Complexes of 3dn Metal Ions with Thiosemicarbazones: Synthesis and Antimicrobial Activity
Abstract
:Introduction
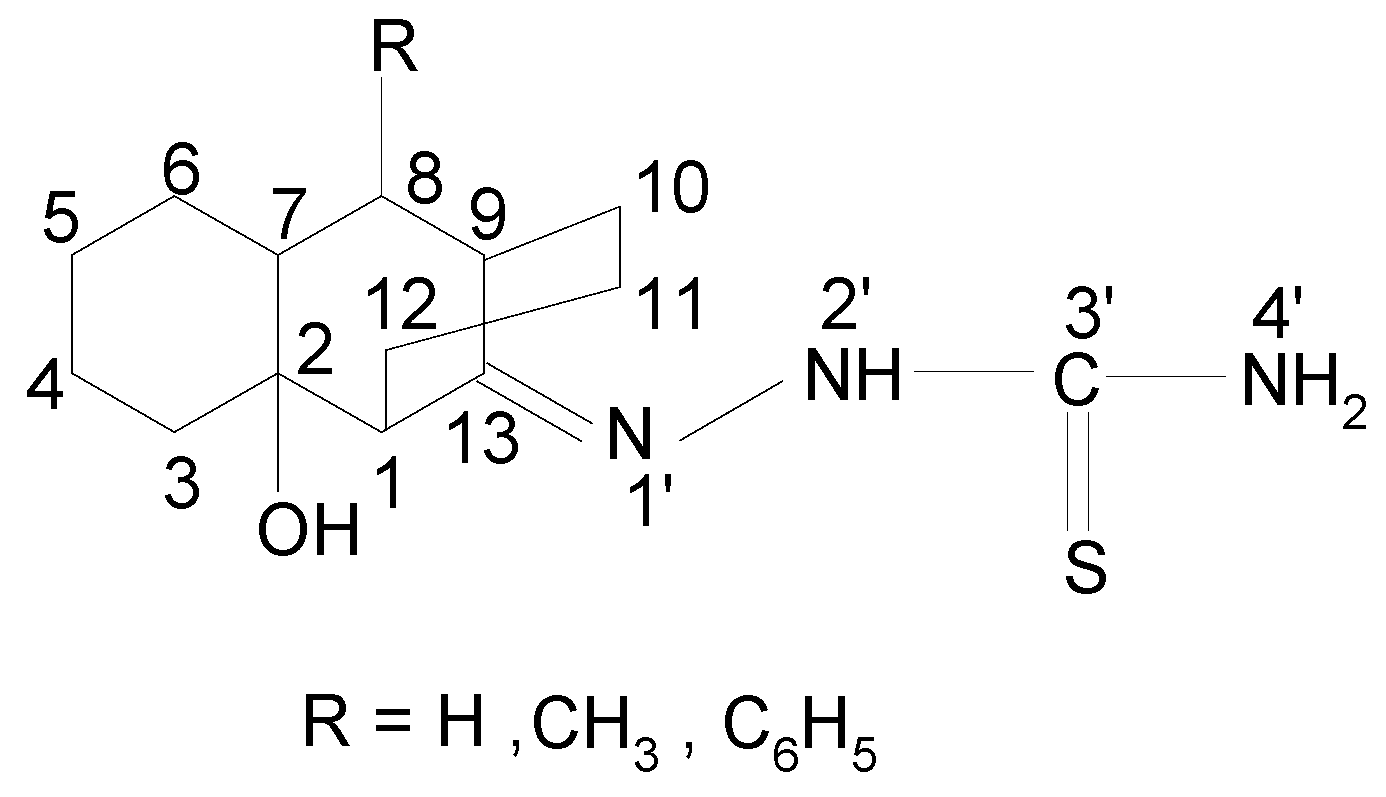
Results and Discussion
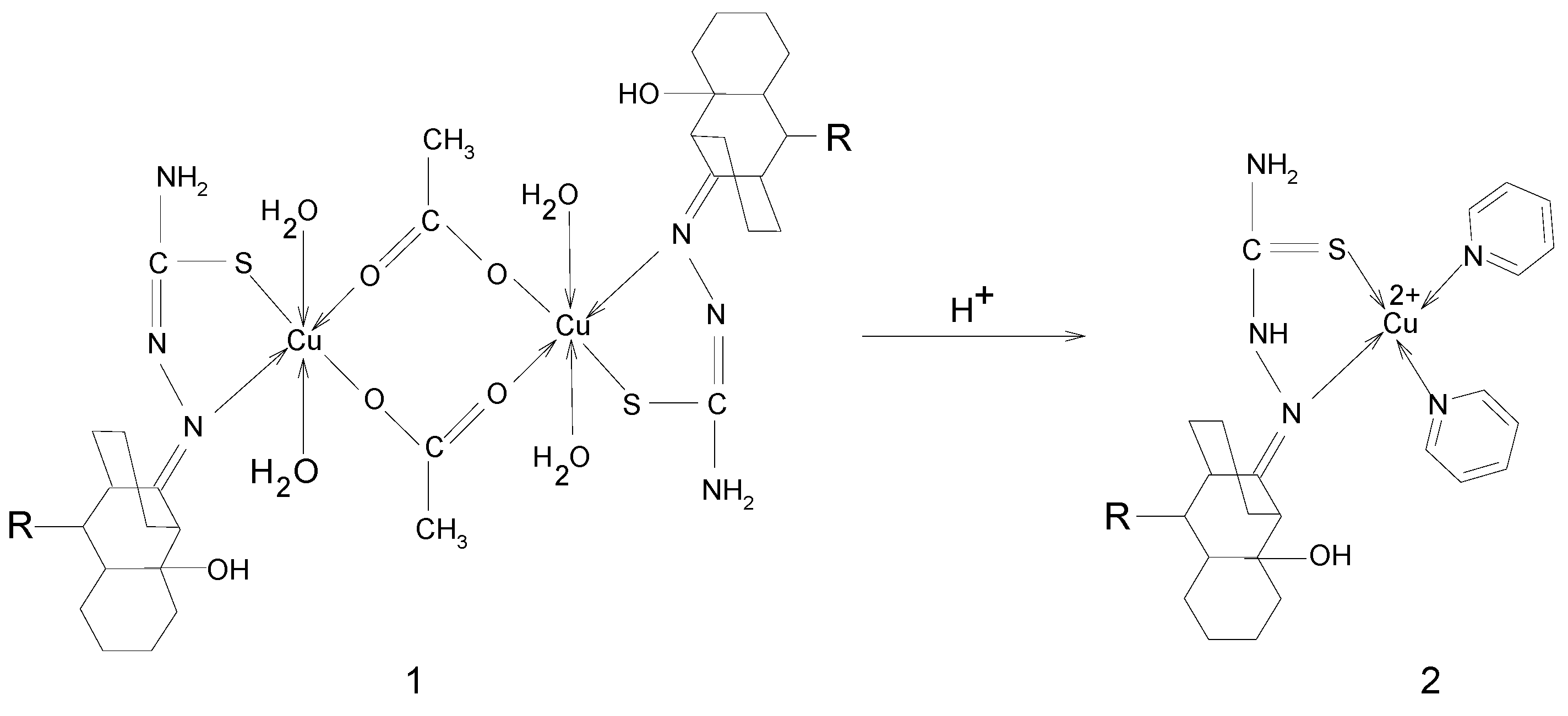
Synthesis
Properties
Thermal Decomposition
IR Spectra
| Compound | Temperature range (oC) | Eliminated fragment | Weight loss % |
|---|---|---|---|
| [M(LH)2]Cl2 | 220-355 | Cl2+LH | 49.50-51.00 |
| 430-680 | LH | 39.00-41.00 | |
| 680 | MO | 10.70-12.50 | |
| M(LCH3)2] | 240-420 | LCH3 | 44.00-46.00 |
| 450-685 | LCH3 | 44.00-45.00 | |
| 685 | MO | 11.00-13.00 | |
| [M(LC6H5)2] | 248-450 | LC6H5 | 45.93-46.00 |
| 453-690 | LC6H5 | 45.93-46.20 | |
| 690 | MO | 9.50-10.25 | |
| [M(LH)(H2O)2ac] | 112-137 | H2O | 7.50-8.50 |
| 174-346 | Ac | 12.93-13.70 | |
| 452-657 | LH | 63.00-64.23 | |
| 657 | MO | 16.90-18.45 | |
| [M(LCH3)(H2O)2ac] | 115-145 | H2O | 7.40-8.50 |
| 180-354 | Ac | 12.65-13.47 | |
| 463-668 | LCH3 | 64.10-65.23 | |
| 668 | MO | 16.43-17.85 | |
| [M(LC6H5)(H2O)2ac] | 119-153 | H2O | 6.73-7.55 |
| 182-357 | Ac | 11.00-12.32 | |
| 467-697 | LC6H5 | 69.86-72.13 | |
| 697 | MO | 14.20-15.69 |
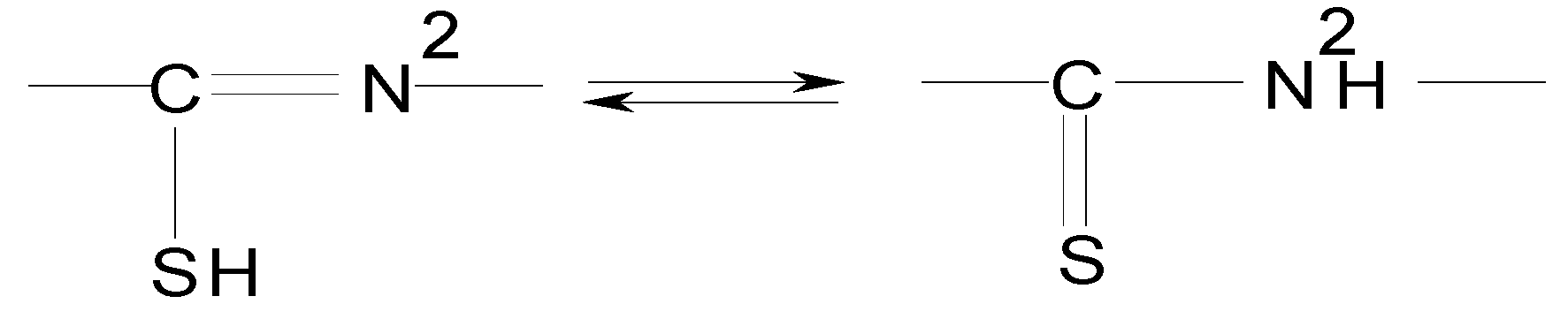
| Compound | νC=N1’ | νN1’-N2’ | νN2’H | νN4’H | νC=S; νC=S + νC=O(s) | νC=O(as) νC=N | νC=N2’ | νC-S | νC-O | νOH |
|---|---|---|---|---|---|---|---|---|---|---|
| LH | 1599 | 1016 | 3230 | 3319 | 740 | - | - | - | - | 3426 |
| 1291 | 1144 | |||||||||
| [M(LH) 2]Cl2 | ||||||||||
| (CuC28H48N6O2S2)Cl2 | 1556 | 1056 | 3232 | 3322 | 712 | - | - | - | - | 3424 |
| 1315 | 1143 | |||||||||
| (CoC28H48N6O2S2)Cl2 | 1564 | 1060 | 3229 | 3318 | 716 | - | - | - | - | 3425 |
| 1320 | 1142 | |||||||||
| (NiC28H48N6O2S2)Cl2 | 1560 | 1058 | 3233 | 3320 | 718 | - | - | - | - | 3420 |
| 1317 | 1140 | |||||||||
| LCH3 | 1621 | 1060 | 3213 | 3330 | 782 | - | - | - | - | 3425 |
| 1296 | 1140 | |||||||||
| [M(LCH3) 2] | ||||||||||
| Cu(C30H50N6O2 S2) | 1580 | 1094 | - | 3329 | - | - | 1595 | 547 | - | 3422 |
| 1142 | ||||||||||
| Co(C30H50N6O2S2) | 1578 | 1103 | - | 3332 | - | - | 1598 | 562 | - | 3420 |
| 1138 | ||||||||||
| Ni(C30H50N6O2S2) | 1575 | 1098 | - | 3328 | - | - | 1593 | 554 | - | 3424 |
| 1138 | ||||||||||
| LC6H5 | 1633 | 1075 | 3191 | 3338 | 798 | - | - | - | - | 3423 |
| 1302 | 1132 | |||||||||
| [M(LC6H5) 2] | ||||||||||
| Cu(C40H54N6S2) | 1591 | 1108 | - | 3340 | - | - | 1615 | 590 | - | 3430 |
| 1138 | ||||||||||
| Co(C40H54N6O2S2) | 1593 | 1113 | - | 3336 | - | - | 1619 | 601 | - | 3426 |
| 1130 | ||||||||||
| Ni(C40H54N6O2S2) | 1586 | 1109 | - | 3340 | - | - | 1613 | 595 | - | 3428 |
| 1136 | ||||||||||
| [MLH (H2O)2ac] | ||||||||||
| Cu(C16H30N3O5S) | 1559 | 1047 | - | 3325 | - | 1532 | 1586 | 552 | - | 3418 |
| 1469 | 1137 | |||||||||
| Co(C16H30N3O5S) | 1568 | 1050 | - | 3321 | - | 1526 | 1589 | 571 | - | 3419 |
| 1452 | 1138 | |||||||||
| Ni(C16H30N3O5S) | 1565 | 1049 | - | 3323 | - | 1517 | 1581 | 564 | - | 3415 |
| 1448 | 1139 | |||||||||
| [MLCH 3(H2O)2ac] | ||||||||||
| Cu(C17H32N3O5S) | 1572 | 1078 | - | 3332 | - | 1528 | 1603 | 562 | - | 3420 |
| 1462 | 1141 | |||||||||
| Co(C17H32N3O5S) | 1569 | 1087 | - | 3336 | - | 1521 | 1598 | 567 | - | 3419 |
| 1448 | 1139 | |||||||||
| Ni(C17H32N3O5S) | 1567 | 1082 | - | 3334 | - | 1514 | 1592 | 561 | - | 3422 |
| 1437 | 1137 | |||||||||
| [MLC6H 5(H2O)2ac] | 1583 | 1098 | - | 3342 | - | 1522 | 1608 | 583 | - | 3425 |
| Cu(C22H34N3O5S) | 1458 | 1135 | ||||||||
| Co(C22H34N3O5S) | 1580 | 1101 | - | 3339 | - | 1518 | 1602 | 594 | - | 3430 |
| 1442 | 1134 | |||||||||
| Ni(C22H34N3O5S) | 1578 | 1097 | - | 3341 | - | 1509 | 1595 | 580 | - | 3428 |
| 1431 | 1137 |
Electronic spectra
| Compound | Transitions d - d | B | β | Dq | Geometry | ||
|---|---|---|---|---|---|---|---|
| [Cu(LH)2] Cl2 | - | - | - | Pseudo-tetrahedral | |||
| [Cu(LCH3)2] | Pseudo-tetrahedral | ||||||
| [Co(LH)2] Cl2 | 4A2 → 4T1(P) | 919 | 0,947 | Pseudo-tetrahedral | |||
| [Co(LCH3)2] | 13030 | 808 | 0,833 | - | Pseudo-tetrahedral | ||
| [Ni(LH)2] Cl2 | ν3 | ||||||
| [Ni(LCH3)2] | 24350 | Sq - pl | |||||
| [CuLR(H2O)2ac] | |||||||
| [CoLR(H2O)2ac] | 4T1 g → 4T1 g (P) | ||||||
| [NiLR(H2O)2ac] | 3A2 g → 4T1 g (P) | ||||||
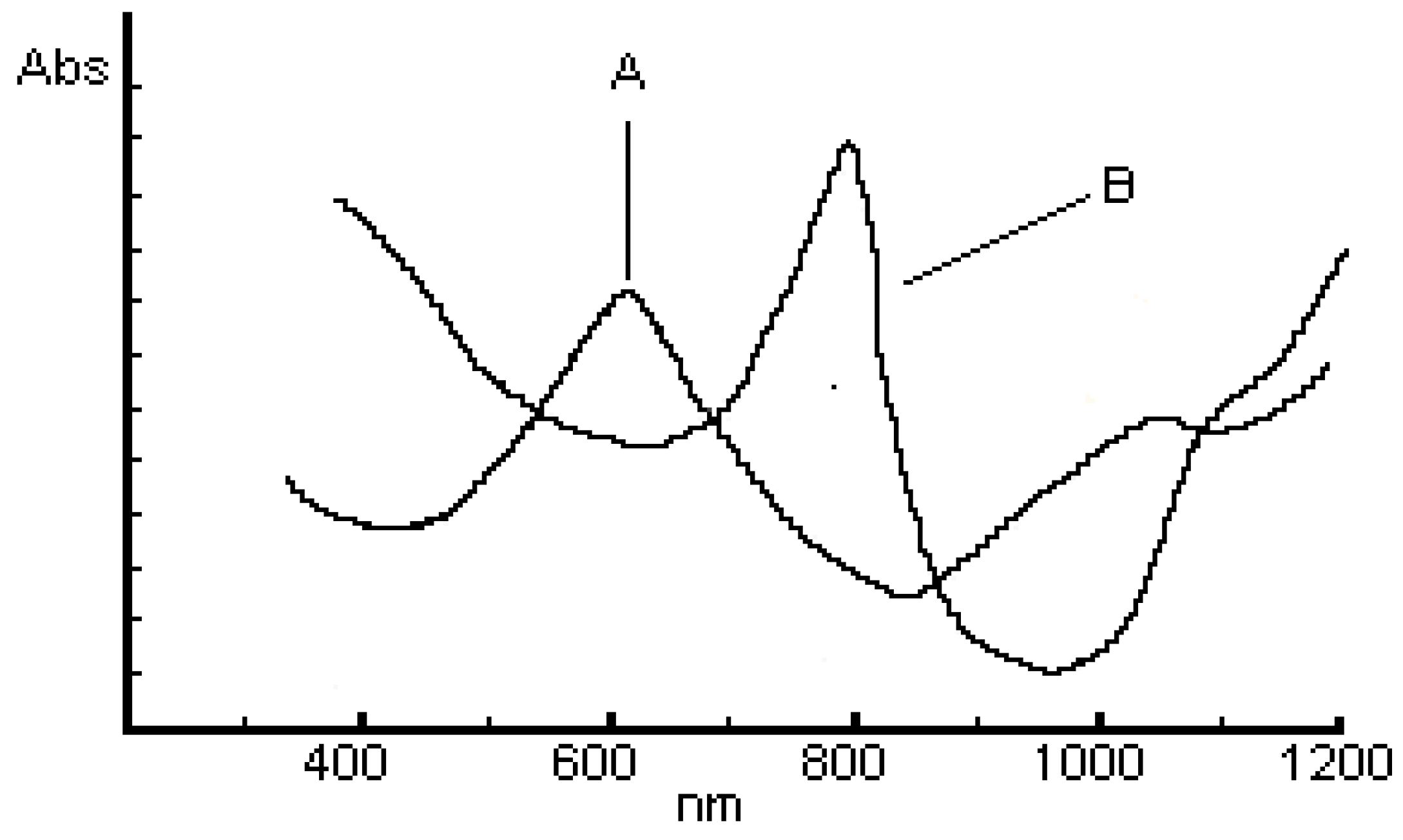
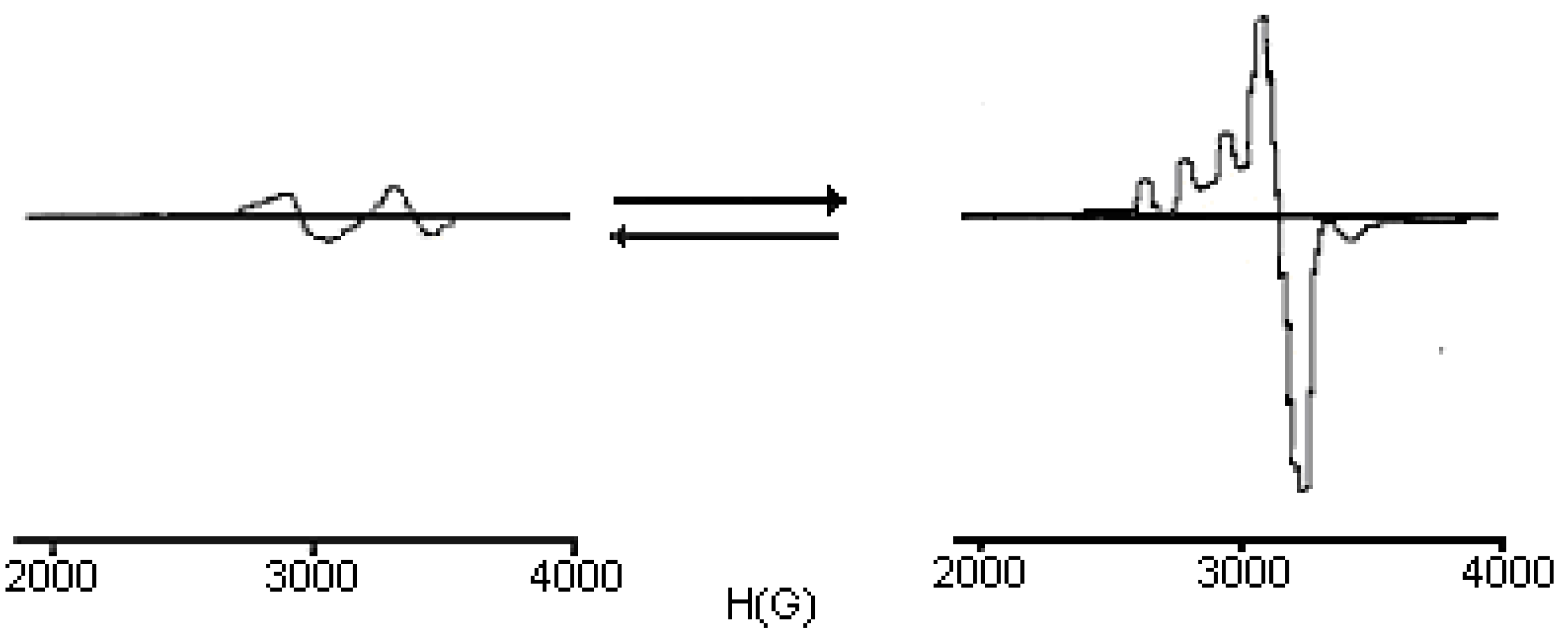
ESR Spectra
| Compound | gx | gy | gz |
|---|---|---|---|
| [Cu(LH)2]Cl2 | 2.054 | 2.107 | 2.211 |
| [Cu(LCH3)2] | 2.035 | 2.115 | 2.203 |
| [Cu(LC6H5)2] | 2.040 | 2.116 | 2.210 |
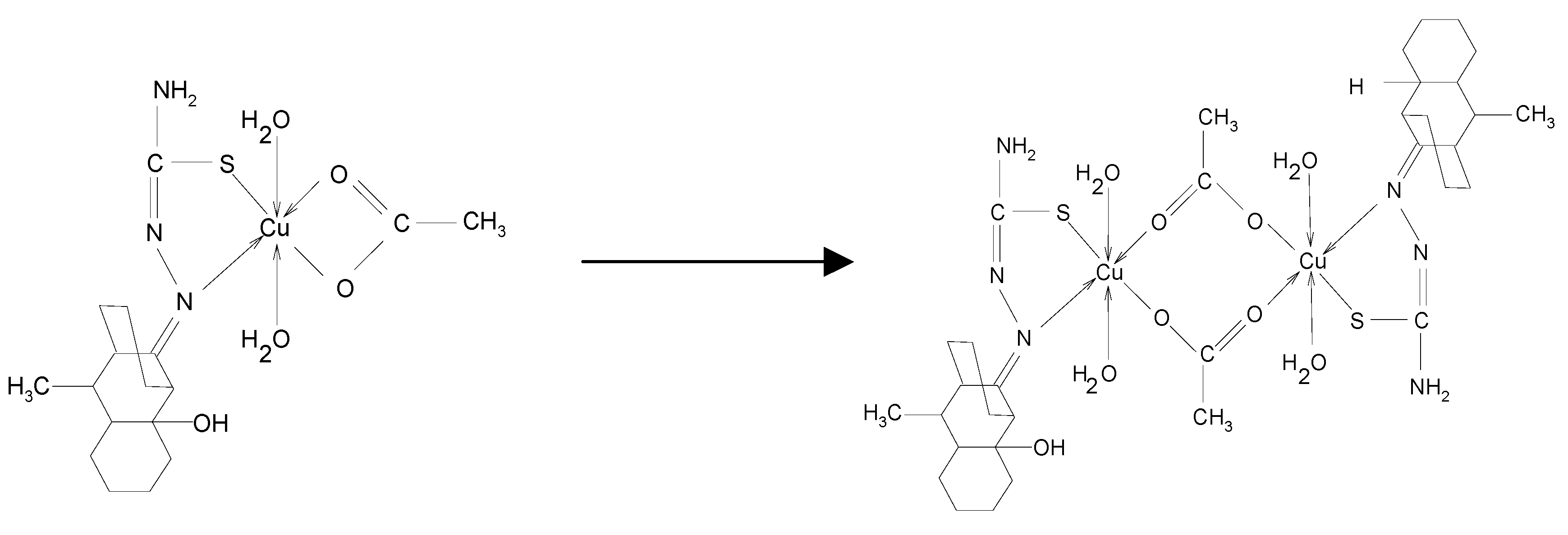
Antimicrobial activity assays
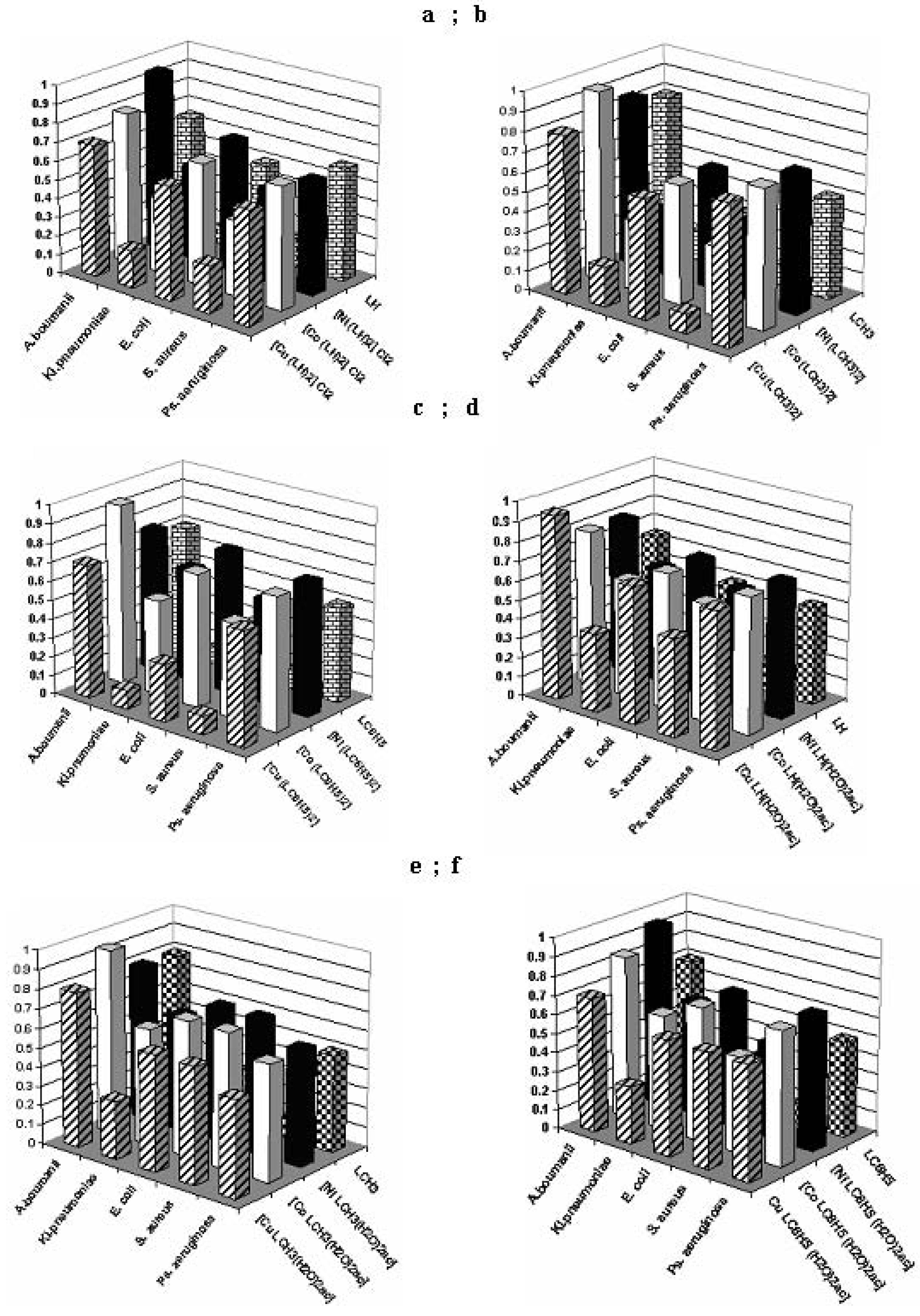
Conclusions
- ■
- the qualitative anti-microbial activity screening results of the tested compounds proved that the most efficient method is the spot method;
- ■
- the quantitative anti-microbial activity test results proved that both the ligands and the complex combinations have specific anti-microbial activity, depending on the microbial species tested;
- ■
- the complex combinations of the [MLR (H2O)2ac] type, except for the [CuLR (H2O)2ac] complex, show very low activity towards the tested bacterial strains.
Experimental
General
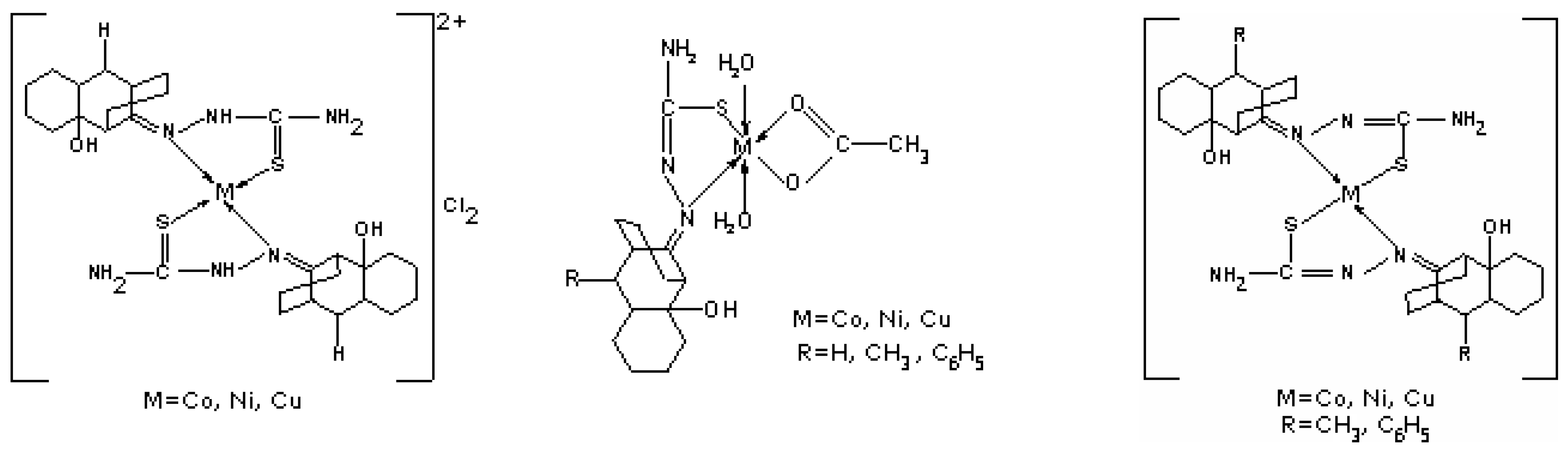
Synthesis of [M(LH)2]Cl2 and [M(LR)2] complexes; M = Cu(II), Ni(II), Co(II); L= 2-hydroxy-8-R-tricyclo[7.3.1.0.2,7]tridecane-13-one-thiosemicarbazone; R = CH3, C6H5
Synthesis of [MLR(H2O)2ac] complexes; M = Cu(II), Ni(II), Co(II)
| Compound | %H | %C | %N | %M | %Cl | Color | Dec. | Yield |
|---|---|---|---|---|---|---|---|---|
| exp. | exp. | exp. | exp. | exp. | Point | |||
| (calc) | (calc) | (calc) | (calc) | (calc) | (0C) | (%) | ||
| [M(LH)2]Cl2 | ||||||||
| (CuC28H48N6O2S2)Cl2 | 6.31 | 48.66 | 12.25 | 9.22 | 10.03 | light-green | 218 | 81 |
| (698.5) | (6.87) | (48.10) | (12.02) | (9.11) | (10.16) | |||
| (CoC28H48N6O2S2)Cl2 | 6.25 | 48.85 | 12.43 | 8.63 | 10.10 | dark-green | 245 | 79 |
| (694) | (6.91) | (48.41) | (12.10) | (8.50) | (10.23) | |||
| (NiC28H48N6O2S2)Cl2 | 6.40 | 48.80 | 12.50 | 8.42 | 10.18 | brown | 231 | 85 |
| (694) | (6.91) | (48.41) | (12.10) | (8.50) | (10.23) | |||
| [M(LCH3) 2] | ||||||||
| Cu(C30H50N6O2S2) | 7.30 | 55.40 | 12.56 | 9.62 | light-green | 239 | 67 | |
| (653.5) | (7.65) | (55.08) | (12.85) | (9.71) | ||||
| Co(C30H50N6O2S2) | 7.28 | 55.87 | 12.72 | 8.90 | dark-green | 261 | 72 | |
| (649) | (7.70) | (55.46) | (12.94) | (9.09) | ||||
| Ni(C30H50N6O2S2) | 7.31 | 55.73 | 12.67 | 8.89 | brown | 259 | 75 | |
| (649) | (7.70) | (55.46) | (12.94) | (9.09) | ||||
| [M(LC6H5) 2] | ||||||||
| Cu(C40H54N6O2 S2) | 6.58 | 62.18 | 10.48 | 8.00 | light-green | 245 | 63 | |
| (777.5) | (6.94) | (61.76) | (10.80) | (8.16) | ||||
| Co(C40H54N6O2S2) | 6.54 | 62.48 | 10.53 | 7.50 | dark- green | 263 | 62 | |
| (773) | (6.98) | (62.09) | (10.86) | (7.63) | ||||
| Ni(C40H54N6O2S2) | 6.57 | 62.57 | 10.66 | 7.52 | brown | 260 | 68 | |
| (773) | (6.98) | (62.09) | (10.86) | (7.63) | ||||
| [MLH (H2O)2ac] | ||||||||
| Cu(C16H30N3O5S) | 6.59 | 43.91 | 9.68 | 14.27 | dark-green | 179 | 87 | |
| (439.5) | (6.83) | (43.68) | (9.55) | (14.44) | ||||
| Co(C16H30N3O5S) | 6.47 | 44.58 | 9.38 | 13.40 | dark-green | 163 | 79 | |
| (435) | (6.89) | (44.13) | (9.65) | (13.56) | ||||
| Ni(C16H30N3O5S) | 6.63 | 44.52 | 9.43 | 13.39 | green | 197 | 81 | |
| (435) | (6.89) | (44.13) | (9.65) | (13.56) | ||||
| [MLCH 3(H2O)2ac] | ||||||||
| Cu(C17H32N3O5S) | 6.80 | 45.31 | 9.51 | 13.87 | green | 198 | 86 | |
| (453.5) | (7.05) | (44.98) | (9.26) | (14.00) | ||||
| Co(C17H32N3O5S) | 6.84 | 45.94 | 9.07 | 13.00 | dark-green | 198 | 83 | |
| (449) | (7.12) | (45.43) | (9.35) | (13.14) | ||||
| Ni(C17H32N3O5S) | 6.75 | 45.87 | 9.11 | 12.98 | green | 208 | 85 | |
| (449) | (7.12) | (45.43) | (9.35) | (13.14) | ||||
| [MLC6H 5(H2O)2ac] | green | |||||||
| Cu(C22H34N3O5S) | 6.27 | 51.63 | 8.37 | 12.02 | 194 | 73 | ||
| (515.5) | (6.59) | (51.21) | (8.14) | (12.13) | ||||
| Co(C22H34N3O5S) | 6.32 | 51.97 | 8.00 | 11.37 | dark-green | 185 | 79 | |
| (511) | (6.65) | (51.66) | (8.21) | (11.54) | ||||
| Ni(C22H34N3O5S) | 6.37 | 51.86 | 7.93 | 11.45 | green | 202 | 82 | |
| (511) | (6.65) | (51.66) | (8.21) | (11.54) |
Biological assays
Supplementary Materials
Supplementary File 1Acknowledgements
References
- Collins, F. M.; Klayman, D. L.; Morrison, N. E. Correlation between structure and anti-mycobacterial activity in a series of 2-acetylpyridine thiosemicarbazones. J. Gen. Microbiol. 1982, 128, 1349–1356. [Google Scholar] [PubMed]
- Williams, D. R. Metals, ligands and cancer. Chem. Rev. 1972, 72, 203–213. [Google Scholar] John, R. P.; Sreekanth, A.; Rajakannan, V.; Ajith, T. A.; Kurup, M. R. P. New copper(II) complexes thiosemicarbazones and polypyridyl co-ligands: structural, electrochemical of 2-hydroxyacetophenone N(4)-substituted and antimicrobial studies. Polyhedron 2004, 23, 2549–2559. [Google Scholar] Belicchi-Ferrari, M.; Bisceglie, F.; Casoli, C.; Durot, S.; Morgenstern-Badarau, I.; Pelosi, G.; Pilotti, E.; Pinelli, S.; Tarasconi, P. Copper(II) and Cobalt(III) Pyridoxal Thiosemicarbazone Complexes with Nitroprusside as Counterion: Syntheses, Electronic Properties, and Antileukemic Activity. J. Med. Chem 2005, 48(5), 1671–1675. [Google Scholar] Gulea, A.; Poirier, D.; Roy, J.; Spinu, S.; Coziri, N.; Birca, M.; Tapcov, V. Anticancer effect of sulfanilamide complexes of Cu (II) with thiosemicarbazones of substituted salicylic aldehydes. COF-rRoCA 2004, Actes du Colloque Franco-Roumain de Chimie Applique, 3rd Bacau, Romania, Sept.22-26 2004, 68(Fr). Rom ISBN: 973-8392-36-5. [Google Scholar] Birca, M.; Tapcov, V.; Prisacari, V.; Gulea, A.; Buraciova, S. Synthesis and antimicrobial properties of Cu complexes with thiosemicarbazones of substituted salicylic aldehydes. COFrRoCA 2004, Actes du Colloque Franco-Roumain de Chimie Applique, 3rdBacau, Romania, sept. 22-26 2004, 44-45(Fr). Rom ISBN: 973-8392-36-5. [Google Scholar]
- Kaska, W. C.; Carrans, C.; Michalowski, J.; Jackson, J.; Levinson, W. Inhibition of the RNA dependent DNA polymerase and the malignant transforming ability of Rous sarcoma virus by Thiosemicarbazone-transition metal complexes. Bioinorg. Chem. 1978, 8, 245–254. [Google Scholar]
- Klayman, D. L.; Bartosevich, J. E.; Griffith, T. S.; Mason, C. J.; Scovill, J. P. 2-Acetylpyridine thiosemicarbazones. 1. A new class of potential antimalarial agents. J. Med. Chem. 1979, 22, 855–862. [Google Scholar] Klayman, D. L.; Scovill, J. P.; Bartosevich, J. P.; Mason, C. J. 2-Acetylpyridine thiosemicarbazones. 2. N4,N4-Disubstituted derivatives as potential antimalarial agents. J. Med. Chem. 1979, 22, 1367–1373. [Google Scholar]
- Chan-Stier, C. H.; Minkel, D. T.; Petering, D. H. Reactions of Bis(thiosemicarbazonato) Copper(II) complexes with tumor cells and mitochondria. Bioinorg. Chem. 1976, 6, 203–217. [Google Scholar]
- Antholine, W.E.; Knight, J.M.; Petering, D. H. Inhibition of tumor cell transplantability by iron and copper complexes of 5-substituted 2-formylpyridine thiosemicarbazones. J. Med. Chem. 1976, 19, 336–341. [Google Scholar]
- Saryan, L. A.; Ankel, E.; Krishanamurti, C.; Petering, D. H.; Elford, H. Comparative cytotoxic and biochemical effects of alpha-N-heterocyclic carboxaldehyde thiosemicarbazones. J. Med. Chem. 1979, 22, 1218–1221. [Google Scholar]
- Karon, M.; Benedict, W.F. Chromatid Breakage: Cytosine Arabinoside-Induced Lesions Inhibited by Ultraviolet Irradiation. Science 1972, 178, 62. [Google Scholar]
- Wengel, A.; Kolind-Andersen, H.; Jacobsen, N.; Svendsen, A. K.; Klemmensen, P. D. A biocidal, particularly fungicidal and/or bactericidal composition and thiocarbazones and metal complexes thereof for use in the composition. PCT Int. Appl. W085/00955, 1985. [Google Scholar]
- West, D. X.; Liberta, E. A.; Padhye, S. B.; Chikate, R. C.; Sonawane, P. B.; Kumbhar, A. S.; Yerande, R. G. Thiosemicarbazone complexes of copper (II): structural and biological studies. Coord. Chem. Rev. 1993, 123, 49–71. [Google Scholar]
- Agarwal, R. K.; Singh, L.; Sharma, D. K. Synthesis, Spectral, and Biological Properties of Copper(II) Complexes of Thiosemicarbazones of Schiff Bases Derived from 4-Aminoantipyrine and Aromatic Aldehydes. Chem. Appl. 2006, 1–10. [Google Scholar]
- Nicolae, A.; Draghici, C. Semicarbazone si tiosemicarbazone in seria 2-hydroxy-8-R Tricycle[7.3.1.0.2,7]tridecane-13-one, substante cu potentiala activitate biologica. Rev. Chim. 2004, 55, 179–182. [Google Scholar]
- Dhumwad, S. D.; Gudasi, K. B.; Goudar, T. R. Synthesis and Structural Characterization of Biologically-Active Metal-Complexes of N(1)-(N-Morpholinoacetyl)-N(4)-Phenyl Thiosemi-carbazide and 3,4-Methylenedioxybenzaldehyde Thiosemicarbazone with Oxovanadium(IV), Chromium(III), Manganese(II), Iron(III), Cobalt(II), Nickel(II), Copper(II), Cadmium(II), Uranium(VI), Thorium(IV) and Silicon(IV). Indian J. Chem. 1994, 33A, 320–324. [Google Scholar]
- Nakamoto, K. Infrared Spectra of Inorganic and Coordination Compounds; J. Wiley and Sons: New York, 1986; p. 233. [Google Scholar]
- Lever, A. B. P. Inorganic Electronic Spectroscopy, 2nd Edn; Elsevier Science: New York, 1984. [Google Scholar]
- Mabbs, F. E.; Collison, D. Electron Paramagnetic Resonance of d Transition Metal Compounds; Elsevier: Amsterdam, 1992. [Google Scholar]
- Koolhaas, G. J. A. A. Cooper Coordination Complexes with poly-imidazole ligands. Ph.D. Thesis, University of Leiden, 1996. [Google Scholar]
- Abragam, A.; Bleaney, B. Electron Paramagnetic Resonance of Transition Ions; Claredon Press: London, 1970; p. 492. [Google Scholar]
- Hathaway, B. J. Comprehensive Coordination Chemistry; Wilkinson, G., Gillard, R.D., McCleverty, J.A., Eds.; Pergamon Press: London, 1987; vol. 5, p. 652. [Google Scholar]
- Rosu, T.; Pasculescu, S.; Lazar, V.; Chifiriuc, C.; Cernat, R. Copper(II) Complexes with Ligands Derived from 4-Amino-2,3-dimethyl-1-phenyl-3-pyrazolin-5-one: Synthesis and Biological Activity. Molecules 2006, 11, 904–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sample availability: Available from MDPI. See the supporting information.
© 2007 by MDPI (http://www.mdpi.org). Reproduction is permitted for noncommercial purposes.
Share and Cite
Rosu, T.; Gulea, A.; Nicolae, A.; Georgescu, R. Complexes of 3dn Metal Ions with Thiosemicarbazones: Synthesis and Antimicrobial Activity. Molecules 2007, 12, 782-796. https://doi.org/10.3390/12040782
Rosu T, Gulea A, Nicolae A, Georgescu R. Complexes of 3dn Metal Ions with Thiosemicarbazones: Synthesis and Antimicrobial Activity. Molecules. 2007; 12(4):782-796. https://doi.org/10.3390/12040782
Chicago/Turabian StyleRosu, Tudor, Aurelian Gulea, Anca Nicolae, and Rodica Georgescu. 2007. "Complexes of 3dn Metal Ions with Thiosemicarbazones: Synthesis and Antimicrobial Activity" Molecules 12, no. 4: 782-796. https://doi.org/10.3390/12040782





