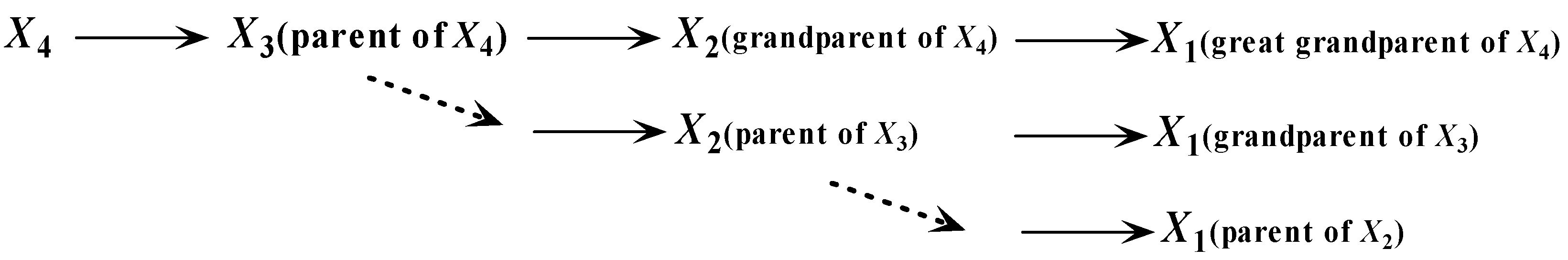1. Introduction
Classification is a basic task in data analysis and pattern recognition that requires the learning of a classifier, which assigns labels or categories to instances described by a set of predictive variables or attributes. The induction of classifiers from datasets of preclassified instances is a central problem in machine learning. Given class label
C and predictive attributes
( capital letters, such as
and
Z, denote attribute names, and lowercase letters, such as
x,
y and
z, denote the specific values taken by those attributes. Sets of attributes are denoted by boldface capital letters, such as
,
and
, and assignments of values to the attributes in these sets are denoted by boldface lowercase letters, such as
and
), discriminative learning [
1,
2,
3,
4] directly models the conditional probability
. Unfortunately,
cannot be decomposed into a separate term for each attribute, and there is no known closed-form solution for the optimal parameter estimates. Generative learning [
5,
6,
7,
8] approximates the joint probability
with different factorizations according to Bayesian network classifiers, which are powerful tools for knowledge representation and inference under conditions of uncertainty. Naive Bayes (NB) [
9], which is the simplest kind of Bayesian network classifier that assumes the attributes are independent given the class label, are surprisingly effective. After the discovery of NB, many state-of-the-art algorithms, for example, tree-augmented naive Bayes (TAN) [
10] and a
k-dependence Bayesian classifier (KDB) [
11], are proposed to relax the independence assumption by allowing conditional dependence between attributes
and
, which is measured by conditional mutual information
. In order to improve predictive accuracy relative to a single model, ensemble methods [
12,
13], for example, averaged one-dependence estimator (AODE) [
14] and averaged tree-augmented naive Bayes (ATAN) [
15] methods, generate multiple global models from a single learning algorithm through randomization (or perturbation).
An ideal Bayesian network classifier should provide the maximum value of mutual information
for classification; that is,
should represent strong mutual dependence between
C and
. However,
The strong conditional dependence between attributes
and
may not help to improve classification performance. As shown in
Figure 1, the proportional distribution of
differs greatly to that of
.
The KDB is a form of a restricted Bayesian network classifier with numerous desirable properties in the context of learning from large quantities of data. It achieves a good trade-off between classification performance and structure complexity with a single parameter, k. KDB uses mutual information to predetermine the order of predictive attributes and conditional mutual information to measure the conditional dependence between predictive attributes.
In this paper, we extend the KDB. The contributions of this paper are as follows:
We propose a new sorting method to predetermine the order of predictive attributes. This sorting method considers not only the dependencies between predictive attributes and the class variable, but also the dependencies between predictive attributes.
We extend the KDB from one single k-dependence tree to a k-dependence forest (KDF). A KDF reflects more dependencies between predictive attributes than the KDB. We show that our algorithm achieves comparable or lower error on University of California at Irvine (UCI) datasets than a range of popular classification learning algorithms.
The rest of this paper is organized as follows.
Section 2 introduces some state-of-the-art Bayesian network classifiers.
Section 3 explains the basic idea of the KDF and introduces the learning procedure in detail.
Section 4 compares experimental results on datasets from the UCI Machine Learning Repository.
Section 5 draws conclusion.
3. The k-Dependence Forest Algorithm
The KDB is supplied with both a database of preclassified instances, a DB, and the k value for the maximum allowable degree of attribute dependence. The structure learning procedure of a KDB can be partitioned into two parts: attribute sorting and dependence analysis. During the sorting procedure, the KDB uses mutual information to predetermine the order of predictive attributes. The KDB ensures that the predictive attributes that are most dependent on the class variable should be considered first and added to the structure. However, mutual information can only measure the dependencies between predictive attributes and the class variable, while it ignores the dependencies between predictive attributes. The sorting process of the KDB only embodies the dependency between each single attribute and class variable, which may result in a suboptimal order. The proposed algorithm, the KDF, uses a new sorting method to address this issue.
According to the chain rule of information theory, mutual information
can be expanded as follows:
In the ideal case, in classification, we would like to obtain the maximum value of
. From Equation (5), we can find that the computational complexity of
grows exponentially as the number of attributes increases. The space to store the conditional probability distribution grows exponentially. How to approximate the probability estimation is challenging. In order to address this issue, we replace
with the following:
Equation (
6) considers both the mutual dependence and the conditional dependence for classification. On the basis of this, we propose a new approach to predetermine the sequence of predictive attributes by comparing the value of
. From Equation (
6), we can find that the first attribute of a sequence does not reflect the conditional dependence. Thus we use each attribute as the root node
in turn. The next attribute, which will be added to the sequence, is the attribute that is most informative about
C conditioned on the first attribute (which is measured by
). Subsequent attributes are chosen to be the most informative about
C conditioned on previously chosen attributes (which is measured by
). Because of the
n different root nodes, we can obtain
n sequences {S
, ⋯, S
}. On the basis of the
n sequences,
n subclassifiers can be generated. The sorting algorithm (Algorithm 1) is depicted below.
| Algorithm 1: KDF: Sorting |
Input: Preclassified dataset DB with n predictive attributes .
Output: Sequences {S, ⋯, S}.
For each sequence , :Let be empty. Let predictive attribute , be the root node. Add the root node to . Repeat until includes all domain attributes: - (a)
Compute for the predictive attribute (), which is not in . - (b)
Select , which has the maximum value of . - (c)
Add to .
|
In order to identify the graphical structure of the resulting classifier, the KDB adopts a greedy search strategy. The weight of conditional dependence between
and its parent
is measured by conditional mutual information
. However, the dependency relationships between
and other parents of
are neglected, whether they are independent or strongly correlated. From Equation (
1), we can see that, for the full Bayesian network classifier, the parent of
is
, the parent of
is
, the parent of
is
, and so forth. Then we can achieve an implicit chain rule, that
is the parent of
,
is the parent of
(or
is the grandparent of
),
is the parent of
(or
is the great grandparent of
), and so forth. Thus, as shown in
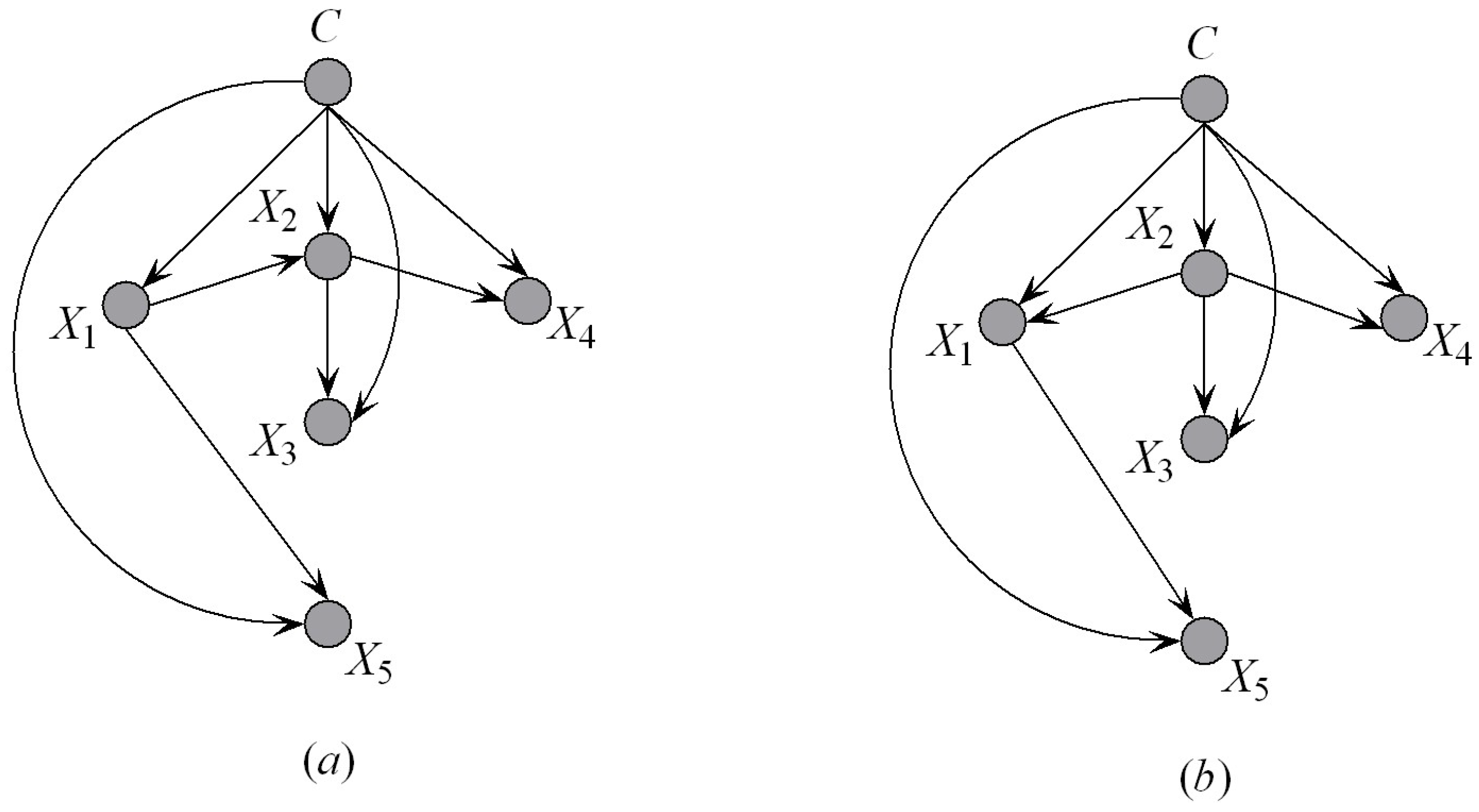
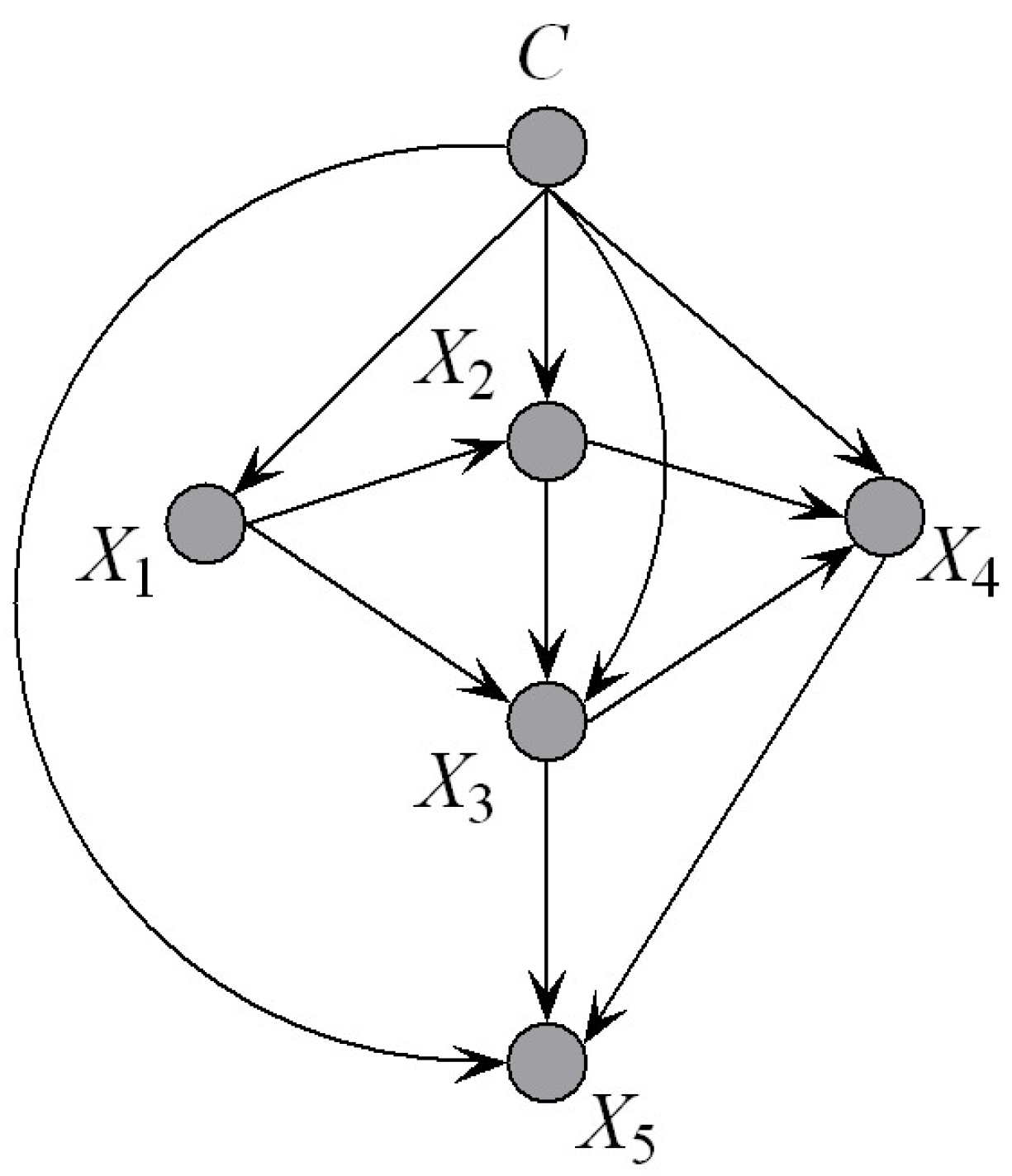
Figure 5, there should exist hierarchical dependency relationships among the parents. If
is one parent of attribute
, we should follow the dotted line shown in
Figure 5 to find the other parents. To make our idea clear, we first introduce the definition of an ancestor node.
Definition 1. Suppose that is the parent of . The ancestor attributes of include ’s parents, grandparents, great grandparents, and so forth.
During the procedure of dependence analysis,
first selects the attribute
that corresponds to the largest value of
as its parent. For the other
parents,
will select among its ancestor attributes.
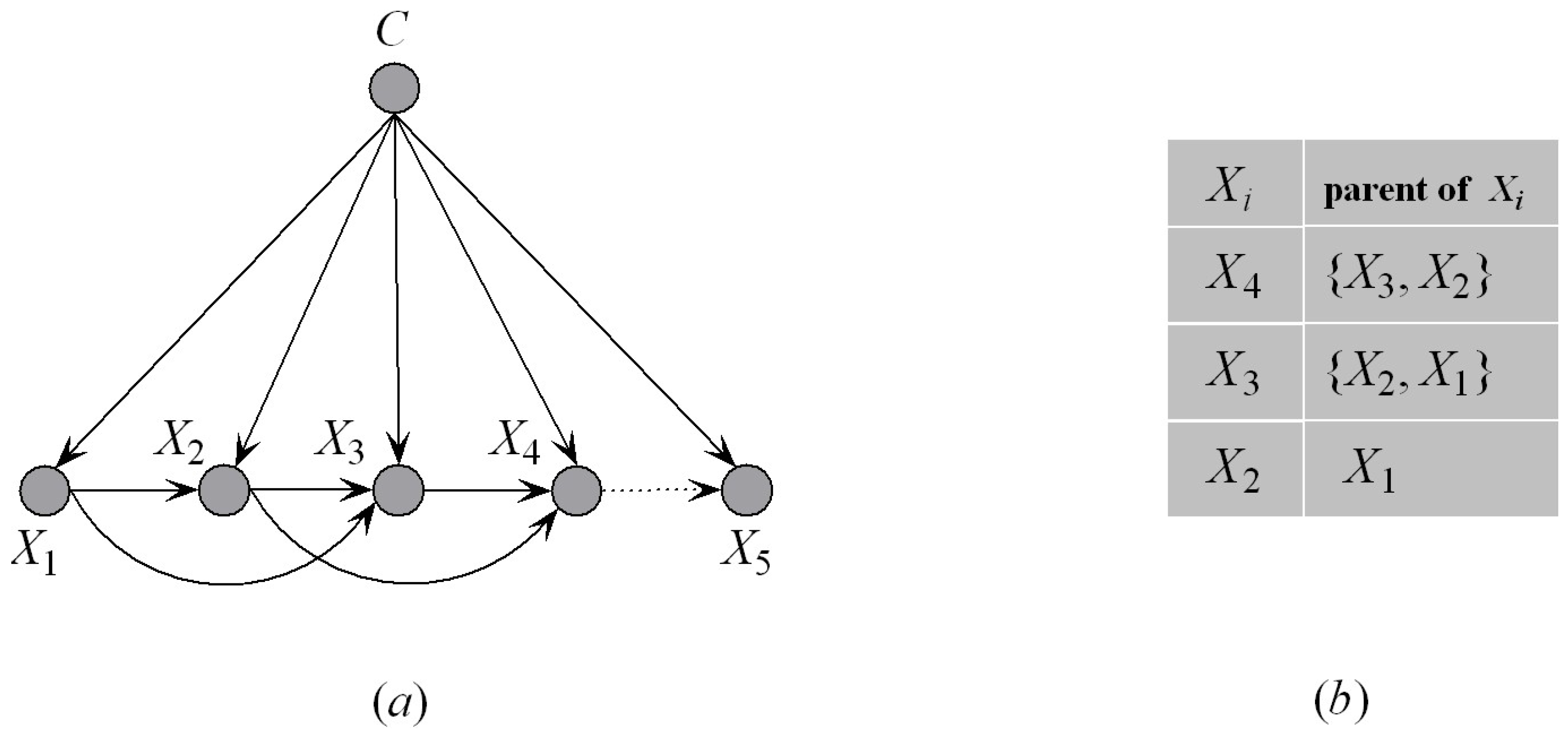
Figure 6a shows an example of the KDF subclassifier, for example, KDF
. We suppose that
. When
is added to KDF
,
will be selected as the first parent of
. The corresponding parent–child relationships are shown in
Figure 6b, from which we can see that the ancestor attributes of
are
, that is,
. Other parents of
will be selected from
by comparing
. This strategy helps to reduce the search space of attribute dependencies. The detailed procedure of dependence analysis (Algorithm 2) is depicted below.
| Algorithm 2: KDF: Dependence Analysis |
Input: Sequences {S, ⋯, S}.
Output: Subclassifiers {KDF, ⋯, KDF}.
Compute , for each pair of attributes and , where . For each sequence , : - (1)
Let the KDF being constructed begin with a single class node, C. - (2)
Repeat until KDF includes all attributes: - (a)
Select the attribute , which is the first attribute in and not in KDF. - (b)
Add a node to KDF representing . - (c)
Add an arc from C to in KDF. - (d)
Select , which is in KDF and has the largest value of , as the first parent of . - (e)
Select other parents from ancestor attributes of by comparing the value of , where is one of the ancestor attributes of , and d is the number of the ancestor attributes of .
Compute the conditional probability tables inferred by the structure of KDF by using counts from DB, and output KDF.
|
After training multiple learning subclassifiers, ensemble learning treats these as a “committee” of decision makers and combines individual predictions appropriately. The decision of the committee should have better overall accuracy, on average, than any individual committee member. There exist numerous methods for model combination, for example, the linear combiner, the product combiner and the voting combiner. For the subclassifier KDF
, an estimate of the probability of class
c given input
is
. The linear combiner is used for models that output real-valued numbers; thus it is applicable for the KDF. The ensemble probability estimate is
If the weights , , this is a simple uniform averaging of the probability estimates. The notation clearly allows for the possibility of a nonuniformly weighted average. If the classifiers have different accuracies on the data, a nonuniform combination could in theory give a lower error than a uniform combination. However, in practice, the difficulty is of estimating the parameters without overfitting and the relatively small gain that is available. Thus, in practice, we use the uniformly rather than nonuniformly weighted average.
The KDF collects the statistics to perform calculations of conditional mutual information of each pair of attributes given the class for structure learning. As an entry must be updated for every training instance and every combination of two attribute values for that instance, the time complexity of forming the three-dimensional probability table is , where m is the number of training instances, n is the number of attributes, c is the number of classes, and v is the maximum number of discrete values that any attribute may take. To calculate the conditional mutual information, the KDF must consider every pairwise combination of their respective values in conjunction with each class value . For each subclassifier KDF, attribute ordering and parent assignment are and , respectively. KDF requires n tables of dimensions, with . Because the KDF needs to average the results of n subclassifiers, the time complexity of classifying a single testing instance is time.
The parameter
k is closely related to the classification performance of a high-dependence classifier. A higher value of
k may result in higher variance and lower bias. Unfortunately there is no a priori means to preselect an appropriate value of
k that can help to achieve the lowest error for a given training set, as this is a complex interplay between the data quantity and the complexity and strength of the interactions between the attributes proved by Martinez et al. [
8]. From the discussion above, we can see that, for each KDF
, the space complexity of the probability table increases exponentially as
k increases; to achieve the trade-off between classification performance and efficiency, we restrict the structure complexity to be two-dependence, which is also adopted by Webb et al. [
26].
4. Experiments and Results
In order to verify the efficiency and effectiveness of the proposed KDF algorithm, experiments were conducted on 40 benchmark datasets from the UCI Machine Learning Repository [
27].
Table 1 summarizes the characteristics of each dataset, including the number of instances, attributes and classes. All the datasets were ordered by dataset scale. Missing values for qualitative attributes were replaced with modes, and those for quantitative attributes were replaced with means from the training data. For each original dataset, we discretized numeric attributes using minimum description length (MDL) discretization [
28]. All experiments were conducted on a desktop computer with an Intel(R) Core(TM) i3-6100 CPU @ 3.70 GHz, 64 bits and 4096 MB of memory. All the experiments for the Bayesian algorithms used C++ software specifically designed to deal with classification methods. The running efficiency of the KDF was good. For example, for a
Poker hand dataset, it took 281 s for the KDF to obtain classification results. The following algorithms were compared:
NB, standard naive Bayes.
TAN, tree-augmented naive Bayes.
AODE, averaged one-dependence estimator.
KDB, k-dependence Bayesian classifier.
KDB, the KDB that only performs the sorting method proposed above.
ATAN, averaged tree-augmented naive Bayes.
RF100, random forest containing 100 trees.
RFn, random forest containing n trees, where n is the number of predictive attributes.
KDF, k-dependence forest.
Kohavi and Wolpert presented a bias-variance decomposition of the expected misclassification rate [
29], which is a powerful tool from sampling theory statistics for analyzing supervised learning scenarios. Supposing
c and
are the true class label and that generated by a learning algorithm, respectively, the zero-one loss function is defined as
The bias term measures the squared difference between the average output of the target and the algorithm. This term is defined as follows:
where
is the combination of any attribute value. The variance term is a real-valued non-negative quantity that equals zero for an algorithm that always makes the same guess regardless of the training set. The variance increases as the algorithm becomes more sensitive to changes in the training set. It is defined as follows:
Given the definite Bayesian network structure,
can be calculated as follows:
The conditional probability
in the bias term can be rewritten as
Given a dataset containing e test instances, the values of zero-one loss, bias and variance for this dataset can be achieved by averaging the result of zero-one loss, bias and variance for all test instances.
In order to clarify the performance of the KDF over datasets of a different scale, we propose a new scoring criterion, which is called goal difference (GD).
Definition 2. Goal difference (GD) is a scoring criterion to compare the performance of two classifiers. Given two classifiers A and B, GD is defined aswhere is the collection of datasets for experimental study, and and represent the number of datasets on which A
outperforms or underperforms B
by comparing the results of the evaluation function (e.g., zero-one loss, bias, and variance), respectively. Diversity has been recognized as a very important characteristic in classifier combination. However, there is no strict definition of what is intuitively perceived as diversity of classifiers. Many measures of the connection between two classifier outputs can be derived from the statistical literature. There is less clarity on the subject when three or more classifiers are concerned. Supposing that each subclassifier votes for a particular class label, given a test instance
and assuming equal weights, the proportion that
n subclassifiers agree on class label
is
where
Entropy is a good measure of dispersion in bootstrap estimation during classification. Given a test set containing
M instances, an appropriate measure to evaluate diversity among ensemble members is
Clearly, when all subclassifiers always vote for the same label, will have a minimum value of 0.
We argue that the KDF benefits from the sorting method, dependence analysis and ensemble mechanism. In the following, we propose experiments for these three aspects.
4.1. Impact of Sorting Method
To illustrate the impact of the sorting method on the performance of classification, we consider another version of the KDB, that is, KDB
. KDB
performs the sorting method proposed above to replace the sorting method of KDB. We note that the root node of KDB
is consistent with that of the KDB to make sure the result is fair.
Table A1 in
Appendix A presents for each dataset the zero-one loss, which is estimated by 10-fold cross-validation to give an accurate estimation of the average performance of an algorithm. The best result is emphasized with bond font. Runs with the various algorithms are carried out on the same training sets and evaluated on the same test sets. In particular, the cross-validation folds are the same for all of the experiments on each dataset. By comparing via a two-tailed binomial sign test with a 95% confidence level, we present summaries of win/draw/loss (W/D/L) records in
Table 2. A win indicates that the algorithm has significantly lower error than the comparator. A draw indicates that the differences in error are not significant. We can easily find that KDB
achieves lower error on 13 datasets over KDB. This proves that the better performance of KDB
on 13 datasets can be attributed to the sorting method.
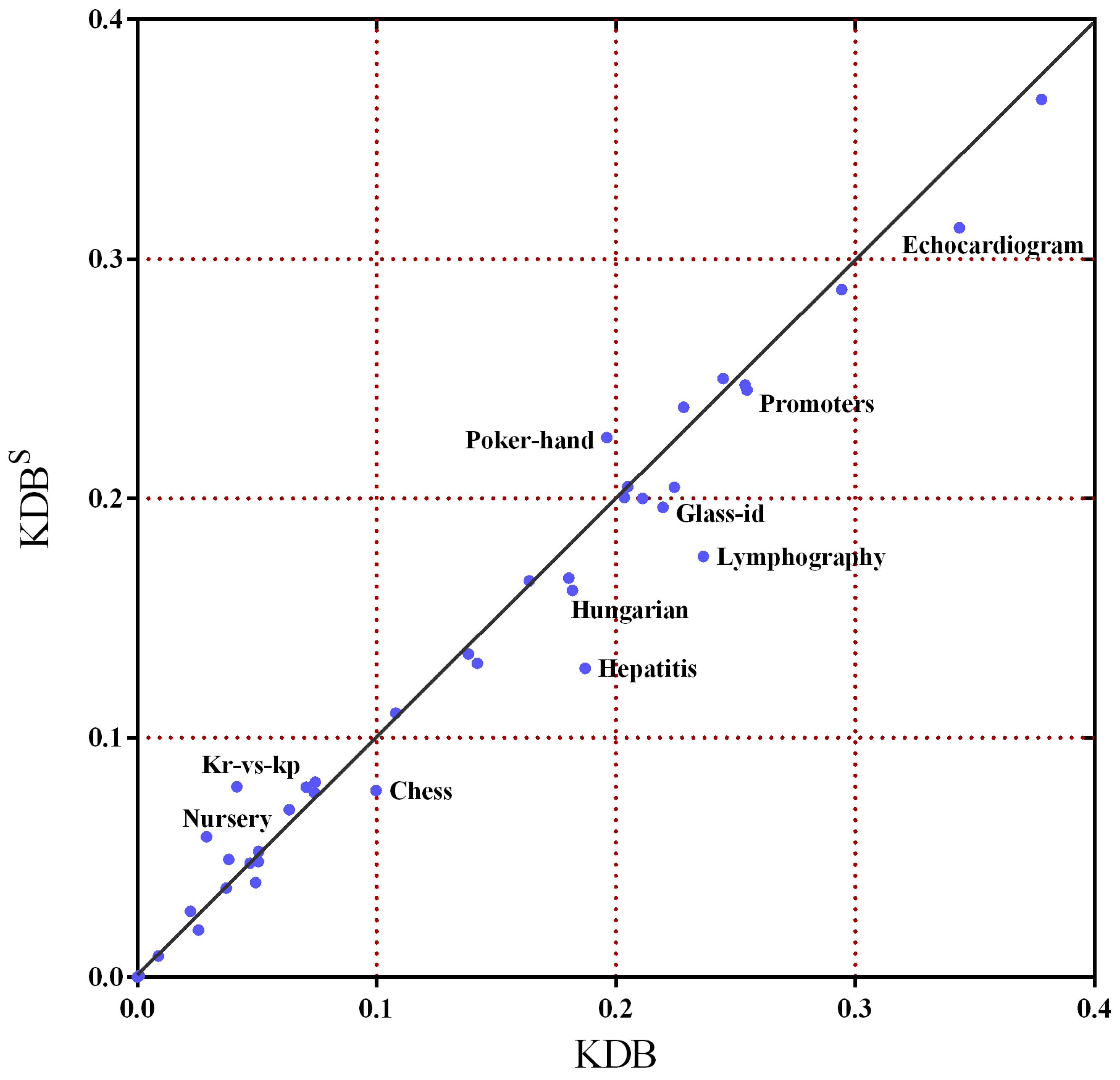
In order to further demonstrate the superiority of this sorting method,
Figure 7 shows the scatter plot of KDB
and KDB in terms of zero-one loss. The
X-axis represents the zero-one loss results of KDB and the
Y-axis represents the zero-one loss results of KDB
. We can see that there are a lot of datasets under the diagonal line, such as
Chess,
Hepatitis,
Lymphography and
Echocardiogram, which means that KDB
has a clear advantage over the KDB. Simultaneously, aside from
Nursery,
Kr vs. kp and
Poker hand, the other datasets fall close to the diagonal line. That means that KDB
has much higher classification error than KDB on only these three datasets. For some datasets, this sorting method did not affect the classification error. However, for many datasets, it substantially reduced the classification error, for example, the reduction from 0.1871 to 0.1290 for the
Hepatitis dataset.
4.2. Impact of Dependence Analysis
To show the superior performance of dependence analysis (i.e., the selection of ancestor attributes) of the KDF, we clarify from the viewpoint of conditional mutual information
, which can be used to quantitatively evaluate the conditional dependence between
and
given
C. We propose the definition of average conditional mutual information, that is,
, to measure the intensity of conditional dependence between predictive attributes for the classifier.
is defined as follows:
where
is the parent of
, and
is the sum of numbers of arcs between predictive attributes. The comparison results of
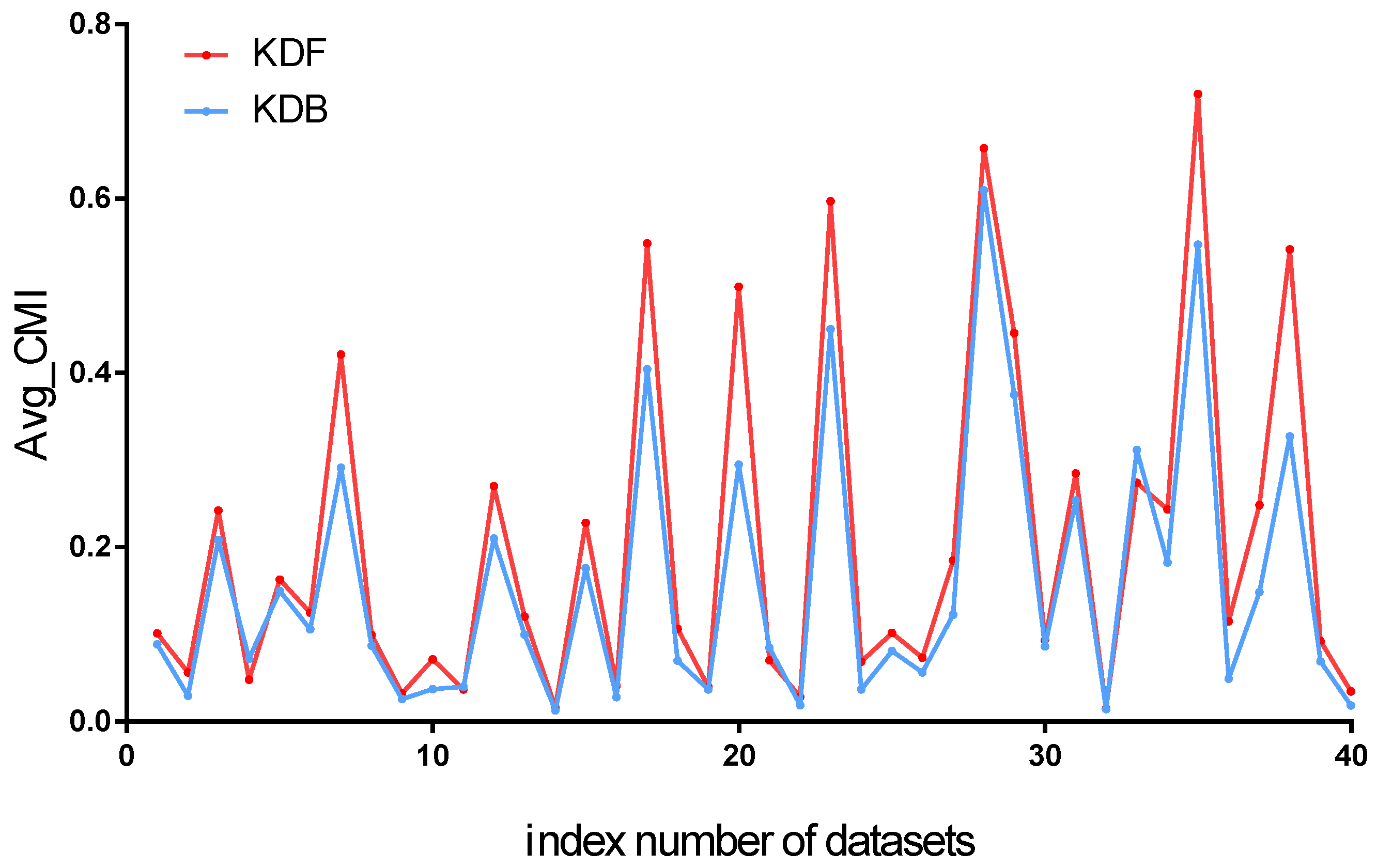
between KDF and KDB are shown in
Figure 8. We can find that KDF has a significant advantage over KDB for almost all the datasets. According to
Figure 8, we can see that the W/D/L of KDF against KDB is 35/1/4. That is to say, KDB has a higher value of
than KDF on only four datasets. The experimental results prove that the selection of ancestor attributes of the KDF can fully demonstrate conditional dependence between predictive attributes; for example, the value of
increases from 0.2947 to 0.4991 for the
Vowel dataset.
4.3. Further Experimental Analysis
This part of the experiments compared the KDF with the out-of-core classifiers described in
Section 4 in terms of zero-one loss. According to the zero-one loss results in
Table A1 in
Appendix A, we present summaries of W/D/L records in
Table 3. When the dependence complexity increases, the performance of TAN and the KDB becomes better than that of NB. The two-dependence relationship helps the KDB to achieve a slightly better performance than TAN (16 wins and 13 losses). It is clear that AODE performs far better than NB (27 wins and 4 losses). However, the ensemble mechanism does not help ATAN to achieve superior performance to TAN (2 wins and 1 loss). The KDF performs the best. For example, when compared with the KDB, the KDF wins on 23 datasets and loses on 5 datasets. This advantage is more apparent when comparing the KDF with ATAN (26 wins and 2 losses). The KDF also provides better classification performance than AODE (26 wins and 5 losses).
To clarify from the viewpoints of the ensemble mechanism and structure complexity, we only compare the KDF with three classifiers, that is, KDB, ATAN and AODE. We present the fitting curve of GD in terms of zero-one loss in
Figure 9. Given datasets
, the
X-axis in
Figure 9 represents the index number of datasets, and the
Y-axis represents the value of
, where
is the collection of datasets
for experimental study. In the following discussion, we first compare the KDF with other two ensemble classifiers, that is, ATAN and AODE. Then, the KDF is compared with the KDB in the case of the same value of
k. As shown in
Figure 9, the KDF only performs a little worse than ATAN when dealing with small datasets with less than 131 instances, for example,
Echocardiogram. This indicates that fewer instances are not enough to support discovering significant dependencies for the KDF. However, as more instances are utilized for the training classifier, the sorting method of the KDF and the higher value of
k will help to ensure that more dependencies will appear and be expressed in the joint probability distribution. This makes the KDF perform much better than ATAN (the maximum value of
is 24). Owing to the same reason, the fitting curve of
has a similar trend compared with the fitting curve of
. When we compare the KDF with the KDB, the fitting curve shows a different trend. It is clear from
Figure 9 that the KDF always performs much better than the KDB on datasets of different scale. This superior performance is due to the ensemble mechanism of the KDF. The KDF has
n subclassifiers, where
n is the number of predictive attributes, and each subclassifier of the KDF reflects almost the same quantities of mutual dependencies and conditional dependencies compared with the KDB. Moreover, diversity among the subclassifiers of the KDF is also a key part in the superior performance of the KDF. In order to prove this point, we show the results of average entropy diversity in the following discussion.
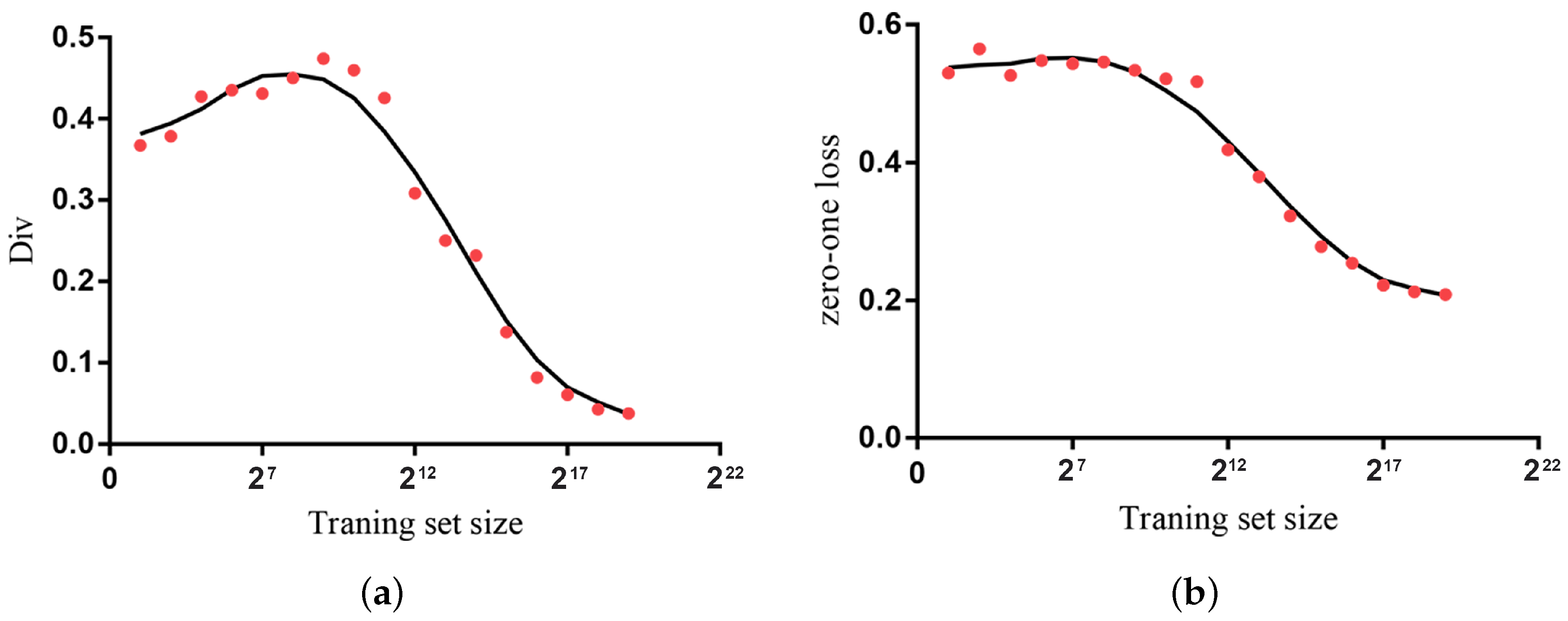
For the purpose of calculating the average entropy diversity of the KDF over datasets of a different scale and simultaneously ensuring the consistency of the data distribution, we take the
Poker hand dataset as an example. Before the segmentation, 200 instances were selected as a test set and the remaining instances were for training. The training set is divided into 17 parts of different sizes. The scale of these 17 parts is in an exponential growth of 2 (from
to
).
Figure 10a shows the fitting curve of average entropy diversity of the KDF on the
Poker hand dataset. As can be seen, there is a strong diversity among the subclassifiers of the KDF, and the maximum value is close to 0.48 when the dataset contains less than
instances (4096 instances). The reason for this result is that fewer training instances make each subclassifier learn diverse mutual dependencies and conditional dependencies. As the quantities of instance increase, each subclassifier can be trained well and tends to vote for the same label. Therefore, the fitting curve of the average entropy diversity has a downward trend. However, the slight decrease in diversity does not produce a bad performance in classification accuracy.
Figure 10b shows the corresponding fitting curve of zero-one loss of the KDF. We can find that as more instances are utilized for training, the KDF still achieves better classification performance in terms of zero-one loss.
4.3.1. Comparison with In-Core Random Forest
A random forest (RF) is a powerful in-core learning algorithm that is state-of-the-art. To further illustrate the performance of the KDF, here we first compare the KDF with the RF, which contains 100 trees (RF100) with respect to zero-one loss. From
Table A1 in
Appendix A, we can see that RF100 seems to perform better than the KDF on several datasets. In order to know how much RF100 wins by, we present the scatter plot in
Figure 11a, where the
X-axis represents the zero-one loss results of RF100 and the
Y-axis represents the zero-one loss results of the KDF. We note that we do not obtain the results for RF100 on such two datasets as
Covtype and
Poker hand because of the limited memory; thus we remove these two points in the plot. We can see that the dataset
Anneal is under the diagonal line, which means the KDF could beat RF100 on the
Anneal dataset. Except for
Vowel,
Tic-tac-toe,
Promoters and
Sign, the other datasets fall close to the diagonal line. This means the performance of the KDF is close to the performance of RF100 on most datasets. It is worthwhile to keep in mind that the number of subclassifiers of the KDF (the maximum number is 64 on the
Optdigits dataset) is much smaller than that of RF100.
It is unfair to make a comparison between the KDF and RF when they have a different number of subclassifiers. Thus we present another experiment that limits the RF with
n trees (RFn), just as for KDF.
Table A1 in
Appendix A presents the zero-one loss in detail. We also present the scatter plot in
Figure 11b, where the X-axis represents the zero-one loss results of RFn and the Y-axis represents the zero-one loss results of the KDF. From
Figure 11b, we can easily find that most datasets are under the diagonal line, for example,
Anneal,
Car,
Chess,
Hungarian,
Promoters, and so on, which means the KDF performs much better than RFn on these datasets. Except for
Vowel and
Sign, the other datasets fall close to the diagonal line, which means the performance of the KDF is close to that of RFn on the remaining datasets. The superior performance of the RF can be partially attributed to the great number of decision trees. The experiment results show that the KDF is competitive with the RF when they contain the same number of subclassifiers.
4.3.2. Bias Results
Bias can be used to evaluate the extent to which the final model learned from training data fits the entire dataset. To further illustrate the performance of the proposed KDF, the experimental results of average bias are shown in
Table A2 in
Appendix A. Only 18 large datasets (size > 2310) are selected for comparison because of statistical significance.
Table 4 shows the corresponding W/D/L records. From
Table 4, we can see that the fitness of NB is the poorest because its structure is definite regardless of the true data distribution. Although the structure of AODE is also definite, it shows a great advantage over NB (17 wins). The main reason may be that it averages all models from a restricted class of one-dependence classifiers and reflects more dependencies between predictive attributes. ATAN and TAN almost have the same bias results (18 draws). The KDF still performs the best, although the advantage is not significant. By sorting attributes and training
n subclassifiers, the ensemble mechanism can help the KDF make full use of the information that is supplied by the training data. The complicated relationship among attributes are measured and depicted from the viewpoint of information theory. Thus, performance robustness can be achieved. The W/D/L records of the KDF compared to AODE show that the advantage is obvious (11 wins and 4 losses) for bias. We can also find that more often than not, the KDF obtains lower bias than ATAN (8 wins and 4 losses) and the KDB (7 wins and 5 losses).
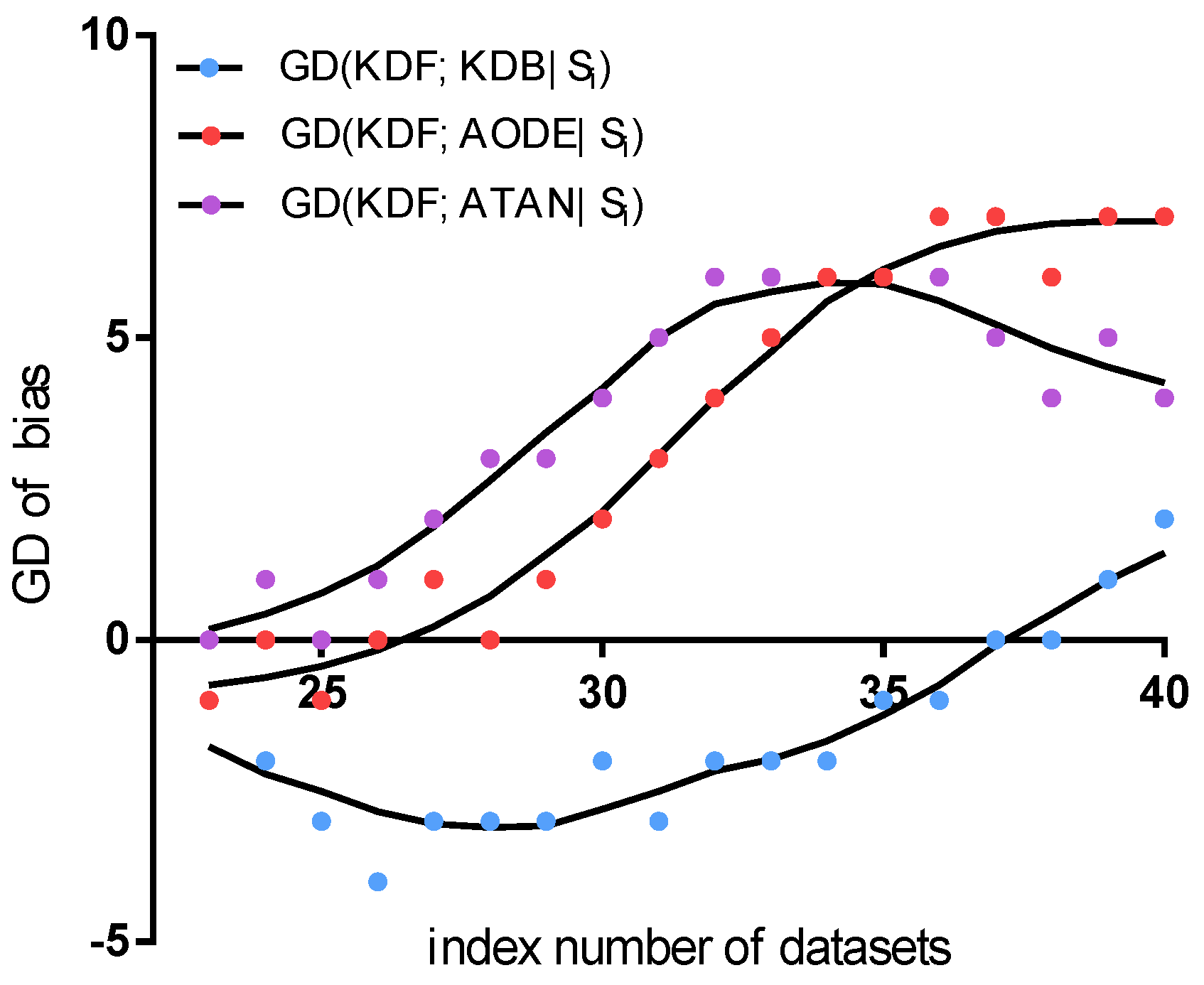
Figure 12 shows the fitting curve of GD in terms of bias. The results indicate that the KDF is competitive to AODE (the minimum value of
is
and the maximum value of
is 7). We believe the reason for the KDF performing better is that the sorting method means it reflect more dependencies than AODE. The KDF performs much better than ATAN (the maximum value of
is 6) when dealing with relatively small datasets containing less than 67,557 instances, for example, the
Connect-4 dataset. As the quantities of instance increase, dependencies between predictive attributes are completely represented, and the final structure of both the KDF and ATAN fits the entire dataset well. Thus, the KDF wins on two out of the last four datasets. The comparison results between the KDF and KDB in terms of
show another trend. From the fitting curve, we can find that the KDB is competitive to the KDF for the first four datasets, which contain less than 4601 instances. The minimum value of
is as low as
. The reason for this result is that KDF cannot discover enough dependencies when the dataset contains lower quantities of data. As the quantities of instance increase, the KDF achieves greater advantage in terms of bias.
4.3.3. Variance Results
Table A3 in
Appendix A shows the experimental results of average variance on 18 large datasets.
Table 5 shows corresponding W/D/L records. A higher degree of attribute dependence means more parameters, which increases the risk of overfitting. An overfitted model does not perform well on data outside the training data. It is clear that NB performs the best among these algorithms, because its network structure is definite and is therefore insensitive to changes in the training set, as shown in
Table 5. Owing to the same reason, AODE also has a competitive performance. ATAN has almost the same performance (17 draws) compared to TAN. By contrast, the KDB performs the worst. When the value of
k increases, the resulting network tends to have a complex structure. The KDF wins on 13 out of 18 datasets compared to the KDB. AODE wins over the KDF, although the advantage is not significant (7 wins and 9 losses).
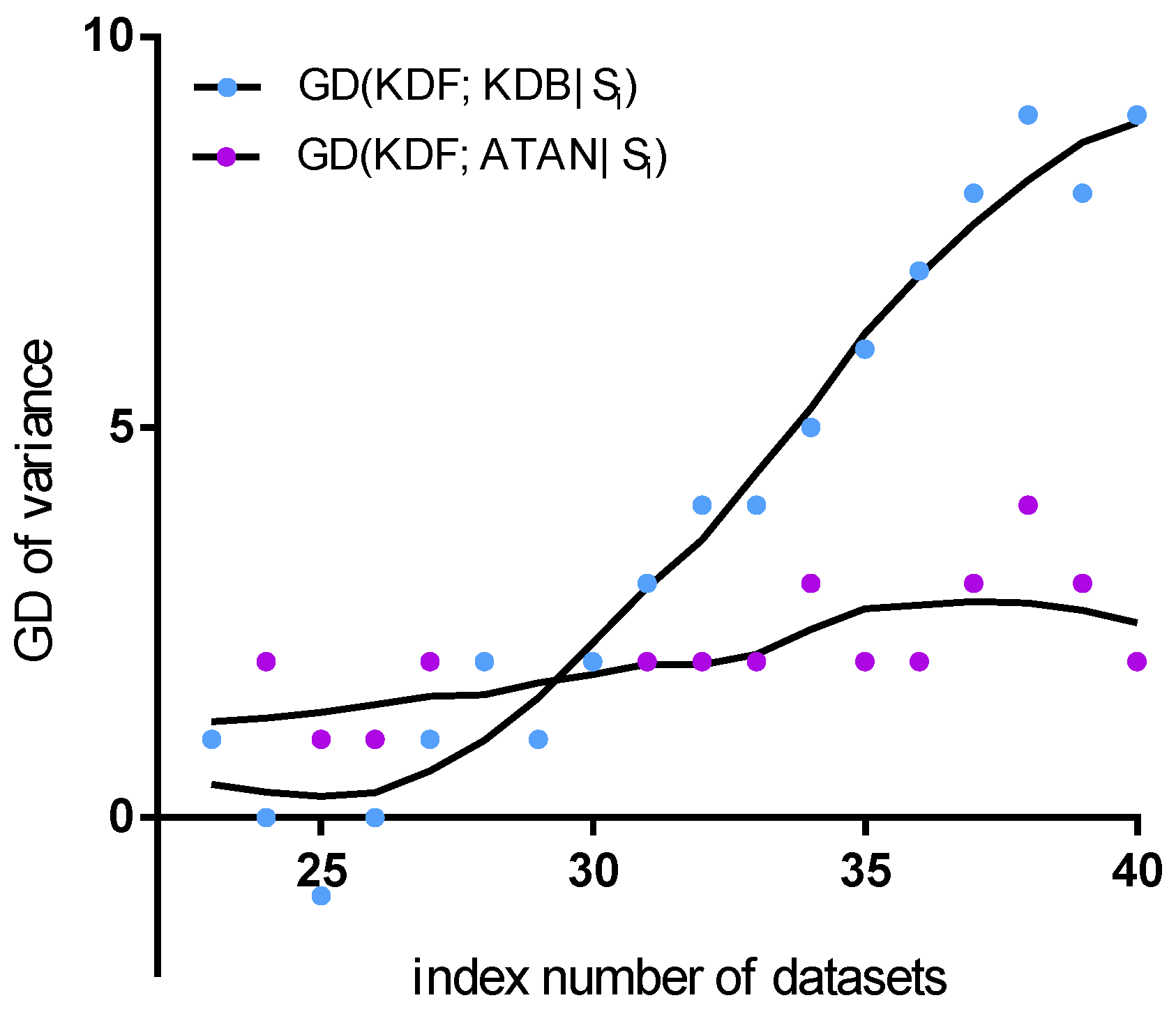
Figure 13 shows the fitting curve of GD of in terms of variance. NB and AODE are neglected, because they are insensitive to the changes in the training set. TAN is not considered, because of almost the same performance as ATAN. The KDF obtains a significant advantage over the KDB, but performs similarly to ATAN. ATAN can only represent the most significant one-dependence relationships between attributes and thus performs similarly to TAN. The ensemble mechanism helps the KDF fully represent many non-significant dependencies. This may be the main reason why ATAN and the KDF are not sensitive to the changes in data distribution. In contrast, although the KDB can also represent significant dependencies, some non-significant dependencies will be affected by the training data, particularly when the dataset size is relatively large.