Thermus thermophilus Strains Active in Purine Nucleoside Synthesis
Abstract
:Introduction
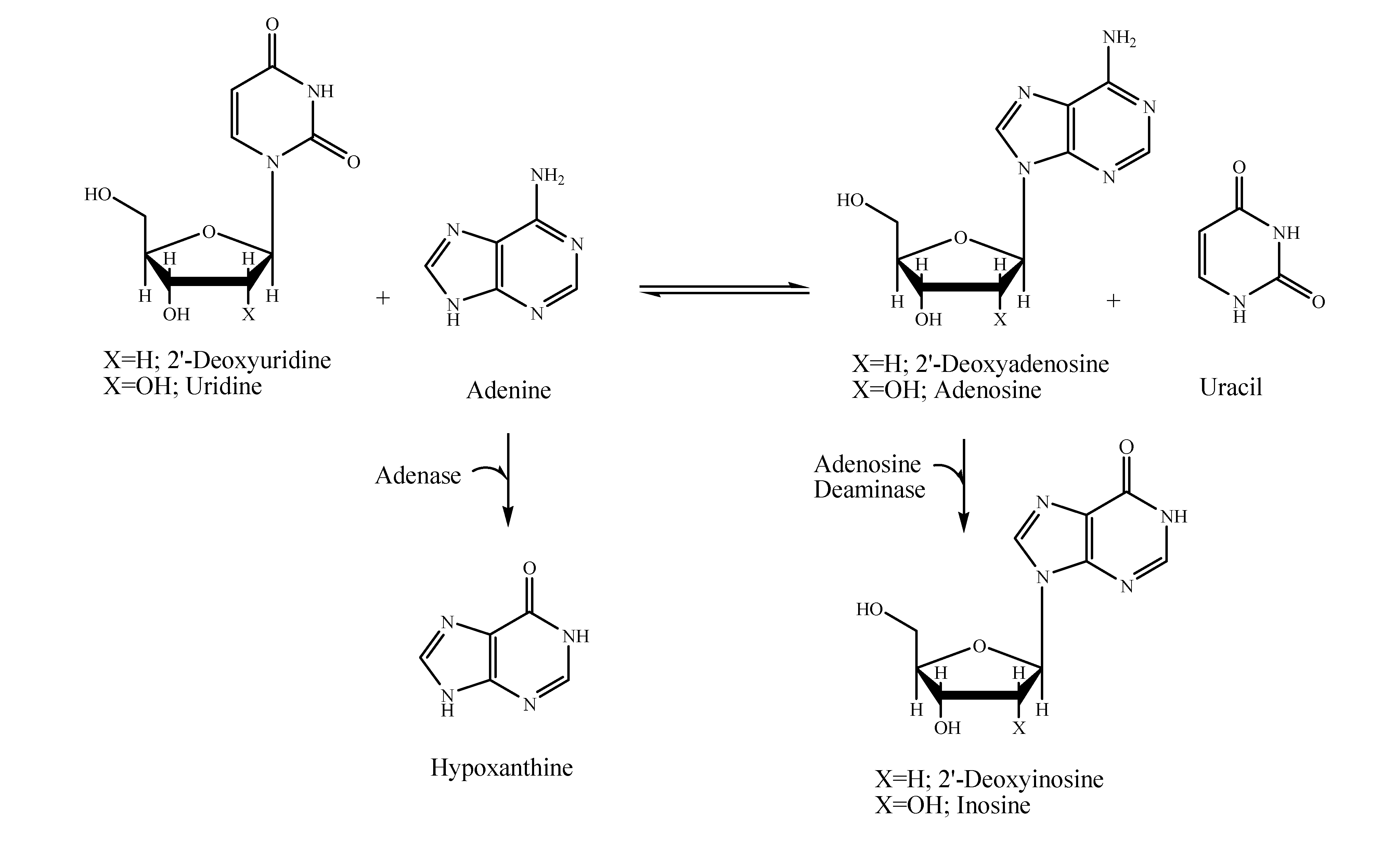
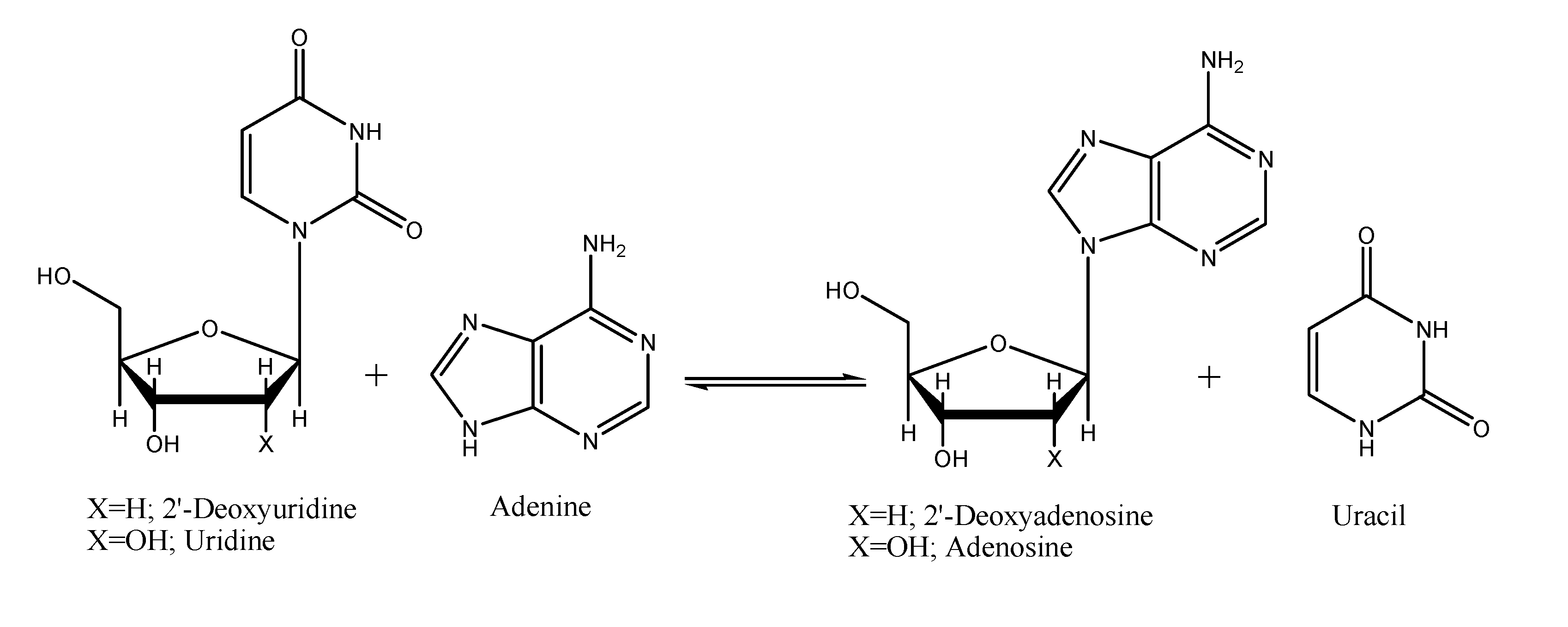
Results and Discussion
| Strain | 2’-Deoxyinosine synthesis | Inosine synthesis | ||||
|---|---|---|---|---|---|---|
| Yield (%) | [cells ]b | Productivityc | Yield (%) | [cells ]b | Productivityc | |
| NAR1 | 24 | 10,000 | 12 x 10-5 | 8.5 | 9,883 | 4.3 x 10-5 |
| HB27 | 28 | 6,364 | 22 x 10-5 | 14 | 6,364 | 11 x 10-5 |
| PRQ-16 | 24 | 6,316 | 19 x 10-5 | 11 | 6,111 | 9.0 x 10-5 |
| PRQ-25 | 25 | 6,250 | 20 x 10-5 | 11 | 5,978 | 9.2 x 10-5 |
| B | 24 | 7,059 | 17 x 10-5 | 11 | 7,142 | 7.7 x 10-5 |
| RQ1 | 20 | 10,417 | 9.6 x 10-5 | 8.7 | 10,610 | 4.1 x 10-5 |
| N17 | 21 | 7,500 | 14 x 10-5 | 10.5 | 7,500 | 7.0 x 10-5 |
| HN1.11 | 12 | 3,529 | 17 x 10-5 | 5.3 | 3,786 | 7.0 x 10-5 |
| Fiji3A1 | 16.5 | 2,750 | 30 x 10-5 | 8.3 | 2,767 | 15 x 10-5 |
| CC16 | 23 | 6,053 | 19 x 10-5 | 10 | 5,952 | 8.4 x 10-5 |
| VG7 | 22 | 2,683 | 41 x 10-5 | 13 | 2,708 | 24 x 10-5 |
| Strain | 2’-Deoxyadenosine synthesis | Adenosine synthesis | ||||
|---|---|---|---|---|---|---|
| Yield (%) | [cells] b | Productivity c | Yield (%) | [cells] b | Productivity c | |
| NAR1 | 30 | 10,714 | 14 x 10-5 | 15 | 10,563 | 7.1 x 10-5 |
| HB27 | 44 | 11,000 | 20 x 10-5 | 21 | 10,938 | 9.6 x 10-5 |
| PRQ-16 | 29 | 6,041 | 24 x 10-5 | 15 | 6,000 | 12.5 x 10-5 |
| PRQ-25 | 28 | 5,600 | 25 x 10-5 | 15 | 5,556 | 13.5 x 10-5 |
| B | 38 | 9,500 | 20 x 10-5 | 21 | 9,545 | 11 x 10-5 |
| RQ1 | 28 | 10,769 | 13 x 10-5 | 15 | 10,870 | 6.9 x 10-5 |
| N17 | 27 | 7,105 | 19 x 10-5 | 12 | 6,771 | 9.6 x 10-5 |
| HN1-11 | 15 | 5,769 | 13 x 10-5 | 6.9 | 5,847 | 5.9 x 10-5 |
| Fiji3A1 | 14 | 5,000 | 14 x 10-5 | 6.3 | 5,250 | 6.0 x 10-5 |
| CC16 | 30 | 6,818 | 22 x 10-5 | 14 | 6,667 | 10.5 x 10-5 |
| VG7 | 23 | 4,423 | 26 x 10-5 | 12 | 4,286 | 14 x 10-5 |
| Strain | Thymidine + hypoxantine | Thymidine + adenine | ||||
|---|---|---|---|---|---|---|
| Yield (%) | [cells]b | Productivityc | Yield (%) | [cells]b | Productivityc | |
| HB27 | 22 | 10,000 | 11 x 10-5 | 33 | 9,706 | 17 x 10-5 |
| PRQ-16 | 14 | 5,385 | 13 x 10-5 | 21 | 5,526 | 19 x 10-5 |
| PRQ-25 | 15 | 5,357 | 14 x 10-5 | 21 | 5,250 | 20 x 10-5 |
| B | 21 | 8,750 | 12 x 10-5 | 39.5 | 8,977 | 22 x 10-5 |
| Fiji3A1 | 12 | 5,455 | 11 x 10-5 | 16 | 5,333 | 15 x 10-5 |
| VG7 | 11.5 | 3,382 | 17 x 10-5 | 19 | 3,519 | 27 x 10-5 |
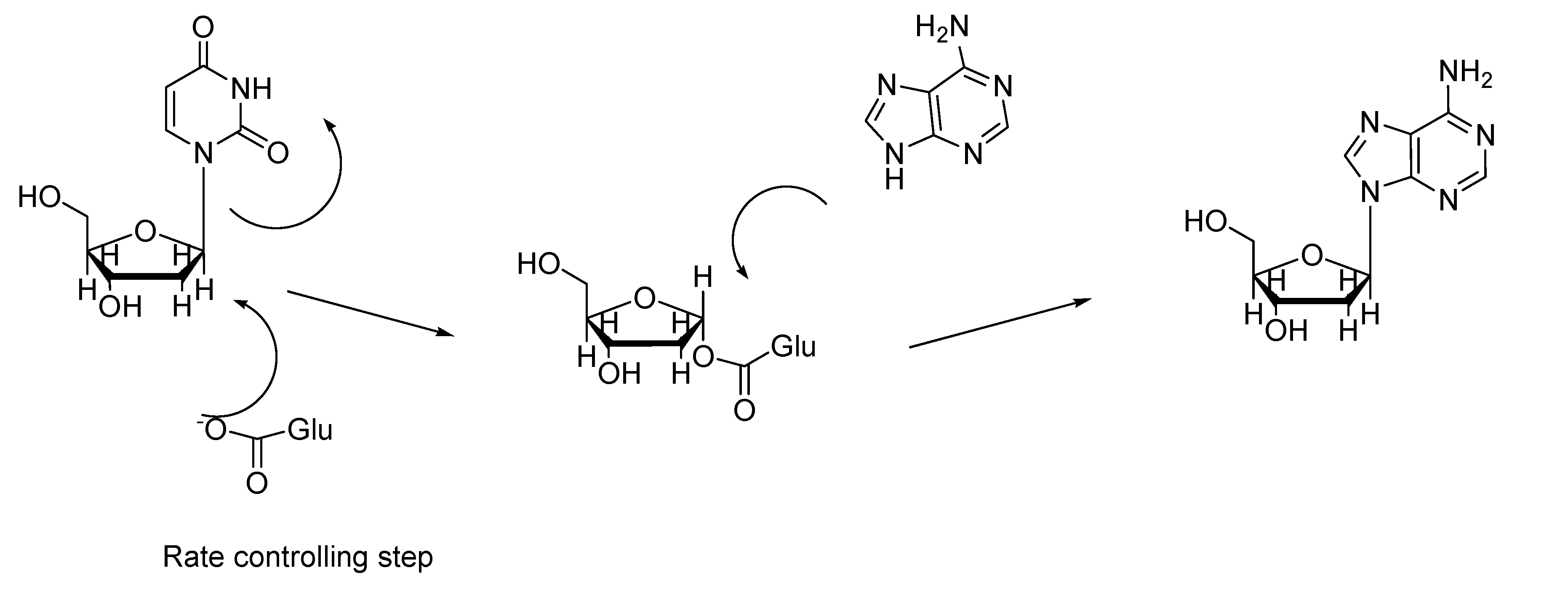
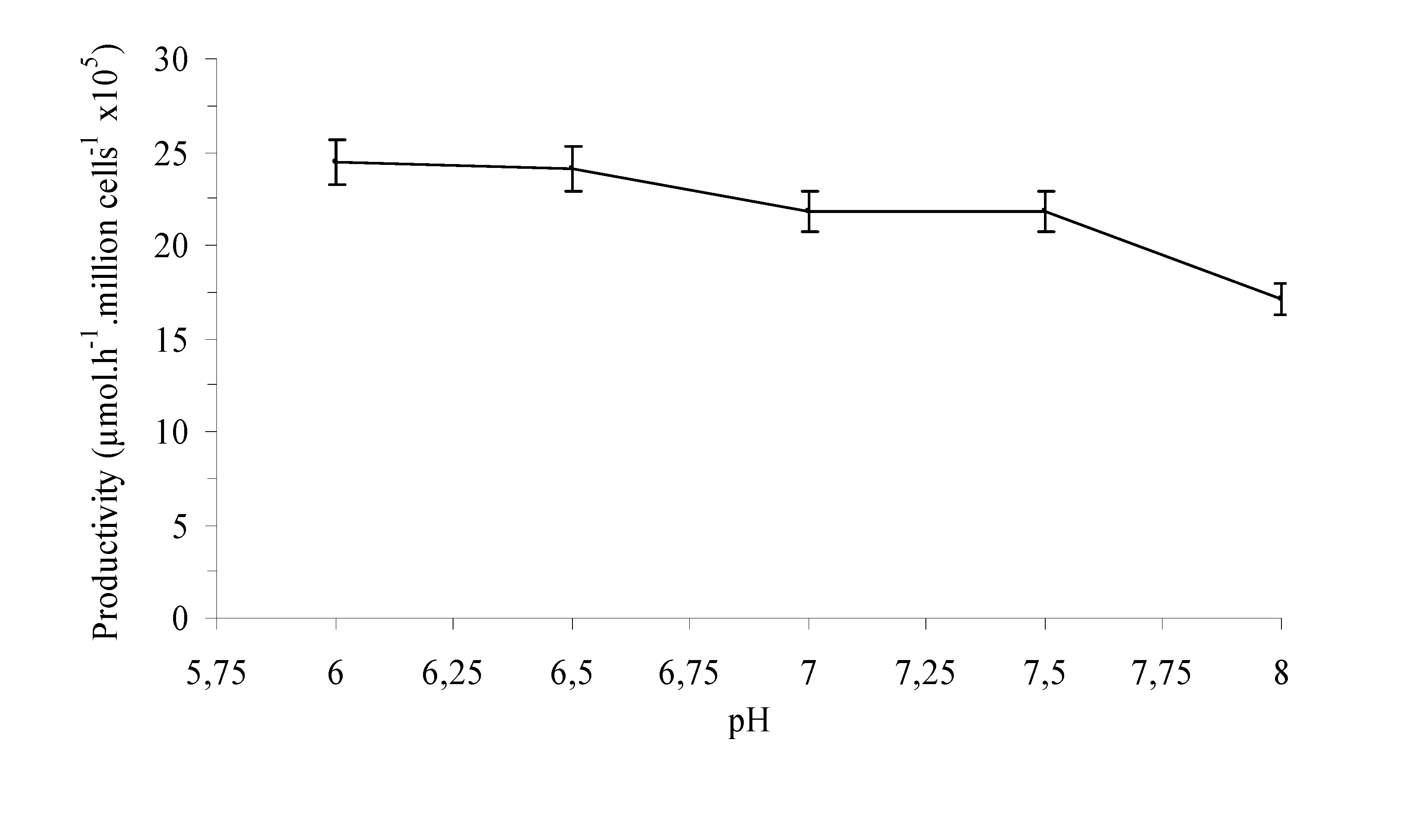

Experimental
Materials and Microorganisms
Standard synthesis of purine nucleosides
Synthesis in Tris/HCl buffer
Sample analysis
Acknowledgements
References and Notes
- Atomi, H. Recent progress towards the application of hyperthermophiles and their enzymes. Curr. Opin. Chem. Biol. 2005, 9, 166–173. [Google Scholar] [CrossRef]
- Domínguez, A.; Fuciños, P.; Rua, M. L; Pastrana, L; Longo, M.A.; Sanromán, M.A. Stimulation of novel thermostable extracellular lipolytic enzyme in cultures of Thermus sp. Enzyme Microb. Thechnol. 2007, 40, 187–194. [Google Scholar] [CrossRef]
- Mikhailopulo, I.A. Biotechnology of nucleic acid constituents- State of the art and perspectives. Curr. Org. Chem. 2007, 11, 317–338. [Google Scholar] [CrossRef]
- Prasad, A.K.; Trikha, S.; Parmar, V.S. Nucleoside synthesis mediated by glycosyl-transferring enzymes. Biorg. Chem. 1999, 27, 135–154. [Google Scholar] [CrossRef]
- Condezo, L.A.; Fernandez-Lucas, J.; Garcia-Burgos, C.A.; Alcántara, A.R.; Sinisterra, J.V. Enzymatic synthesis of modified nucleosides. In Biocatalysis in the Pharmaceutical and Biotechnology Industrie; Patel, R.N., Ed.; CRC Press: Boca Raton, FL, USA, 2006; Chapter 14; pp. 401–423. [Google Scholar]
- Fernandez-Lucas, J.; Condezo, L.A.; Martinez-Lagos, F.; Sinisterra, J.V. Synthesis of 2’-deoxyribosylnucleosides using new 2’-deoxyribosyltransferase microorganisms producers. Enzyme Microb. Technol. 2007, 40, 1147–1155. [Google Scholar]
- Bzowska, A.; Kulikowska, E.; Shugar, D. Purine nucleoside phosphorylases: properties, functions, and clinical aspects. Pharm. Ther. 2000, 88, 349–425. [Google Scholar] [CrossRef]
- Medici, R.; Lewkowicz, E.S.; Iribarren, A.M. Microbial synthesis of 2,6-diaminopurine nucleosides. J. Mol. Catal. B-Enzym. 2006, 39, 40–44. [Google Scholar] [CrossRef]
- Utagawa, T. Enzymatic preparation of nucleoside antibiotics. J. Mol. Catal. B Enzym. 1999, 6, 215–222. [Google Scholar]
- Ramírez-Arcos, S.; Fernandez-Herrero, L.A.; Berenguer, J. A thermophilic nitrate reductase is responsible for the strain specific anaerobic growth of Thermus thermophilus HB8. Biochim. Biophys. Acta 1998, 1396, 215–227. [Google Scholar] [CrossRef]
- Pugmiere, M.J.; Ealick, S.E. Structural analysis reveal two distinct families of nucleoside phosphorylases. Biochem. J. 2002, 361, 1–25. [Google Scholar] [CrossRef]
- Zimmerman, M.; Seidenberg, J. Thymidine phosphorylase and nucleoside deoxy-ribosyltransferase in normal and malignal tissues. J. Biol. Chem. 1964, 239, 2618–2621. [Google Scholar]
- Fukushima, M.; Suzuki, N.; Emura, T.; Yano, S.; Kazuno, H.; Tada, Y.; Yamada, Y.; Asao, T. Structure and activity of specific inhibitors of thymidine phosphorylase to potentiate the function of antitumor 2'-deoxyribonucleosides. Biochem. Pharmacol. 2000, 59, 1227–1236. [Google Scholar] [CrossRef]
- Holguin, J.; Cardinaud, R.; Salemnik, C.A. Trans-N-doxyriboxylase: substrate specificity studies. Purine base acceptors. Eur. J. Biochem. 1975, 54, 515–520. [Google Scholar] [CrossRef]
- Danzin, C.; Cardinaud, R. Deoxyribosyltransfer catalysis with trans-N-deoxyriboxylase: kinetic study of purine (pyrimidine) to pyrimidine (purine) trans-N-deoxyriboxylase. Eur. J. Biochem. 1976, 62, 365–372. [Google Scholar] [CrossRef]
- Cardinaud, R.; Holguin, J. Nucleoside deoxyribosyltransfrerase II from Lactobacillus helveticus. Biochim. Biophys. Acta 1979, 568, 339–347. [Google Scholar] [CrossRef]
- Chawdhri, R.F.; Hutchinson, D.W.; Richards, A.O’L. Nucleoside deoxyriboxyltransferase and inosine phosphorylase activity in lactic acid bacteria. Arch. Microbiol. 1991, 155, 409–411. [Google Scholar]
- Trelles, J.A.; Fernandez, M.; Lewkowicz, E.S.; Iribarrren, A.M.; Sinisterra, J.V. Purine nucleoside synthesis from uridine using immobilized Enterobacter gergoviae CECT 875 whole cells. Tetrahedron Lett. 2003, 44, 2605–2609. [Google Scholar] [CrossRef]
- Fernandez-Lucas, J.; Condezo, L.A.; Quezada, M.A.; Sinisterra, J.V. Low-temperature synthesis of 2’-deoxyadenosine using immobilized psychrotrophic microorganisms. Biotechnol. Bioeng. 2008, 100, 213–222. [Google Scholar] [CrossRef]
- Shimizu, K.; Kunishima, N. Purification, crystallization and preliminary X-ray diffraction study on pyrimidine nucleoside phosphorylase TTHA1771 from Thermus thermophilus HB8. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 2007, 63, 308–310. [Google Scholar] [CrossRef]
- Sample availability: Not available.
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Almendros, M.; Gago, J.-V.S.; Carlos, J.B. Thermus thermophilus Strains Active in Purine Nucleoside Synthesis. Molecules 2009, 14, 1279-1287. https://doi.org/10.3390/molecules14031279
Almendros M, Gago J-VS, Carlos JB. Thermus thermophilus Strains Active in Purine Nucleoside Synthesis. Molecules. 2009; 14(3):1279-1287. https://doi.org/10.3390/molecules14031279
Chicago/Turabian StyleAlmendros, Marcos, José-Vicente Sinisterra Gago, and José Berenguer Carlos. 2009. "Thermus thermophilus Strains Active in Purine Nucleoside Synthesis" Molecules 14, no. 3: 1279-1287. https://doi.org/10.3390/molecules14031279




