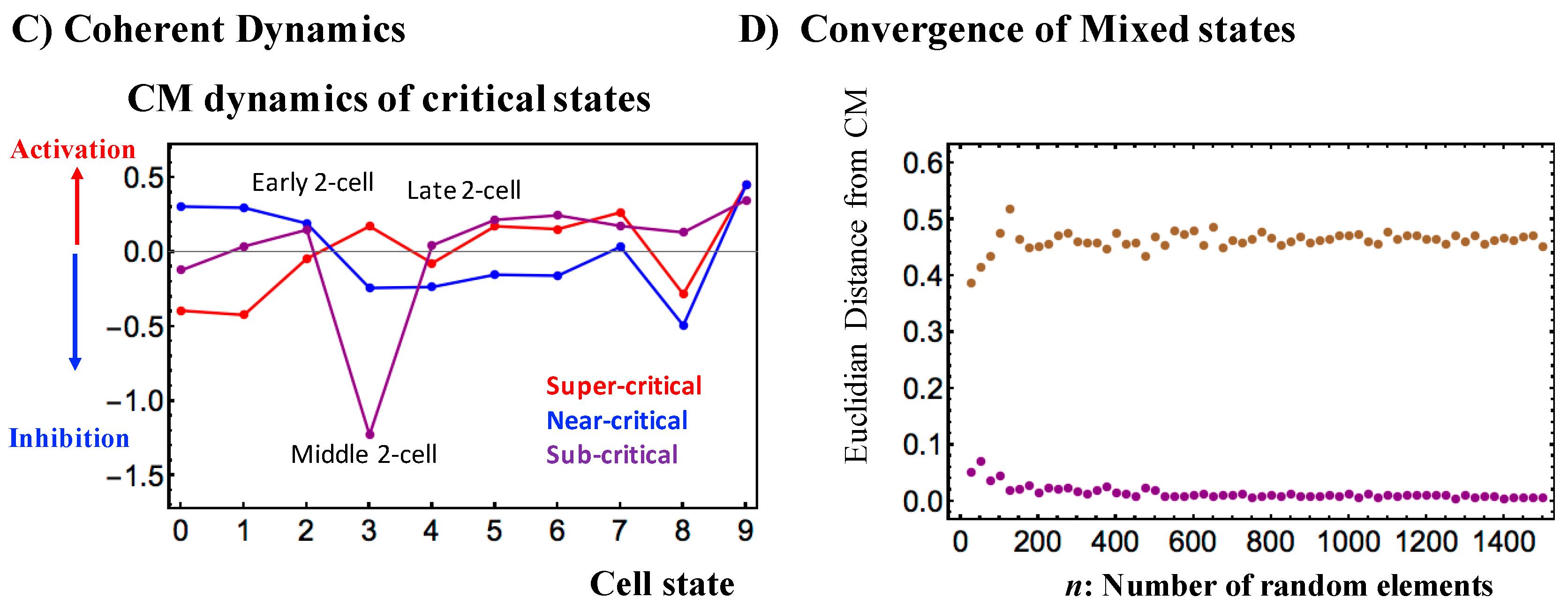
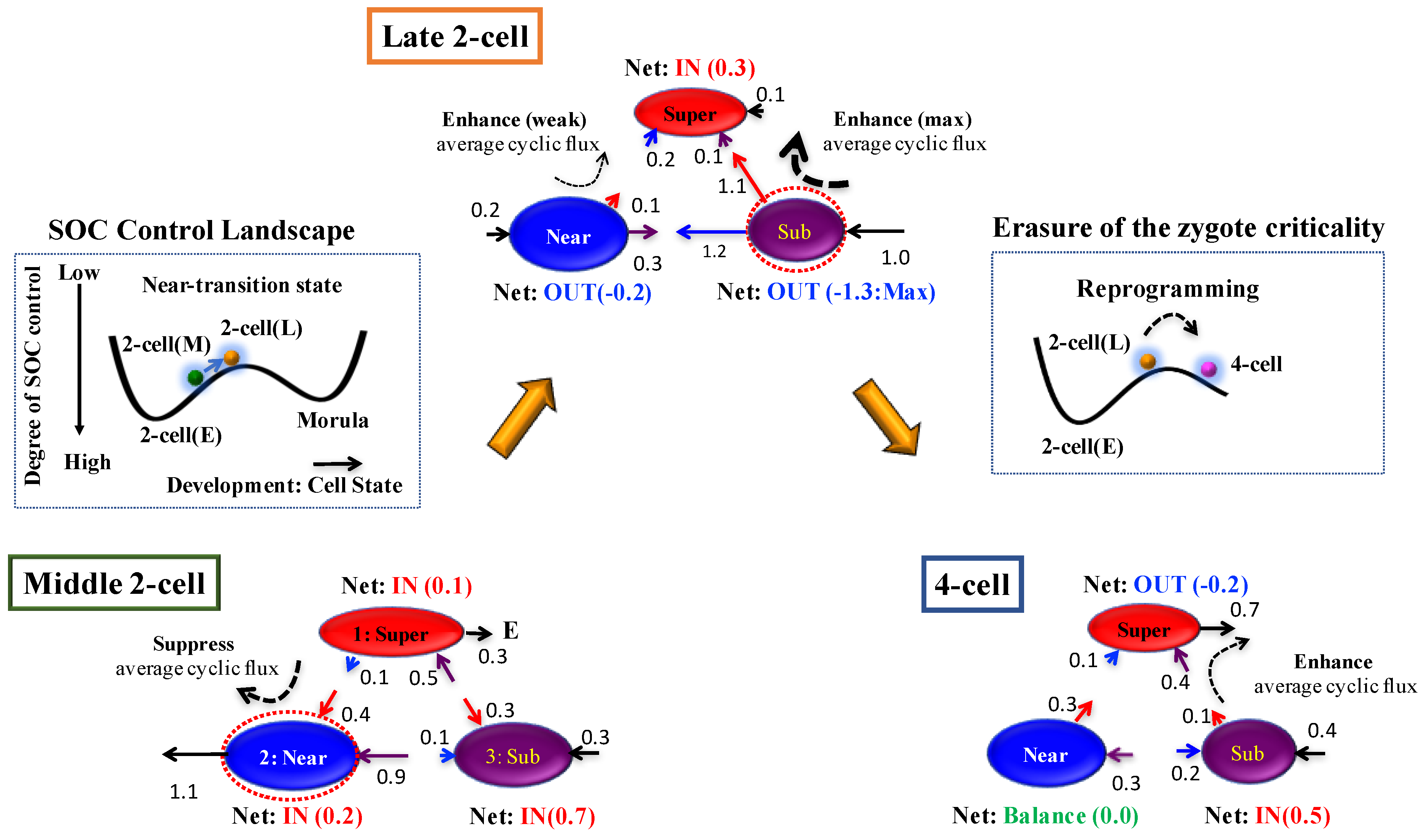
- Before reprogramming: In the middle 2-cell state, interaction flux (Super—Near) suppresses cyclic flux, where the near-critical state acts as a major suppressor. All critical states receive the net IN flux, which indicates that inside the nucleus, the mouse genome system is activated through the cell milieu (environment).
- (ii)
- Right before reprogramming: A substantial change in the net interaction flux at critical states occurs beginning in the middle 2-cell state and into the late 2-cell state. These changes induce a major change in the two cyclic fluxes: a change from suppression to the enhancement of cyclic flux (Super—Near), and strongest enhancement in the dominant cyclic flux (Super—Sub). This leads to the reverse change in the net interaction fluxes at the near-critical state (IN to OUT and vice versa) to enhance the cyclic flux (Super—Near) from the major suppressor at the early 2-cell state. Thus, these reverse cyclic enhancements make the genome system pass through the critical transition state. This interaction model reveals that
- (a)
- A major biological event in genome reprogramming through a stochastic pattern is guided by the sub-critical state at the late 2-cell state, and
- (b)
- There is a detailed open thermodynamic mechanism regarding the erasure of criticality of the zygote state in the late 2-cell state.
- (c)
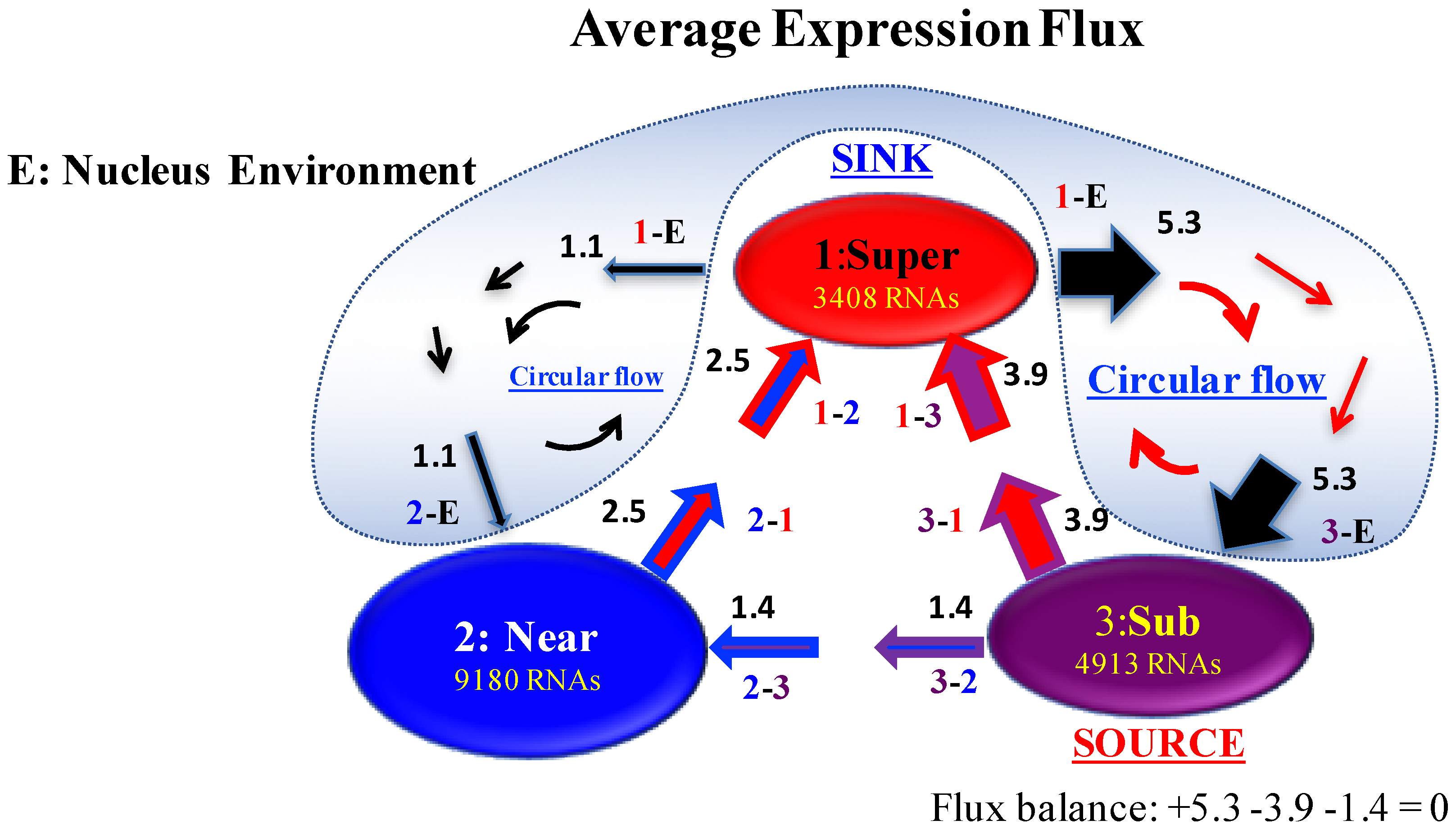
- The asymmetry becomes most significant for the IN-and-OUT fluxes between the super- and sub-critical states at the late 2-cell state, i.e., the greatest time-reversal symmetry breaking is caused.
In this respect, it is worth noting that, with no time-reversal symmetry breaking, interaction fluxes between the super- and sub-critical states should be balanced (equal), corresponding to the fundamental framework of a detailed balance in equilibrium statistical physics, while detailed balance should be violated in a thermodynamically open system [18]. - (iii)
- After reprogramming: In the 4-cell state, while the enhancement of cyclic flux (Super—Sub) becomes weak, the perturbation of average flux activity almost disappears due to passage through the transition state.
2.3. Discussion and Conclusions
3. Methods
3.1. Biological Data Sets
3.2. Normalized Root Mean Square Fluctuation (nrmsf)
3.3. Bimodality Coefficient
3.4. SOC Control Mechanism of Overall Expression
- (i)
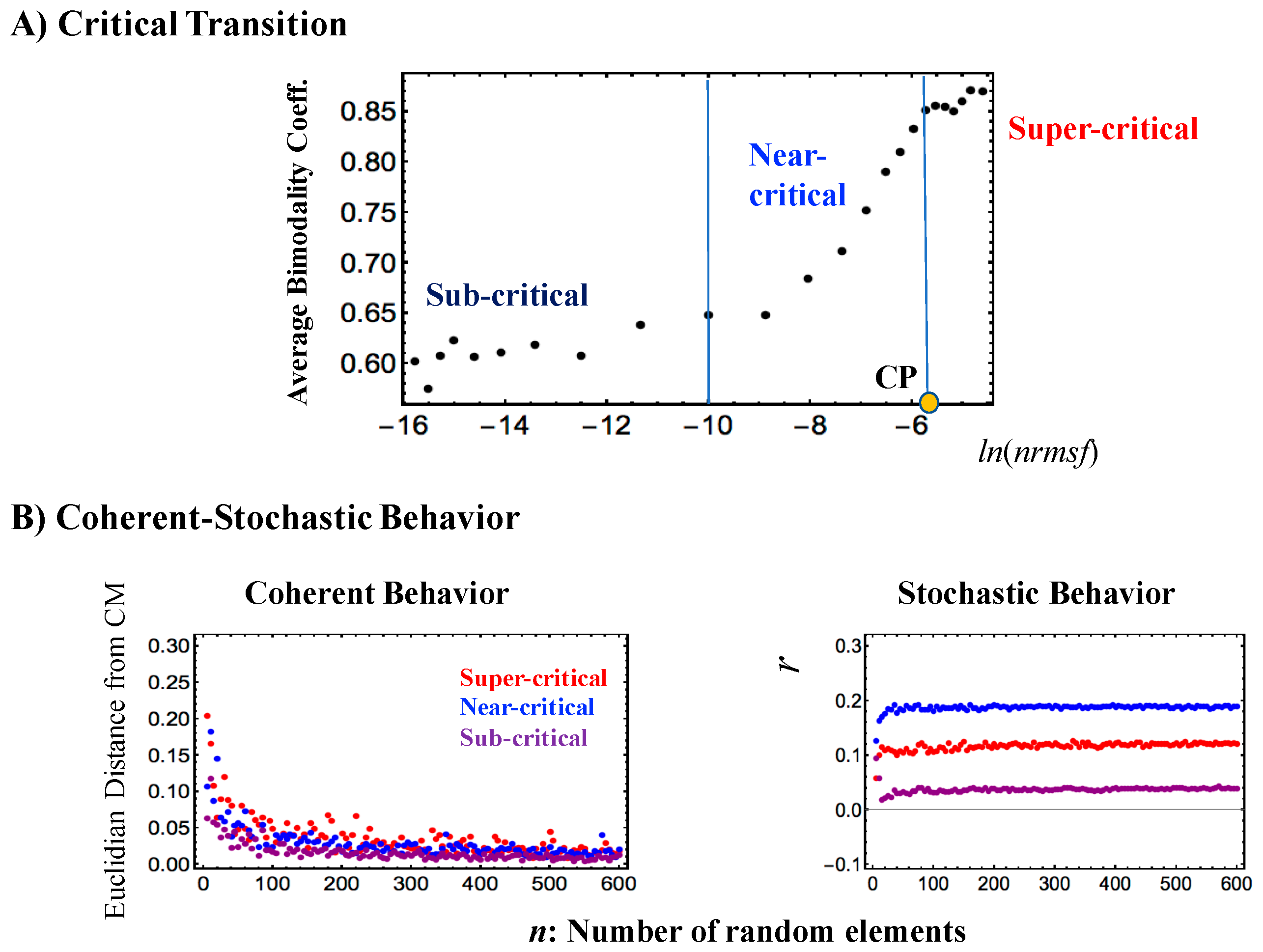
- SOC of overall expression does not correspond to a phase transition from one critical state to another. Instead, it represents the self-organization of the coexisting critical states through a critical transition, i.e., SOC consolidates critical states into a genome expression system (called “SOC control of overall expression”) according to temporal expression variance (nrmsf). Nrmsf acts as an order parameter in self-organization. In critical states, distinct coherent (collective) behaviors emerge in ensembles of stochastic expression (coherent-stochastic behavior), where the coherent dynamics of high-variance gene expression (super-critical state) is anti-phase to that of low-variance gene expression (sub-critical state).
- (ii)
- The characteristics of the self-organization through SOC become apparent only in the collective behaviors of groups with an average of more than around 50 genes (mean-field approach). The same value of 50 genes (or around this value) as the threshold for the onset of coherent ensemble behavior was previously recognized in a completely different context and by different analytical techniques [6,8,9]. This effect is clearly a statistical one, but not a “simple statistical one” in the sense that it is a proxy of the underlying gene regulation network (GRN).
- (iii)
- Self-organization occurs through distinctive critical behaviors: sandpile-type criticality and scaling-divergent behavior:
- (a)
- Sandpile-type critical behavior (criticality) based on the grouping of expression according to the fold-change in expression. The summit of the sandpile represents the critical point (CP); as the distance from the CP increases, two different regulatory behaviors diverge, which represent up-regulation and down-regulation, respectively. Furthermore, in the vicinity of the CP according to nrmsf, in terms of coherent expression, self-similar bifurcation of overall expression occurs to show a unimodal-bimodal transition (Figures 1 and 3A in [6]: MCF-7 cancer cells) and a step function like transition (Figure 3A in [4]: HL-60 cancer cells); these results indicate the existence of cell-type-specific transitions. Thus, since a critical behavior and a critical transition occur at the CP, we can characterize it as a sandpile-type transition.
- (b)
- Scaling-divergent behavior (genomic avalanche) based on the grouping of expression according to nrmsf: a nonlinear correlation trend between the ensemble averages of nrmsf and gene expression at each time point, which has both linear (scaling) and divergent domains in a log-log plot; the onset of divergence occurs at the CP (in cell-differentiation): order (scaling) and disorder (divergence) are balanced at the CP, which presents a genomic avalanche. The scaling-divergent behavior reflects the co-existence of distinct response domains (critical states) in overall expression. Distinct critical behaviors from different averaging behaviors occur (numerically) at around the same nrmsf.
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Reik, W.; Dean, W.; Walter, J. Epigenetic reprogramming in mammalian development. Science 2001, 10, 1089–1093. [Google Scholar] [CrossRef] [PubMed]
- Santos, F.; Dean, W. Epigenetic reprogramming during early development in mammals. Reproduction 2004, 127, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Cantone, I.; Fisher, A.G. Epigenetic programming and reprogramming during development. Nat. Struct. Mol. Biol. 2013, 20, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, M.; Giuliani, A.; Hashimoto, M.; Erenpreisa, J.; Yoshikawa, K. Self-organizing global gene expression regulated through criticality: Mechanism of the cell-fate change. PLoS ONE 2016, 11, e0167912. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, M.; Hashimoto, M.; Takenaka, Y.; Motoike, I.N.; Yoshikawa, K. Global genetic response in a cancer cell: Self-organized coherent expression dynamics. PLoS ONE 2014, 9, e97411. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, M.; Giuliani, A.; Hashimoto, M.; Erenpreisa, J.; Yoshikawa, K. Emergent Self-Organized Criticality in gene expression dynamics: Temporal development of global phase transition revealed in a cancer cell line. PLoS ONE 2015, 11, e0128565. [Google Scholar] [CrossRef] [PubMed]
- Hollerbach, R.; Kim, E. Information Geometry of Non-Equilibrium Processes in a Bistable System with a Cubic Damping. Entropy 2017, 19, 268. [Google Scholar] [CrossRef]
- Censi, F.; Giuliani, A.; Bartolini, P.; Calcagnini, G. A multiscale graph theoretical approach to gene regulation networks: A case study in atrial fibrillation. IEEE Trans. Biomed. Eng. 2011, 58, 2943–2946. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, M.; Selvarajoo, K.; Piras, V.; Tomita, M.; Giuliani, A. Local and global responses in complex gene regulation networks. Physica A 2009, 388, 1738–1746. [Google Scholar] [CrossRef]
- Scheffer, M.; Carpenter, S.R.; Lenton, T.M.; Bascompte, J.; Brock, W.; Dakos, V.; van de Koppe, J.; van de Leemput, I.A.; Levin, S.A.; van Nes, E.H.; et al. Anticipating critical transitions. Science 2012, 338, 344–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, P.W. More is different. Science 1972, 177, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Laughlin, R.B.; Pines, D.; Schmalian, J.; Stojković, B.P.; Wolynes, P. The middle way. Proc. Natl. Acad. Sci. USA 2000, 97, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, A.; Tsuchiya, M.; Yoshikawa, K. Single-cell genome dynamics in early embryo development: A statistical thermodynamics approach. bioRxiv 2017. Available online: https://www.biorxiv.org/content/early/2017/04/13/123554 (accessed on 02 November 2017). [CrossRef]
- Mirny, L.A. The fractal globule as a model of chromatin architecture in the cell. Chromosome Res. 2011, 19, 37–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.; Ramsköld, D.; Reinius, B.; Sandberg, R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 2014, 343, 193–196. [Google Scholar] [CrossRef] [PubMed]
- Marković, D.; Gros, C. Power laws and self-organized criticality in theory and nature. Phys. Rep. 2014, 536, 41–74. [Google Scholar] [CrossRef]
- Tomita, K.; Tomita, H. Irreversible Circulation of Fluctuation. Prog. Theor. Phys. 1974, 51, 1731–1749. [Google Scholar] [CrossRef]
- Tsuchiya, M.; Wong, S.T.; Yeo, Z.X.; Colosimo, A.; Palumbo, M.C.; Farina, L.; Crescenzi, M.; Mazzola, A.; Negri, R.; Bianchi, M.M.; et al. Gene expression waves: Cell cycle independent collective dynamics in cultured cells. FEBS J. 2007, 274, 2874–2886. [Google Scholar] [CrossRef] [PubMed]
- Einstein, A.; Schilpp, P.A. Autobiographical Notes; Open Court Publishing Company: Chicago, IL, USA, 1949. [Google Scholar]
- Nagashima, T.; Shimodaira, H.; Ide, K.; Nakakuki, T.; Tani, Y.; Takahashi, K.; Yumoto, N.; Hatakeyama, M. Quantitative transcriptional control of ErbB receptor signaling undergoes graded to biphasic response for cell differentiation. J. Biol. Chem. 2007, 282, 4045–4056. [Google Scholar] [CrossRef] [PubMed]
- Mojtahedi, M.; Skupin, A.; Zhou, J.; Castaño, I.G.; Leong-Quong, R.Y.Y.; Chang, H.; Trachana, K.; Giuliani, A.; Huang, S. Cell Fate Decision as High-Dimensional Critical State Transition. PLoS Biol. 2016, 14, e2000640. [Google Scholar] [CrossRef] [PubMed]
- Moris, N.; Pina, C.; Arias, A.M. Transition states and cell fate decisions in epigenetic landscapes. Nat. Rev. Genet. 2016, 17, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Yang, M.; Guo, H.; Yang, L.; Wu, J.; Li, R.; Liu, P.; Lian, Y.; Zheng, X.; Yan, J.; et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013, 20, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Ciofani, M.; Madar, A.; Galan, C.; Sellars, M.; Mace, K.; Pauli, F.; Agarwal, A.; Huang, W.; Parkhurst, C.N.; Muratet, M.; et al. A validated regulatory network for Th17 cell specification. Cell 2012, 151, 289–303. [Google Scholar] [CrossRef] [PubMed]
- Knapp, T.R. Bimodality Revisited. J. Mod. Appl. Stat. Methods 2007, 6. [Google Scholar] [CrossRef]
- Kim, K.Y.; Wang, J. Potential energy landscape and robustness of a gene regulatory network: Toggle switch. PLoS Comp. Biol. 2007, 3, e60. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsuchiya, M.; Giuliani, A.; Yoshikawa, K. Single-Cell Reprogramming in Mouse Embryo Development through a Critical Transition State. Entropy 2017, 19, 584. https://doi.org/10.3390/e19110584
Tsuchiya M, Giuliani A, Yoshikawa K. Single-Cell Reprogramming in Mouse Embryo Development through a Critical Transition State. Entropy. 2017; 19(11):584. https://doi.org/10.3390/e19110584
Chicago/Turabian StyleTsuchiya, Masa, Alessandro Giuliani, and Kenichi Yoshikawa. 2017. "Single-Cell Reprogramming in Mouse Embryo Development through a Critical Transition State" Entropy 19, no. 11: 584. https://doi.org/10.3390/e19110584





