Changes in the Complexity of Heart Rate Variability with Exercise Training Measured by Multiscale Entropy-Based Measurements
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Protocol
2.2. Data Acquisition and Processing
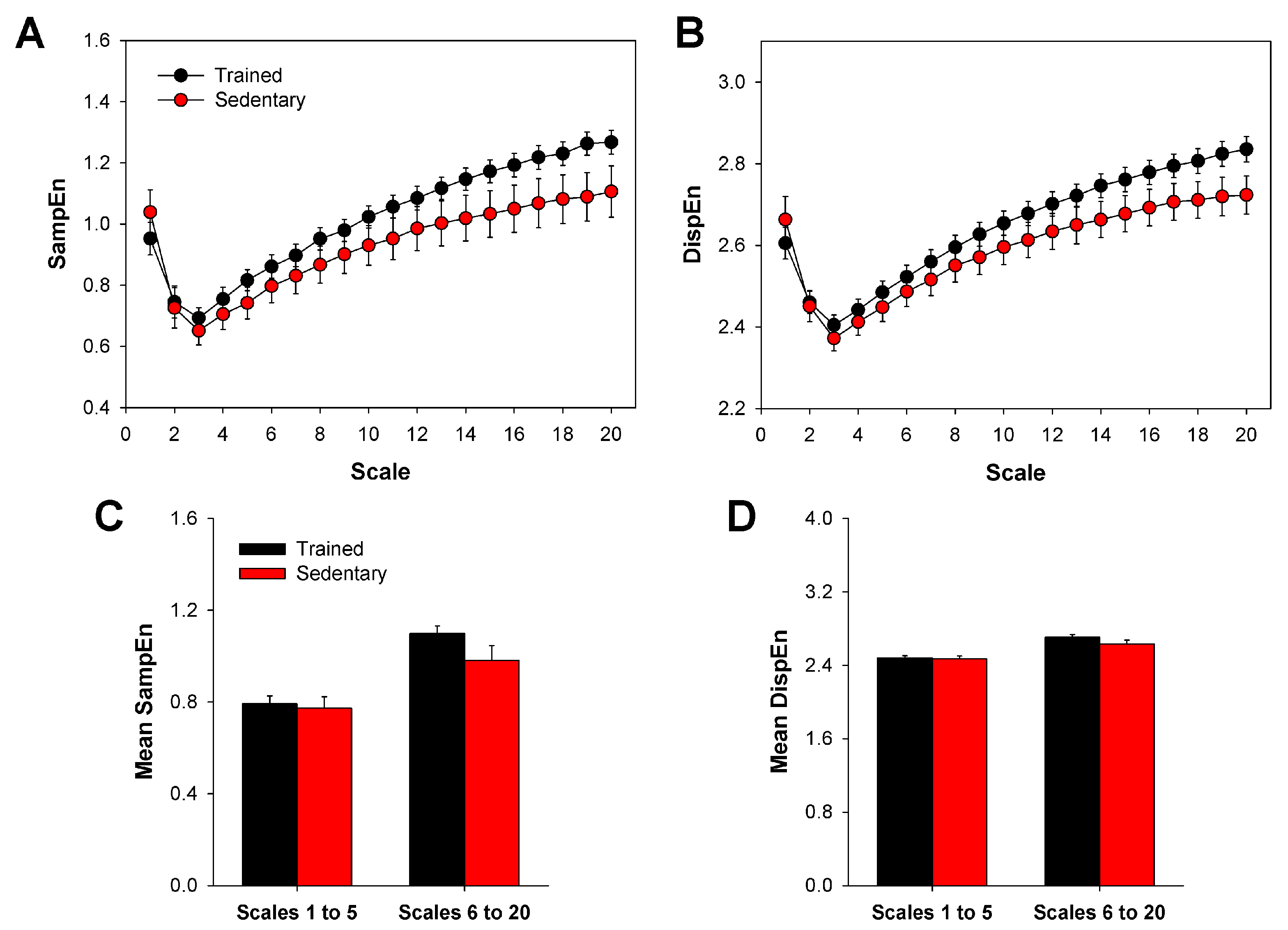
2.3. Multiscale Sample Entropy
2.4. Multiscale Dispersion Entropy
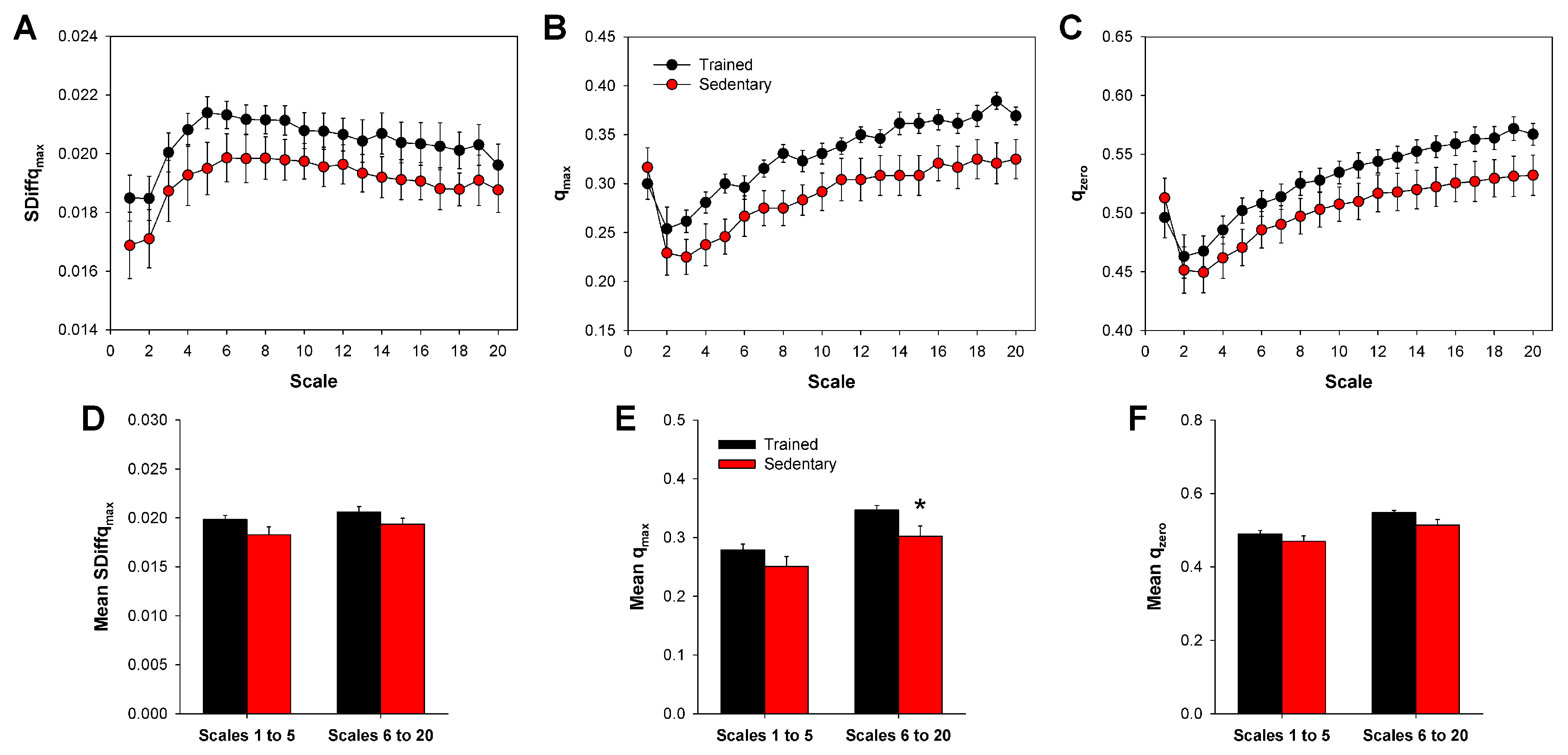
2.5. Multiscale SDiff
- From a given time series u, S surrogate series are generated from u. The surrogate series is obtained by simply shuffling u [44];
- Next, values and are calculated from u;
- Values of and are also calculated from each surrogate instance, obtaining their mean values and ;
- Finally, SDiff is defined by Equation (11) below:
2.6. Statistical Analysis
3. Results
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Boccara, N. Modeling Complex Systems; Springer: New York, NY, USA, 2004. [Google Scholar]
- Baranger, M. Chaos, Complexity, and Entropy: A Physics Talk for Non-Physicists; New England Complex Systems Institute: Cambridge, MA, USA, 2001. [Google Scholar]
- Goldberger, A. Giles f. Filley lecture. Complex systems. Proc. Am. Thorac. Soc. 2006, 3, 467–471. [Google Scholar] [CrossRef] [PubMed]
- Goldberger, A.L.; Peng, C.K.; Lipsitz, L.A. What is physiologic complexity and how does it change with aging and disease? Neurobiol. Aging 2002, 23, 23–26. [Google Scholar] [CrossRef]
- Seely, A.J.E.; Macklem, P.T. Complex systems and the technology of variability analysis. Crit. Care 2004, 8, R367–R384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, S.; Ramsay, T.; Huebsch, L.; Flanagan, S.; McDiarmid, S.; Batkin, I.; McIntyre, L.; Sundaresan, S.R.; Maziak, D.E.; Shamji, F.M.; et al. Continuous multi-parameter heart rate variability analysis heralds onset of sepsis in adults. PLoS ONE 2009, 4, e6642. [Google Scholar] [CrossRef] [PubMed]
- Arab, C.; Dias, D.P.M.; de Almeida Barbosa, R.T.; de Carvalho, T.D.; Valenti, V.E.; Crocetta, T.B.; Ferreira, M.; de Abreu, L.C.; Ferreira, C. Heart rate variability measure in breast cancer patients and survivors: A systematic review. Psychoneuroendocrinology 2016, 68, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.K.; Havlin, S.; Stanley, H.E.; Goldberger, A.L. Quantification of Scaling Exponents and Crossover Phenomena In Nonstationary Heartbeat Time-series. Chaos 1995, 5, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, P.C.; Amaral, L.A.N.; Goldberger, A.L.; Havlin, S.; Rosenblum, M.G.; Struzik, Z.R.; Stanley, H.E. Multifractality in human heartbeat dynamics. Nature 1999, 399, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Porta, A.; Casali, K.R.; Casali, A.G.; Gnecchi-Ruscone, T.; Tobaldini, E.; Montano, N.; Lange, S.; Geue, D.; Cysarz, D.; Van Leeuwen, P. Temporal asymmetries of short-term heart period variability are linked to autonomic regulation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008, 295, R550–R557. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.D.; Peng, C.K.; Goldberger, A.L. Multiscale analysis of heart rate dynamics: Entropy and time irreversibility measures. Cardiovasc. Eng. 2008, 8, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Porta, A.; Tobaldini, E.; Guzzetti, S.; Furlan, R.; Montano, N.; Gnecchi-Ruscone, T. Assessment of cardiac autonomic modulation during graded head-up tilt by symbolic analysis of heart rate variability. Am. J. Physiol. Heart Circ. Physiol. 2007, 293, H702–H708. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.D.; Davis, R.; Goldberger, A. Heart Rate Fragmentation: A Symbolic Dynamical Approach. Front. Physiol. 2017, 8, 827. [Google Scholar] [CrossRef] [PubMed]
- Bashan, A.; Bartsch, R.P.; Kantelhardt, J.W.; Havlin, S.; Ivanov, P.C. Network physiology reveals relations between network topology and physiological function. Nat. Commun. 2012, 3, 702. [Google Scholar] [CrossRef] [PubMed]
- Hou, F.Z.; Wang, J.; Wu, X.C.; Yan, F.R. A dynamic marker of very short-term heartbeat under pathological states via network analysis. EPL (Europhys. Lett.) 2014, 107, 58001. [Google Scholar] [CrossRef]
- Chen, C.; Jin, Y.; Lo, I.L.; Zhao, H.; Sun, B.; Zhao, Q.; Zheng, J.; Zhang, X.D. Complexity Change in Cardiovascular Disease. Int. J. Biol. Sci. 2017, 13, 1320–1328. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Faes, L.; Ivanov, P.C. Entropy measures, entropy estimators, and their performance in quantifying complex dynamics: Effects of artifacts, nonstationarity, and long-range correlations. Phys. Rev. E 2017, 95, 062114. [Google Scholar] [CrossRef] [PubMed]
- Humeau-Heurtier, A. The Multiscale Entropy Algorithm and Its Variants: A Review. Entropy 2015, 17, 3110–3123. [Google Scholar] [CrossRef] [Green Version]
- Bandt, C.; Pompe, B. Permutation entropy: A natural complexity measure for time series. Phys. Rev. Lett. 2002, 88, 174102. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Wang, Z.; Xie, H.; Yu, W. Characterization of surface EMG signal based on fuzzy entropy. IEEE Trans. Neural Syst. Rehabil. Eng. 2007, 15, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Liu, C.; Li, K.; Zheng, D.; Liu, C.; Hou, Y. Assessing the complexity of short-term heartbeat interval series by distribution entropy. Med. Biol. Eng. Comput. 2015, 53, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Rostaghi, M.; Azami, H. Dispersion Entropy: A Measure for Time-Series Analysis. IEEE Signal Process. Lett. 2016, 23, 610–614. [Google Scholar] [CrossRef]
- Rényi, A. On measures of entropy and information. In Proceedings of the Fourth Berkeley Symposium on Mathematical Statistics and Probability, Volume 1: Contributions to the Theory of Statistics; The Regents of the University of California: Berkeley, CA, USA, 1961. [Google Scholar]
- Manis, G.; Aktaruzzaman, M.; Sassi, R. Bubble Entropy: An Entropy Almost Free of Parameters. IEEE Trans. Biomed. Eng. 2017, 64, 2711–2718. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.F.; Wei, S.Y.; Huang, H.P.; Hsu, L.; Chi, S.; Peng, C.K. Entropy of Entropy: Measurement of Dynamical Complexity for Biological Systems. Entropy 2017, 19, 550. [Google Scholar] [CrossRef]
- Silva, L.E.V.; Cabella, B.C.T.; Neves, U.P.d.C.; Murta Junior, L.O. Multiscale entropy-based methods for heart rate variability complexity analysis. Phys. A Stat. Mech. Its Appl. 2015, 422, 143–152. [Google Scholar] [CrossRef]
- Amaral, L.S.d.B.; Silva, F.A.; Correia, V.B.; Andrade, C.E.F.; Dutra, B.A.; Oliveira, M.V.; de Magalhães, A.C.M.; Volpini, R.A.; Seguro, A.C.; Coimbra, T.M.; et al. Beneficial effects of previous exercise training on renal changes in streptozotocin-induced diabetic female rats. Exp. Biol. Med. 2016, 241, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Faleiros, C.M.; Francescato, H.D.C.; Papoti, M.; Chaves, L.; Silva, C.G.A.; Costa, R.S.; Coimbra, T.M. Effects of previous physical training on adriamycin nephropathy and its relationship with endothelial lesions and angiogenesis in the renal cortex. Life Sci. 2017, 169, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Weippert, M.; Behrens, K.; Rieger, A.; Kumar, M.; Behrens, M. Effects of breathing patterns and light exercise on linear and nonlinear heart rate variability. Appl. Physiol. Nutr. Metab. 2015, 40, 762–768. [Google Scholar] [CrossRef] [PubMed]
- Soares-Miranda, L.; Sandercock, G.; Vale, S.; Silva, P.; Moreira, C.; Santos, R.; Mota, J. Benefits of achieving vigorous as well as moderate physical activity recommendations: Evidence from heart rate complexity and cardiac vagal modulation. J. Sports Sci. 2011, 29, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Karavirta, L.; Costa, M.D.; Goldberger, A.L.; Tulppo, M.P.; Laaksonen, D.E.; Nyman, K.; Keskitalo, M.; Häkkinen, A.; Häkkinen, K. Heart rate dynamics after combined strength and endurance training in middle-aged women: Heterogeneity of responses. PLoS ONE 2013, 8, e72664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goulopoulou, S.; Fernhall, B.; Kanaley, J.A. Hemodynamic responses and linear and non-linear dynamics of cardiovascular autonomic regulation following supramaximal exercise. Eur. J. Appl. Physiol. 2009, 105, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Platisa, M.M.; Mazic, S.; Nestorovic, Z.; Gal, V. Complexity of heartbeat interval series in young healthy trained and untrained men. Physiol. Meas. 2008, 29, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Kuipers, H.; Verstappen, F.; Keizer, H.; Geurten, P.; Van Kranenburg, G. Variability of aerobic performance in the laboratory and its physiologic correlates. Int. J. Sports Med. 1985, 6, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Silva, K.A.d.S.; Luiz, R.S.; Rampaso, R.R.; de Abreu, N.P.; Moreira, E.D.; Mostarda, C.T.; De Angelis, K.; Teixeira, V.d.P.C.; Irigoyen, M.C.; Schor, N. Previous exercise training has a beneficial effect on renal and cardiovascular function in a model of diabetes. PLoS ONE 2012, 7, e48826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, M.; Goldberger, A.L.; Peng, C.K. Multiscale Entropy Analysis of Complex Physiologic Time Series. Phys. Rev. Lett. 2002, 89, 068102. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.; Goldberger, A.L.; Peng, C.K. Multiscale entropy analysis of biological signals. Phys. Rev. E 2005, 71, 021906. [Google Scholar] [CrossRef] [PubMed]
- Richman, J.S.; Moorman, J.R. Physiological time-series analysis using approximate entropy and sample entropy. Am. J. Physiol. Heart Circ. Physiol. 2000, 278, H2039–H2049. [Google Scholar] [CrossRef] [PubMed]
- Azami, H.; Rostaghi, M.; Abasolo, D.; Escudero, J. Refined Composite Multiscale Dispersion Entropy and Its Application to Biomedical Signals. IEEE Trans. Biomed. Eng. 2017, 64, 2872–2879. [Google Scholar] [CrossRef] [PubMed]
- Tsallis, C. Possible generalization of Boltzmann-Gibbs statistics. J. Stat. Phys. 1988, 52, 479–487. [Google Scholar] [CrossRef]
- Tsallis, C. Introduction to Nonextensive Statistical Mechanics; Springer: New York, NY, USA, 2009. [Google Scholar]
- Silva, L.E.V.; Murta, L.O. Evaluation of physiologic complexity in time series using generalized sample entropy and surrogate data analysis. Chaos 2012, 22, 043105. [Google Scholar] [CrossRef] [PubMed]
- Borges, E.P. A possible deformed algebra and calculus inspired in nonextensive thermostatistics. Phys. A Stat. Mech. Its Appl. 2004, 340, 95–101. [Google Scholar] [CrossRef]
- Theiler, J.; Eubank, S.; Longtin, A.; Galdrikian, B.; Doyne Farmer, J. Testing for nonlinearity in time series: The method of surrogate data. Phys. D Nonlinear Phenom. 1992, 58, 77–94. [Google Scholar] [CrossRef]
- Silva, L.E.V.; Lataro, R.M.; Castania, J.A.; da Silva, C.A.A.; Valencia, J.F.; Murta, L.O.; Salgado, H.C.; Fazan, R.; Porta, A. Multiscale entropy analysis of heart rate variability in heart failure, hypertensive, and sinoaortic-denervated rats: Classical and refined approaches. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2016, 311, R150–R156. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.E.V.; Silva, C.A.A.; Salgado, H.C.; Fazan, R. The role of sympathetic and vagal cardiac control on complexity of heart rate dynamics. Am. J. Physiol. Heart Circ. Physiol. 2017, 312, H469–H477. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.E.V.; Rodrigues, F.L.; de Oliveira, M.; Salgado, H.C.; Fazan, R. Heart rate complexity in sinoaortic-denervated mice. Exp. Physiol. 2015, 100, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.L.; Lin, C.; Lin, Y.H.; Lo, M.T. The Prognostic Value of Non-Linear Analysis of Heart Rate Variability in Patients with Congestive Heart Failure—A Pilot Study of Multiscale Entropy. PLoS ONE 2011, 6, e18699. [Google Scholar] [CrossRef] [PubMed]
- Bari, V.; Valencia, J.F.; Vallverdú, M.; Girardengo, G.; Marchi, A.; Bassani, T.; Caminal, P.; Cerutti, S.; George, A.L.; Brink, P.A.; et al. Multiscale complexity analysis of the cardiac control identifies asymptomatic and symptomatic patients in long QT syndrome type 1. PLoS ONE 2014, 9, e93808. [Google Scholar] [CrossRef] [PubMed]
- Pardo, Y.; Merz, N.; Bairey, C.; Velasquez, I.; Paul-Labrador, M.; Agarwala, A.; Peter, C.T. Exercise conditioning and heart rate variability: Evidence of a threshold effect. Clin. Cardiol. 2000, 23, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Mueller, P.J. Exercise training and sympathetic nervous system activity: evidence for physical activity dependent neural plasticity. Clin. Exp. Pharmacol. Physiol. 2007, 34, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Gao, R. Multiscale Entropy Analysis of the Differential RR Interval Time Series Signal and Its Application in Detecting Congestive Heart Failure. Entropy 2017, 19, 251. [Google Scholar]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fazan, F.S.; Brognara, F.; Fazan Junior, R.; Murta Junior, L.O.; Virgilio Silva, L.E. Changes in the Complexity of Heart Rate Variability with Exercise Training Measured by Multiscale Entropy-Based Measurements. Entropy 2018, 20, 47. https://doi.org/10.3390/e20010047
Fazan FS, Brognara F, Fazan Junior R, Murta Junior LO, Virgilio Silva LE. Changes in the Complexity of Heart Rate Variability with Exercise Training Measured by Multiscale Entropy-Based Measurements. Entropy. 2018; 20(1):47. https://doi.org/10.3390/e20010047
Chicago/Turabian StyleFazan, Frederico Sassoli, Fernanda Brognara, Rubens Fazan Junior, Luiz Otavio Murta Junior, and Luiz Eduardo Virgilio Silva. 2018. "Changes in the Complexity of Heart Rate Variability with Exercise Training Measured by Multiscale Entropy-Based Measurements" Entropy 20, no. 1: 47. https://doi.org/10.3390/e20010047



