Synthesis of Functionalised Nucleosides for Incorporation into Nucleic Acid-Based Serine Protease Mimics
Abstract
:Introduction
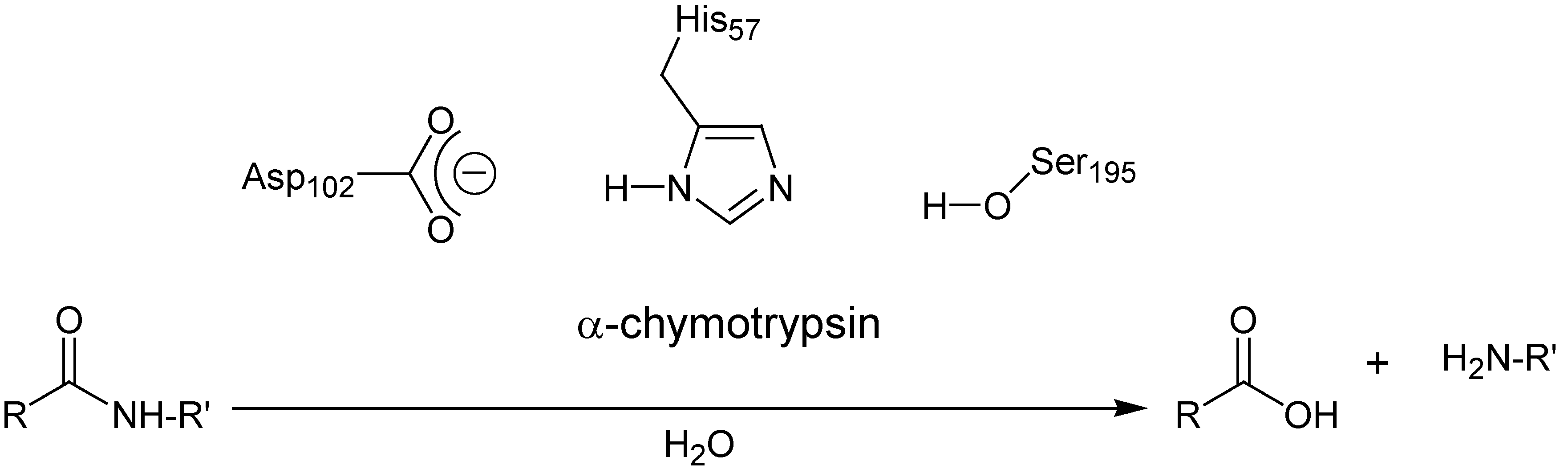
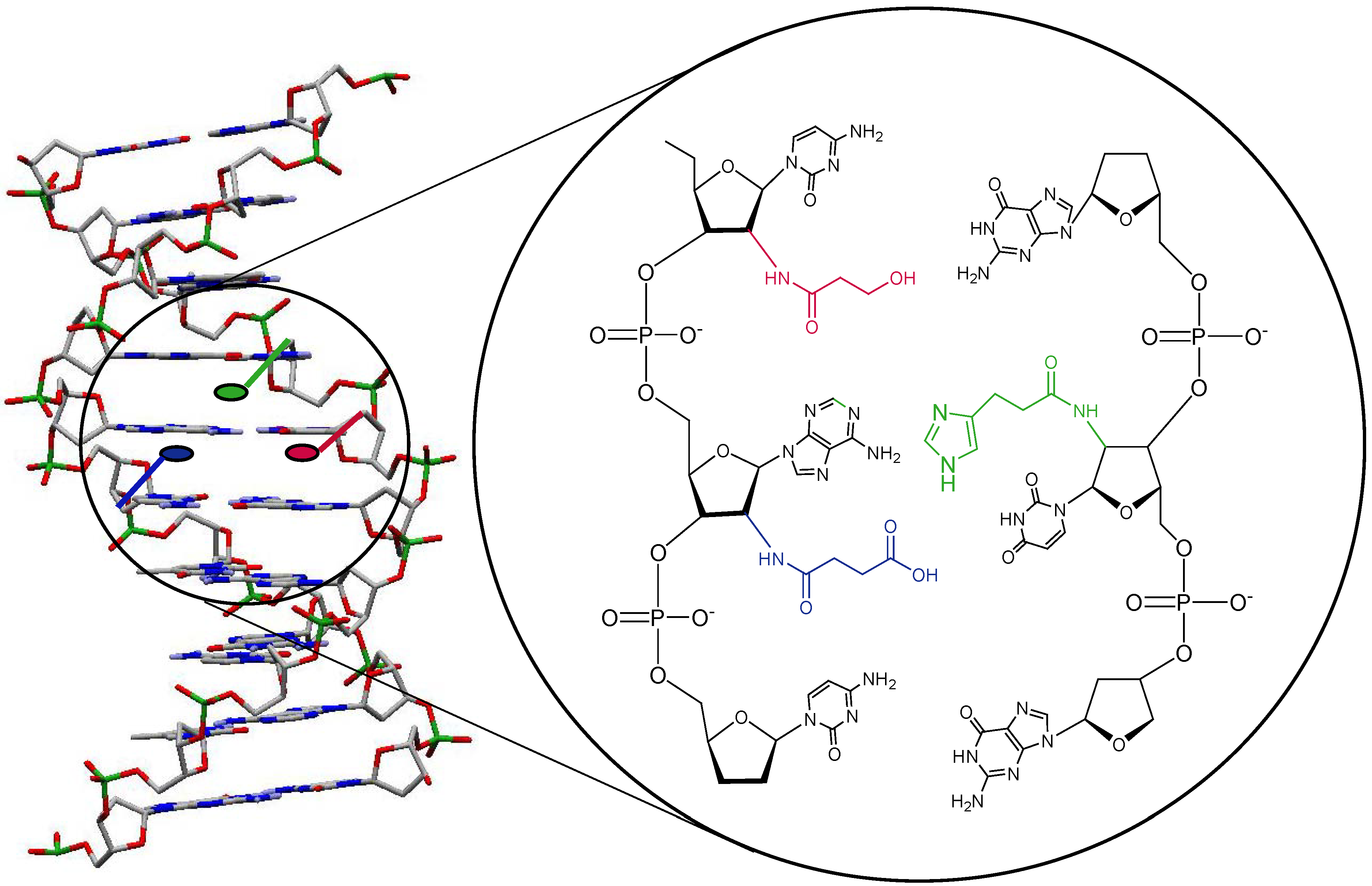
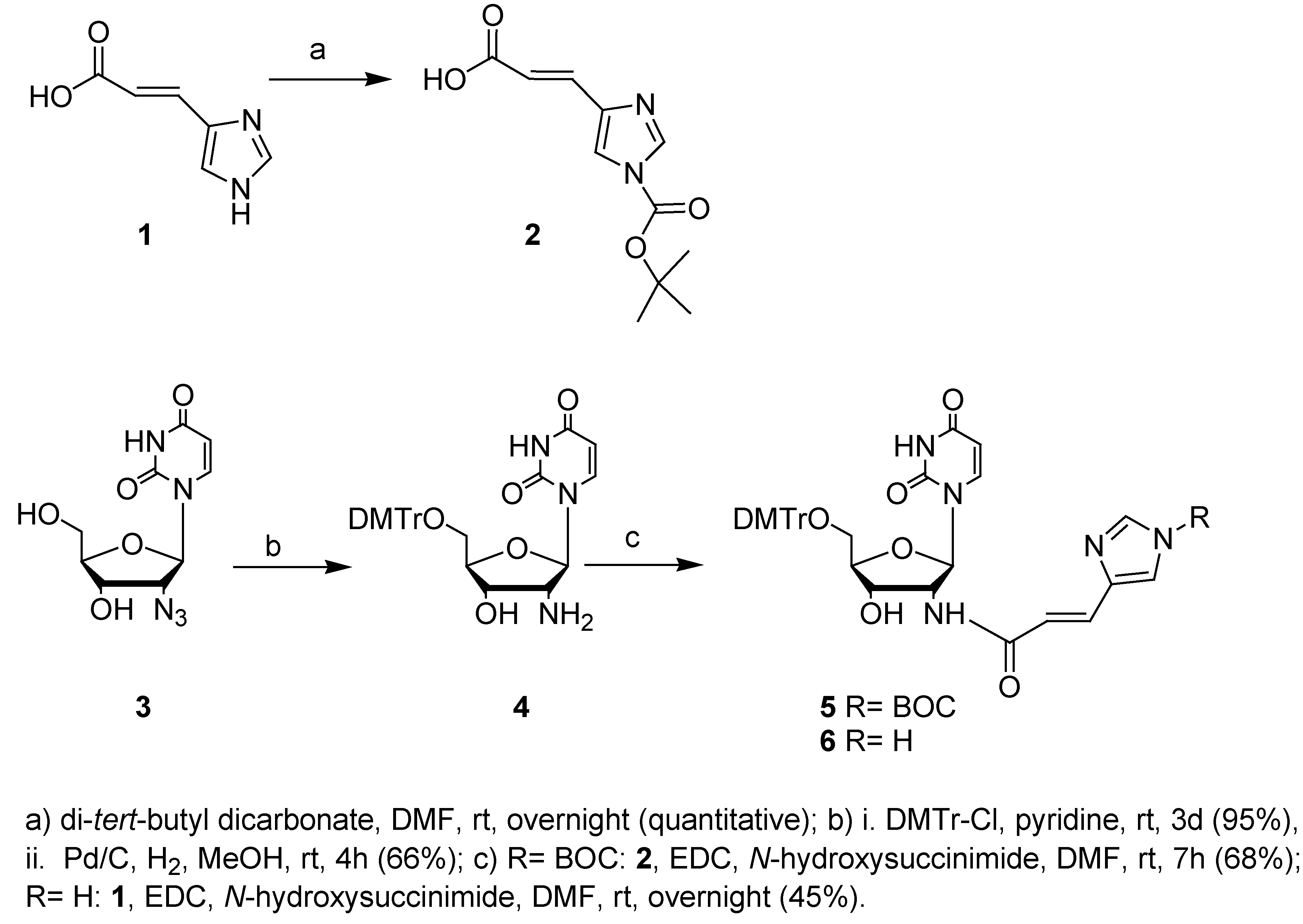
Results and Discussion
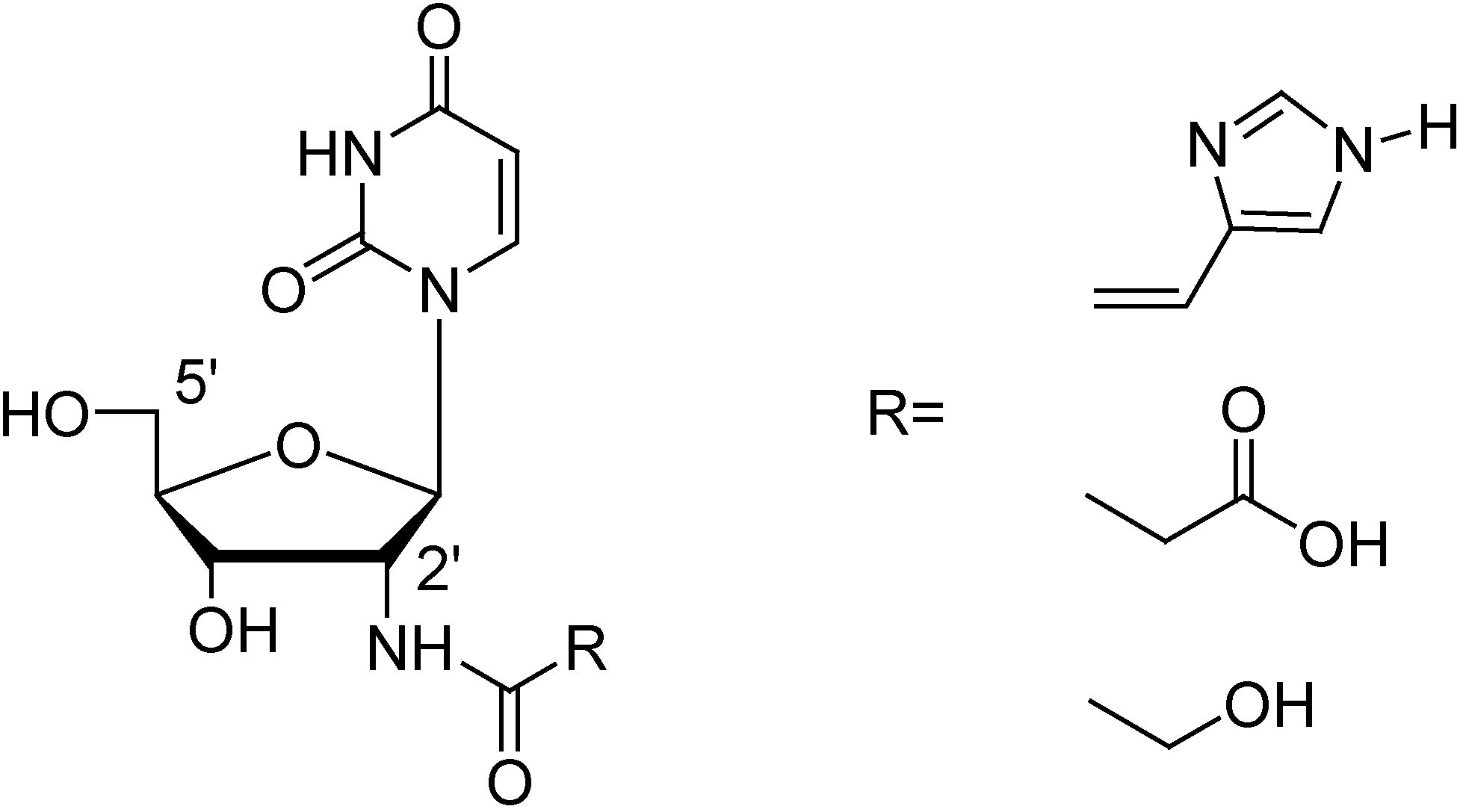

Conclusions
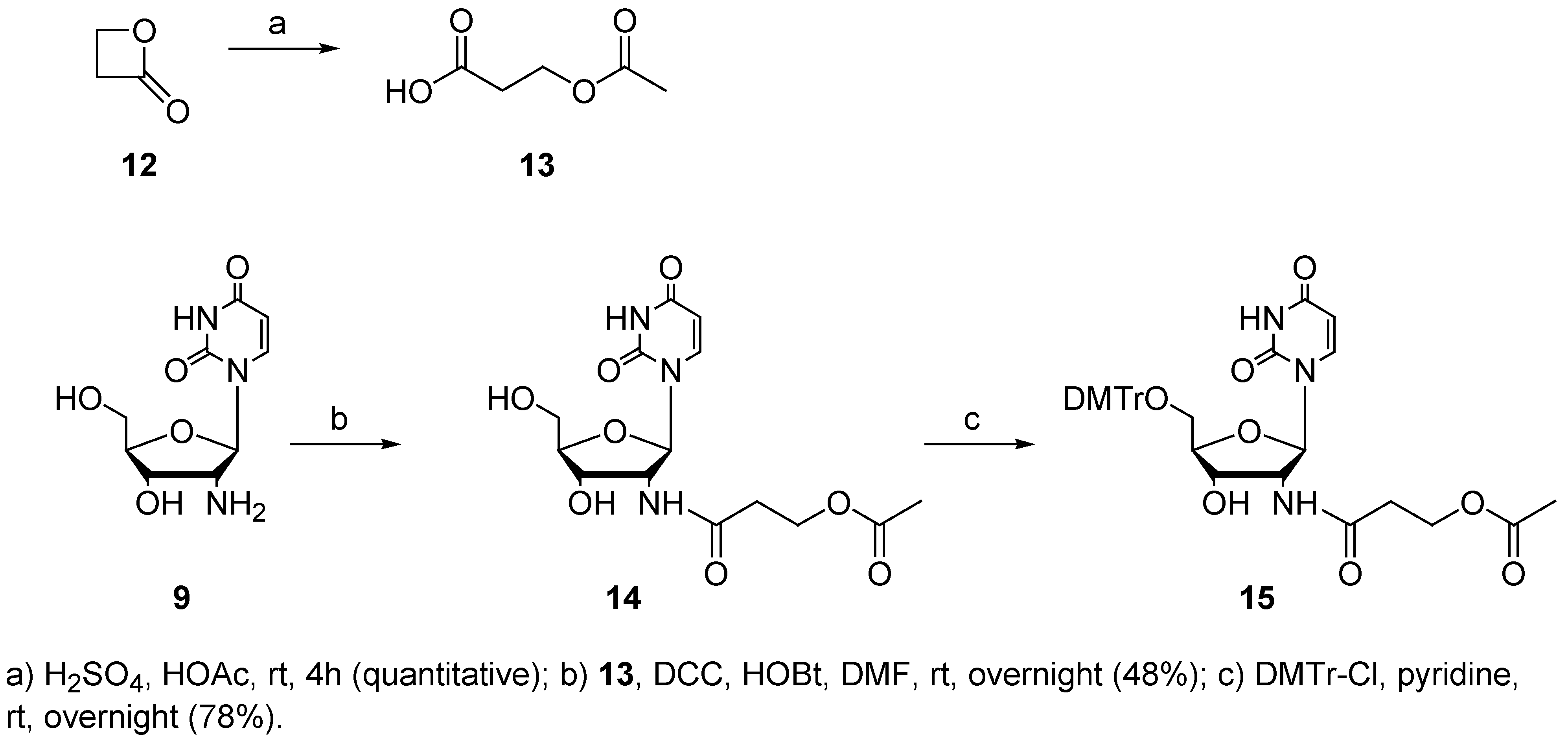
Experimental Section
General
2’-Amino-5’-O-(4,4’-dimethoxytrityl)-2’-deoxyuridine (4)
3-(1-tertbutoxycarbonyl-imidazol-4-yl)-acrylic acid (2)
2’-[3-(1-tertbutoxycarbonyl-imidazol-4-yl)acryloyl]-amido-5’-O-(4,4’-dimethoxytrityl)-2’-deoxy-uridine (5)
2’-[3-(imidazol-4-yl]acryloyl]-amido-5’-O-(4,4’-dimethoxytrityl)-2’-deoxyuridine (6)
4-allyloxy-4-oxo-butanoic acid (8)
2’-[(4-allyloxy-4-oxo)-butanoyl]-amido-2’-deoxyuridine (10)
2’-[(4-allyloxy-4-oxo)-butanoyl]-amido-5’-O-(4,4’-dimethoxytrityl)-2’-deoxyuridine (11)
3-Acetoxy-propanoic acid (13)
2’-(3-acetoxypropanoyl)-amido-2’-deoxyuridine (14)
2’-(3-acetoxypropanoyl)-amido-5’-O-(4,4’-dimethoxytrityl)-2’-deoxyuridine (15)
Acknowledgements
References
- Matthews, B. W.; Sigler, P. B.; R., H.; Blow, D. M. Three-dimensional structure of tosyl-chymotrypsin. Nature 1967, 214, 652–656. [Google Scholar] [CrossRef]
- Blow, D. M.; Birktoft, J. J.; Hartley, B. S. Role of a buried acid group in the mechanism of action of chymotryspin. Nature 1969, 221, 337–340. [Google Scholar] [CrossRef]
- Steitz, T. A.; Henderson, R.; Blow, D. M. Structure of crystalline chymotrypsin. J. Mol. Biol. 1969, 46, 337–348. [Google Scholar]
- Blow, D. M. Structure and Mechanism of Chymotrypsin. Acc. Chem. Res. 1976, 9, 145–152. [Google Scholar] [CrossRef]
- Hedstrom, L. Serine Protease Mechanism and Specificity. Chem. Rev. 2002, 102, 4501–4523. [Google Scholar] [CrossRef]
- Hahn, K. W.; Klis, W. A.; Stewart, J. M. Design and Synthesis of a Peptide Having Chymotrypsin-Like Esterase-Activity. Science 1990, 248, 1544–1547. [Google Scholar]
- Atassi, M. Z.; Manshouri, T. Design of Peptide Enzymes (Pepzymes) - Surface-Simulation Synthetic Peptides That Mimic the Chymotrypsin and Trypsin Active-Sites Exhibit the Activity and Specificity of the Respective Enzyme. Proc. Nat. Acad. Sci. USA 1993, 90, 8282–8286. [Google Scholar] [CrossRef]
- Corey, M. J.; Hallakova, E.; Pugh, K.; Stewart, J. M. Studies on Chymotrypsin-Like Catalysis by Synthetic Peptides. Appl. Biochem. Biotechnol. 1994, 47, 199–212. [Google Scholar] [CrossRef]
- Corey, D. R.; Phillips, M. A. Cyclic-Peptides as Proteases - a Reevaluation. Proc. Nat. Acad. Sci. USA 1994, 91, 4106–4109. [Google Scholar] [CrossRef]
- Wells, J. A.; Fairbrother, W. J.; Otlewski, J.; Laskowski, M.; Burnier, J. A Reinvestigation of a Synthetic Peptide (Trpepz) Designed to Mimic Trypsin. Proc. Nat. Acad. Sci. USA 1994, 91, 4110–4114. [Google Scholar]
- Walse, B.; Ullner, M.; Lindbladh, C.; Bulow, L.; Drakenberg, T.; Teleman, O. Structure of a cyclic peptide with a catalytic triad, determined by computer simulation and NMR spectroscopy. J. Comput.-Aided Mol. Des. 1996, 10, 11–22. [Google Scholar] [CrossRef]
- Fukushima, Y. Enantioselectivity enhancement of ester cleavage by a beta-sheet polypeptide containing catalytic triads in a serine protease. B. Chem. Soc. Jpn. 1996, 69, 2269–2274. [Google Scholar] [CrossRef]
- Stavrakoudis, A.; Demetropoulos, I.; Sakarellos, C.; Sakarellos-Daaitsiotis, M.; Tsikaris, V. Design, synthesis and catalytic activity of a serine protease synthetic model. Lett. Pept. Sci. 1997, 4, 481–487. [Google Scholar]
- Broo, K. S.; Brive, L.; Ahlberg, P.; Baltzer, L. Catalysis of hydrolysis and transesterification reactions of p-nitrophenyl esters by a designed helix-loop-helix dimer. J. Am. Chem. Soc. 1997, 119, 11362–11372. [Google Scholar]
- Li, Y. S.; Zhao, Y. F.; Hatfield, S.; Wan, R.; Zhu, Q.; Li, X. H.; McMills, M.; Ma, Y.; Li, J.; Brown, K. L.; He, C.; Liu, F.; Chen, X. Z. Dipeptide seryl-histidine and related oligopeptides cleave DNA, protein, and a carboxyl ester. Bioorg. Med. Chem. 2000, 8, 2675–2680. [Google Scholar] [CrossRef]
- Delort, E.; Darbre, T.; Reymond, J. A strong positive dendritic effect in a peptide dendrimer-catalyzed ester hydrolysis reaction. J. Am. Chem. Soc. 2004, 126, 15642–15643. [Google Scholar] [CrossRef]
- Dsouza, V. T.; Bender, M. L. Miniature Organic-Models of Enzymes. Accounts Chem. Res. 1987, 20, 146–152. [Google Scholar] [CrossRef]
- Zimmerman, S. C. On the Evaluation of a Small Molecule Mimic of Chymotrypsin. Tetrahedron Lett. 1989, 30, 4357–4358. [Google Scholar]
- Rao, K. R.; Srinivasan, T. N.; Bhanumathi, N.; Sattur, P. B. Artificial Enzymes - Synthesis of Imidazole Substituted at C(2) of Beta-Cyclodextrin as an Efficient Enzyme Model of Chymotrypsin. J. Chem. Soc. Chem. Comm. 1990, 10–11. [Google Scholar]
- Breslow, R. Biomimetic Chemistry and Artificial Enzymes - Catalysis by Design. Acc. Chem. Res. 1995, 28, 146–153. [Google Scholar] [CrossRef]
- Wulff, G. Enzyme-like catalysis by molecularly imprinted polymers. Chem. Rev. 2002, 102, 1–27. [Google Scholar] [CrossRef]
- Madder, A.; De Clercq, P.; Declercq, J.P. Stepwise Approach toward First Generation Nonenzymatic Hydrolases. J. Org. Chem. 1998, 63, 2548–2559. [Google Scholar] [CrossRef]
- De Muynck, H.; Madder, A.; Farcy, N.; De Clercq, P.; Pérez-Payan, M.; Öhberg, L. ; Davis, A. Application of Combinatorial Procedures in the Search for Serine-Protease-Like Activity with Focus on the Acyl Transfer Step. Angew. Chem. Int. Ed. 2000, 39, 145–148. [Google Scholar] [CrossRef]
- Madder, A.; Li, L.; De Muynck, H.; Farcy, N; Van Haver, D.; Fant, F.; Vanhoenacker, G.; Sandra, P.; Davis, A.; De Clercq, P. Evaluation of a Two-Stage Screening Procedure in the Combinatorial Search for Serine Protease-Like Activity. J. Comb. Chem. 2002, 4, 552–562. [Google Scholar] [CrossRef]
- Gea, A.; Farcy, N.; Roqué i Rossell, N.; Martins, J.; De Clercq, P.; Madder, A. Solid-Supported Synthesis of Highly Functionalized Tripodal Peptides with flexible but Preorganized Geometry: Towards Potential Serine Protease Mimics. Eur. J. Org. Chem. 2006, 4135–4146. [Google Scholar]
- Beaucage, S. L.; Caruthers, M. H. Deoxynucleoside Phosphoramidites - a New Class of Key Intermediates for Deoxypolynucleotide Synthesis. Tetrahedron Lett. 1981, 22, 1859–1862. [Google Scholar] [CrossRef]
- Verheyden, J. P.; Wagner, D.; Moffatt, J. G. Synthesis of some pyrimidine 2'-amino-2'-deoxynucleosides. J. Org. Chem. 1971, 36, 250–254. [Google Scholar] [CrossRef]
- Kirschenheuter, G. P.; Zhai, Y.; Pieken, W. A. An improved synthesis of 2'-azido-2'deoxyuridine. Tetrahedron Lett. 1994, 35, 8517–8520. [Google Scholar] [CrossRef]
- Kohgo, S.; Shinozuka, K.; Ozaki, H.; Sawai, H. Synthesis of a novel 2 '-deoxyuridine derivative bearing a cyanomethoxycarbonylmethyl group at C-5 position and its use for versatile post-synthetic functionalization of oligodeoxyribonucleotides. Tetrahedron Lett. 1998, 39, 4067–4070. [Google Scholar] [CrossRef]
- Beloglazova, N. G.; Fabani, M. M.; Zenkova, M. A.; Bichenkova, E. V.; Polushin, N. N.; Sil'nikov, V. V.; Douglas, K. T.; Vlassov, V. V. Sequence-specific artificial ribonucleases. I. Bis-imidazole-containing oligonucleotide conjugates prepared using precursor-based strategy. Nucleic Acids Res. 2004, 32, 3887–3897. [Google Scholar] [CrossRef]
- Beban, M.; Miller, P. S. Preparation of an imidazole-conjugated oligonucleotide. Bioconjugate Chem. 2000, 11, 599–603. [Google Scholar] [CrossRef]
- Wang, G. Y.; Bergstrom, D. E. Synthesis of Oligonucleotides Containing N2-[2-(Imidazol-4-Ylacetamido)Ethyl]-2'-Deoxyguanosine. Tetrahedron Lett. 1993, 34, 6725–6728. [Google Scholar]
- Heeb, N. V.; Benner, S. A. Guanosine Derivatives Bearing an N-2-3-Imidazolepropionic Acid. Tetrahedron Lett. 1994, 35, 3045–3048. [Google Scholar] [CrossRef]
- Truffert, J. C.; Asseline, U.; Brack, A.; Thuong, N. T. Synthesis, purification and characterization of two peptide-oligonucleotide conjugates as potential artificial nucleases. Tetrahedron 1996, 52(8), 3005–3016. [Google Scholar] [CrossRef]
- Holmes, S. C.; Gait, M. J. Syntheses and oligonucleotide incorporation of nucleoside analogues containing pendant imidazolyl or amino functionalities - The search for sequence-specific artificial ribonucleases. Eur. J. Org. Chem. 2005, 5171–5183. [Google Scholar] [CrossRef]
- Bashkin, J. K.; Gard, J. K.; Modak, A. S. Synthesis and Characterization of Nucleoside Peptides - toward Chemical Ribonucleases .1. J. Org. Chem. 1990, 55, 5125–5132. [Google Scholar] [CrossRef]
- Bashkin, J. K.; Mcbeath, R. J.; Modak, A. S.; Sample, K. R.; Wise, W. B. Synthesis and Characterization of Oligonucleotide Peptides. J. Org. Chem. 1991, 56, 3168–3176. [Google Scholar] [CrossRef]
- Zaramella, S.; Stromberg, R.; Yeheskiely, E. Stability studies of N-acylimidazoles. Eur. J. Org. Chem. 2002, 2633–2639. [Google Scholar] [CrossRef]
- Verbeure, B.; Lacey, C. J.; Froeyen, M.; Rozenski, J.; Herdewijn, P. Synthesis and cleavage experiments of oligonucleotide conjugates with a diimidazole-derived catalytic center. Bioconj. Chem. 2002, 13, 333–350. [Google Scholar]
- Lermer, L.; Roupioz, Y.; Ting, R.; Perrin, D. M. Toward an RNaseA mimic: a DNAzyme with imidazoles and cationic amines. J. Am. Chem. Soc. 2002, 124, 9960–9961. [Google Scholar]
- Santoro, S. W.; Joyce, G. F.; Sakthivel, K.; Gramatikova, S.; Barbas, C. F. RNA cleavage by a DNA enzyme with extended chemical functionality. J. Am. Chem. Soc. 2000, 122, 2433–2439. [Google Scholar]
- Lapidot, Y.; Rappoport, S.; Wolman, Y. Use of esters of N-hydroxysuccinimide in the synthesis of N-acylamino acids. J. Lipid Res. 1967, 8, 142–145. [Google Scholar]
- Purwanto, M. G. M.; Lengeler, D.; Weisz, K. Nucleosides derived from urocanic acid: potential ligands for CG base pairs. Tetrahedron Lett. 2002, 43, 61–64. [Google Scholar] [CrossRef]
- Winkler, J.; Urban, E.; Losert, D.; Wacheck, V.; Pehamberger, H.; Noe, C. A novel concept for ligand attachment to oligonucleotides via a 2'-succinyl linker. Nucleic Acids Res. 2004, 32, 710–718. [Google Scholar] [CrossRef]
- Berthod, T.; Petillot, Y.; Guy, A.; Cadet, J.; Molko, D. Synthesis of oligonucleotides containing 5-carboxy-2'-deoxyuridine at defined sites. J. Org. Chem. 1996, 61, 6075–6078. [Google Scholar] [CrossRef]
- Guerniou, V.; Gasparutto, D.; Sauvaigo, S.; Favier, A.; Cadet, J. New synthesis of 5-carboxy-2 '-deoxyuridine and its incorporation into synthetic oligonucleotides. Nucleos. Nucleot. Nuc. 2003, 22, 1073–1075. [Google Scholar] [CrossRef]
- Kachalova, A.; Zubin, E.; Stetsenko, D.; Gait, M.; Oretskaya, T. Oligonucleotides with 2 '-O-carboxymethyl group: synthesis and 2 '-conjugation via amide bond formation on solid phase. Org. Biomol. Chem. 2004, 2, 2793–2797. [Google Scholar] [CrossRef]
- Grandas, A.; Marchan, V.; Debéthune, L.; Pedroso, E. Stepwise solid-phase synthesis of nucleopeptides. Curr. Prot. Nucl. Acid Chem. 2004, 4.22.1–4.22.54. [Google Scholar]
- Lioy, E.; Suarez, J.; Guzman, F.; Siegrist, S.; Pluschke, G.; Patarroyo, M. E. Synthesis, biological, and immunological properties of cyclic peptides from Plasmodium falciparum merozoite surface protein-1. Angew. Chem. Int. Ed. 2001, 40, 2631–2635. [Google Scholar] [CrossRef]
- Kawai, K.; Kawabata, K.; Tojo, S.; Majima, T. Synthesis of ODNs containing 4-methylamino-1,8-naphthalimide as a fluorescence probe in DNA. Bioorg. Med. Chem. Lett. 2002, 12, 2363–2366. [Google Scholar] [CrossRef]
- Zatsepin, T. S.; Stetsenko, D. A.; Gait, M. J.; Oretskaya, T. S. Synthesis of DNA conjugates by solid-phase fragment condensation via aldehyde-nucleophile coupling. Tetrahedron Lett. 2005, 46(18), 3191–3195. [Google Scholar] [CrossRef]
- Casimir, J. R.; Turetta, C.; Ettouati, L.; Paris, J. First Application of the Dakin-West Reaction to Fmoc Chemistry - Synthesis of the Ketomethylene Tripeptide Fmoc-N-Alpha-Asp(Tbu)-(R,S)Tyr(Tbu)Psi(Co-Ch2)Gly-Oh. Tetrahedron Lett. 1995, 36, 4797–4800. [Google Scholar]
- Chenna, A.; Singer, B. Large-Scale Synthesis of P-Benzoquinone-2'-Deoxycytidine and P-Benzoquinone-2'-Deoxyadenosine Adducts and Their Site-Specific Incorporation into DNA Oligonucleotides. Chem. Res. Toxicol. 1995, 8, 865–874. [Google Scholar] [CrossRef]
- Chenna, A.; Singer, B. Synthesis of a benzene metabolite adduct, 3''-hydroxy-1,N-2-benzetheno-2'-deoxyguanosine, and its site-specific incorporation into DNA oligonucleotides. Chem. Res. Toxicol. 1997, 10, 165–171. [Google Scholar] [CrossRef]
- Lawrence, A. J.; Pavey, J. B. J.; Cosstick, R.; ONeil, I. A. Synthesis and properties of 2'-deoxy-2'-alpha-C-branched nucleosides and nucleotides. J. Org. Chem. 1996, 61, 9213–9222. [Google Scholar]
- Pavey, J. B. J.; Cosstick, R.; O'Neill, I. A. The synthesis of a novel 2 ',3 '-cyclic phosphate derived from 2 '-homouridine. Tetrahedron Lett. 1998, 39, 8923–8924. [Google Scholar] [CrossRef]
- Pavey, J. B. J.; Lawrence, A. J.; Potter, A. J.; Cosstick, R.; O'Neil, I. A. The synthesis of 2 '-homouridine, its incorporation into a dinucleoside monophosphate and hydrolytic behaviour of the dimer. Tetrahedron Lett. 1998, 39, 6967–6970. [Google Scholar] [CrossRef]
- Lawrence, A.; Pavey, J.; O’Neil, I.; Cosstick, R. Synthesis of Functionalised 2’-C-Branched Nucleosides via Their γ-Butyrolactones. Tetrahedron Lett. 1995, 36, 6341–6344. [Google Scholar] [CrossRef]
- Asseline, U.; Chassignol, M.; Draus, J.; Durand, M.; Maurizot, J. Synthesis and Properties of Oligo-2’-deoxyribonucleotides Containing Internucleotidic Phosphoramidate Linkages Modified with Pendant Groups Ending with either Two Amino or Two Hydroxyl Fucntions. Bioorg. Med. Chem. 2003, 11, 3499–3511. [Google Scholar] [CrossRef]
- Gefflaut, T.; Blonski, C.; Perie, J. Slow reversible inhibitions of rabbit muscle aldolase with substrate analogues: Synthesis, enzymatic kinetics and UV difference spectroscopy studies. Bioorg. Med. Chem. 1996, 4, 2043–2054. [Google Scholar] [CrossRef]
- Sample Availability: No samples available
© 2007 by MDPI (http://www.mdpi.org). Reproduction is permitted for noncommercial purposes.
Share and Cite
Catry, M.A.; Madder, A. Synthesis of Functionalised Nucleosides for Incorporation into Nucleic Acid-Based Serine Protease Mimics. Molecules 2007, 12, 114-129. https://doi.org/10.3390/12010114
Catry MA, Madder A. Synthesis of Functionalised Nucleosides for Incorporation into Nucleic Acid-Based Serine Protease Mimics. Molecules. 2007; 12(1):114-129. https://doi.org/10.3390/12010114
Chicago/Turabian StyleCatry, Mieke A., and Annemieke Madder. 2007. "Synthesis of Functionalised Nucleosides for Incorporation into Nucleic Acid-Based Serine Protease Mimics" Molecules 12, no. 1: 114-129. https://doi.org/10.3390/12010114



