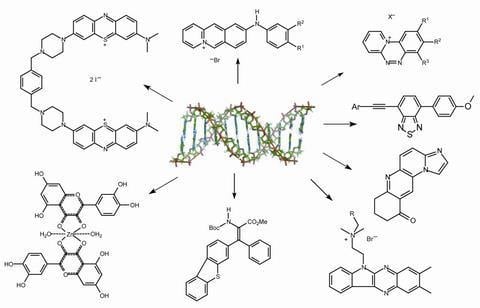
Recent Developments in the Chemistry of Deoxyribonucleic Acid (DNA) Intercalators: Principles, Design, Synthesis, Applications and Trends
Abstract
:1. Introduction
2. DNA structure: a basic background


3. Principles of intercalation

4. Synthesis, intercalation of small fluorescent molecules and possible applications






5. Clinical applications



6. Conclusions and Trends

Acknowledgements
References and Notes
- Hannon, M.J. Supramolecular DNA recognition. Chem. Soc. Rev. 2007, 36, 280–295. [Google Scholar] [CrossRef]
- MacMillan, A.M. Fifty years of "Watson-Crick". Pure Appl. Chem. 2004, 76, 1521–1524. [Google Scholar] [CrossRef]
- MacMillan, A.M. Chemistry of nucleic acids - Part 3 - Preface. Pure Appl. Chem. 2004, 76. [Google Scholar] [CrossRef]
- Jelly, R.; Lewis, S.W.; Lennard, C.; Lim, K.F.; Almog, J. Lawsone: a novel reagent for the detection of latent fingermarks on paper surfaces. Chem. Commun. 2008, 3513–3515. [Google Scholar]
- Kranaster, R.; Marx, A. Increased single-nucleotide discrimination in allele-specific polymerase chain reactions through primer probes bearing nucleobase and 2'-deoxyribose modifications. Chem. Eur. J. 2007, 13, 6115–6122. [Google Scholar] [CrossRef]
- Marras, S.A.E.; Tyagi, S.; Kramer, F.R. Real-time assays with molecular beacons and other fluorescent nucleic acid hybridization probes. Clin. Chim. Acta 2006, 363, 48–60. [Google Scholar] [CrossRef]
- Werder, S.; Malinovskii, V.L.; Haner, R. Triazolylpyrenes: Synthesis, fluorescence properties, and incorporation into DNA. Org. Lett. 2008, 10, 2011–2014. [Google Scholar]
- Yang, Q.; Yang, P.; Qian, X.H.; Tong, L.P. Naphthalimide intercalators with chiral amino side chains: Effects of chirality on DNA binding, photodamage and antitumor cytotoxicity. Bioorg. Med. Chem. Lett. 2008, 18, 6210–6213. [Google Scholar] [CrossRef]
- McKnight, R.E.; Onogul, B.; Polasani, S.R.; Gannon, M.K.; Detty, M.R. Substituent control of DNA binding modes in a series of chalcogenoxanthylium photosensitizers as determined by isothermal titration calorimetry and topoisomerase I DNA unwinding assay. Bioorg. Med. Chem. 2008, 16, 10221–10227. [Google Scholar]
- Timko, M.P.; Rushton, P.J.; Laudeman, T.W.; Bokowiec, M.T.; Chipumuro, E.; Cheung, F.; Town, C.D.; Chen, X. F. Sequencing and analysis of the gene-rich space of cowpea. BMC Genomics 2008, 9, 103. [Google Scholar]
- Shen, X-C.; Zhang, Z-L.; Zhou, B.; Peng, J.; Xie, M.; Zhang, M.; Pang, D-W. Visible light-induced plasmid DNA damage catalyzed by a CdSe/ZnS-photosensitized nano-TiO2 film. Environ. Sci. Technol. 2008, 42, 5049–5054. [Google Scholar] [CrossRef]
- Varma-Basil, M.; El-Hajj, H.; Marras, S.A.E.; Hazbon, M.H.; Mann, J.M.; Connell, N.D.; Kramer, F.R.; Alland, D. Molecular beacons for multiplex detection of four bacterial bioterrorism agents. Clin. Chem. 2004, 50, 1060–1063. [Google Scholar]
- Silverman, A.P.; Kool, E.T. Detecting RNA and DNA with templated chemical reactions. Chem. Rev. 2006, 106, 3775–3789. [Google Scholar] [CrossRef]
- Cosnier, S.; Mailley, P. Recent advances in DNA sensors. Analyst 2008, 133, 984–991. [Google Scholar] [CrossRef]
- Marras, S.A.E.; Kramer, F.R.; Tyagi, S. Efficiencies of fluorescence resonance energy transfer and contact-mediated quenching in oligonucleotide probes. Nucleic Acids Res. 2002, 30, e122–e130. [Google Scholar] [CrossRef]
- Tyagi, S.; Marras, S.A.E.; Kramer, F.R. Wavelength-shifting molecular beacons. Nat. Biotechnol. 2000, 18, 1191–1196. [Google Scholar] [CrossRef]
- Shchyolkina, A.K.; Borisova, O.F. Stabilizing and destabilizing effects of intercalators on DNA triplexes. FEBS Lett. 1997, 419, 27–31. [Google Scholar] [CrossRef]
- Silverman, A.P.; Kool, E.T. Quenched probes for highly specific detection of cellular RNAs. Trends Biotechnol. 2005, 23, 225–230. [Google Scholar] [CrossRef]
- Marras, S.A.E.; Gold, B.; Kramer, F.R.; Smith, I.; Tyagi, S. Real-time measurement of in vitro transcription. Nucleic Acids Res. 2004, 32, e72–e77. [Google Scholar] [CrossRef]
- Marti, A.A.; Jockusch, S.; Stevens, N.; Ju, J.Y.; Turro, N.J. Fluorescent hybridization probes for sensitive and selective DNA and RNA detection. Acc. Chem. Res. 2007, 40, 402–409. [Google Scholar] [CrossRef]
- Wheate, N.J.; Brodie, C.R.; Collins, J.G.; Kemp, S.; Aldrich-Wright, J.R. DNA intercalators in cancer therapy: Organic and inorganic drugs and their spectroscopic tools of analysis. Med. Chem. 2007, 7, 627–648. [Google Scholar]
- Ma, S.G.; Zhu, M.S. Recent advances in applications of liquid chromatography-tandem mass spectrometry to the analysis of reactive drug metabolites. Chem. Biol. Interact. 2009, 179, 25–37. [Google Scholar] [CrossRef]
- He, P.A.; Xu, Y.; Fang, Y.Z. A review: Electrochemical DNA biosensors for sequence recognition. Anal. Lett. 2005, 38, 2597–2623. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H.C. Molecular Structure of Nucleic Acids - A Structure for Deoxyribose Nucleic Acid. Nature 1953, 171, 737. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H.C. Genetical Implications of the Structure of Deoxyribonucleic Acid. Nature 1953, 171, 964–967. [Google Scholar] [CrossRef]
- Crick, F.H.C.; Watson, J.D. Structure of Small Viruses. Nature 1956, 177, 473–475. [Google Scholar] [CrossRef]
- Franklin, R.E.; Gosling, R.G. Evidence for 2-Chain Helix in Crystalline Structure of Sodium Deoxyribonucleate. Nature 1953, 172, 156–157. [Google Scholar] [CrossRef]
- Franklin, R.E.; Gosling, R.G. Molecular Configuration in Sodium Thymonucleate. Nature 1953, 171, 740–741. [Google Scholar] [CrossRef]
- Wilkins, M.H.F.; Seeds, W.E.; Stokes, A.R.; Wilson, H.R. Helical Structure of Crystalline Deoxypentose Nucleic Acid. Nature 1953, 172, 759–762. [Google Scholar]
- Feughelman, M.; Langridge, R.; Seeds, W.E.; Stokes, A.R.; Wilson, H.R.; Hooper, C.W.; Wilkins, M.H.F.; Barclay, R.K.; Hamilton, L.D. Molecular Structure of Deoxyribose Nucleic Acid and Nucleoprotein. Nature 1955, 175, 834–838. [Google Scholar]
- Drew, H.R.; Wing, R.M.; Takano, T.; Broka, C.; Tanaka, S.; Itakura, K.; Dickerson, R.E. Structure of A B-DNA Dodecamer - Conformation And Dynamics. Proc. Natl. Acad. Sci. 1981, 78, 2179–2183. [Google Scholar] [CrossRef]
- Wing, R.; Drew, H.; Takano, T.; Broka, C.; Tanaka, S.; Itakura, K.; Dickerson, R.E. Crystal-Structure Analysis of a Complete Turn of B-DNA. Nature 1980, 287, 755–758. [Google Scholar] [CrossRef]
- Dickerson, R.E.; Drew, H.R. Structure of a B-DNA Dodecamer Influence of Base Sequence on Helix Structure. J. Mol. Biol. 1981, 149, 761–786. [Google Scholar] [CrossRef]
- Drew, H.R.; Dickerson, R.E. Structure of a B-DNA Dodecamer .3. Geometry of Hydration. J. Mol. Biol. 1981, 151, 535–556. [Google Scholar] [CrossRef]
- Dickerson, R.E.; Drew, H.R.; Conner, B.N.; Wing, R.M.; Fratini, A.V.; Kopka, M.L. The Anatomy of A-DNA, B-DNA, and Z-DNA. Science 1982, 216, 475–485. [Google Scholar]
- Drew, H.R.; Samson, S.; Dickerson, R.E. Structure of a B-DNA Dodecamer at 16-K. Proc. Natl. Acad. Sci. 1982, 79, 4040–4044. [Google Scholar] [CrossRef]
- Dickerson, R.E.; Drew, H.R. Kinematic Model for B-DNA. Proc. Natl. Acad. Sci. 1981, 78, 7318–7322. [Google Scholar] [CrossRef]
- Corradini, R.; Sforza, S.; Tedeschi, T.; Marchelli, R. Chirality as a tool in nucleic acid recognition: Principles and relevance in biotechnology and in medicinal chemistry. Chirality 2007, 19, 269–294. [Google Scholar] [CrossRef]
- Pu, L. Fluorescence of organic molecules in chiral recognition. Chem. Rev. 2004, 104, 1687–1716. [Google Scholar] [CrossRef]
- Elliott, S.L.; Brazier, J.; Cosstick, R.; Connolly, B.A. Mechanism of the Escherichia coli DNA T : G-mismatch endonuclease (Vsr protein) thiophosphate-containing probed with oligodeoxynucleotides. J. Mol. Biol. 2005, 353, 692–703. [Google Scholar] [CrossRef]
- Qu, X.G.; Trent, J.O.; Fokt, I.; Priebe, W.; Chaires, J.B. Allosteric, chiral-selective drug binding to DNA. Proc. Natl. Acad. Sci. 2000, 97, 12032–12037. [Google Scholar] [CrossRef]
- Yang, X.L.; Wang, A.H.J. Structural studies of atom-specific anticancer drugs acting on DNA. Pharmacol. Ther. 1999, 83, 181–215. [Google Scholar] [CrossRef]
- Mukherjee, A.; Lavery, R.; Bagchi, B.; Hynes, J.T. On the molecular mechanism of drug intercalation into DNA: A simulation study of the intercalation pathway, free energy, and DNA structural changes. J. Am. Chem. Soc. 2008, 130, 9747–9755. [Google Scholar] [CrossRef]
- Ihmels, H.; Faulhaber, K.; Vedaldi, D.; Dall'Acqua, F.; Viola, G. Intercalation of oganic dye molecules into double-stranded DNA. Part 2: The annelated quinolizinium ion as a structural motif in DNA intercalators. Photochem. Photobiol. 2005, 81, 1107–1115. [Google Scholar] [CrossRef]
- Dervan, P.B.; Poulin-Kerstien, A.T.; Fechter, E.J.; Edelson, B.S. Regulation of gene expression by synthetic DNA-binding ligands. In DNA Binders and Related Subjects; Springer: Berlin, Heidelberg, German, 2005; Volume 253, pp. 1–31. [Google Scholar]
- Hurley, L.H. DNA and its associated processes as targets for cancer therapy. Nature Reviews Cancer 2002, 2, 188–200. [Google Scholar] [CrossRef]
- Graves, D.E.; Velea, L.M. Intercalative binding of small molecules to nucleic acids. Curr. Org. Chem. 2000, 4, 915–929. [Google Scholar]
- Ihmels, H.; Otto, D. Intercalation of organic dye molecules into double-stranded DNA general principles and recent developments. Top. Curr. Chem. 2005, 258, 161–204. [Google Scholar] [CrossRef]
- Zeglis, B.M.; Pierre, V.C.; Barton, J.K. Metallo-intercalators and metallo-insertors. Chem. Commun. 2007, 4565–4579. [Google Scholar] [CrossRef]
- Greguric, I.; AldrichWright, J.R.; Collins, J.G. A H-1 NMR study of the binding of Delta-[Ru(phen)(2)DPQ](2+) to the hexanucleotide d(GTCGAC)(2). Evidence for intercalation from the minor groove. J. Am. Chem. Soc. 1997, 119, 3621–3622. [Google Scholar] [CrossRef]
- Lerman, L.S. Structure of DNA-Acridine Complex. Proc. Natl. Acad. Sci. 1963, 49, 94–102. [Google Scholar] [CrossRef]
- Ren, J.S.; Chaires, J.B. Sequence and structural selectivity of nucleic acid binding ligands. J. Inorg. Biochem. 2003, 96, 82–82. [Google Scholar]
- Herzyk, P.; Neidle, S.; Goodfellow, J.M. Conformation and Dynamics of Drug-DNA Intercalation. J. Biomol. Struct. Dyn. 1992, 10, 97–139. [Google Scholar] [CrossRef]
- Rao, S.N.; Kollman, P.A. Molecular Mechanical Simulations on Double Intercalation of 9-Amino Aridcine Into D(CGCGCGC).D(GCGCGCG) - Analysis of the Physical Basis for the Neighbor-Exclusion Principle. Proc. Natl. Acad. Sci. 1987, 84, 5735–5739. [Google Scholar] [CrossRef]
- Biver, T.; Secco, F.; Venturini, M. Mechanistic aspects of the interaction of intercalating metal complexes with nucleic acids. Coord. Chem. Rev. 2008, 252, 1163–1177. [Google Scholar] [CrossRef]
- Lippard, S.J. Platinum Complexes Probes of Polynucleotide Structure and Anti-Tumor Drugs. Acc. Chem. Res. 1978, 11, 211–217. [Google Scholar] [CrossRef]
- Waring, M.J. Drugs Which Affect Structure and Function of DNA. Nature 1968, 219, 1320–1325. [Google Scholar] [CrossRef]
- Medhi, C.; Mitchell, J.B.O.; Price, S.L.; Tabor, A.B. Electrostatic factors in DNA intercalation. Biopolymers 1999, 52, 84–93. [Google Scholar] [CrossRef]
- Reha, D.; Kabelac, M.; Ryjacek, F.; Sponer, J.; Sponer, J.E.; Elstner, M.; Suhai, S.; Hobza, P. Intercalators Nature of stacking interactions between intercalators (ethidium, daunomycin, ellipticine, and 4 ',6-diaminide-2-phenylindole) and DNA base pairs Ab initio quantum chemical, density functional theory, and empirical potential study. J. Am. Chem. Soc. 2002, 124, 3366–3376. [Google Scholar]
- Garbett, N.C.; Chaires, J.B. Binding: A polemic and rough guide. In Biophysical Tools for Biologists, Volume 1: in Vitro Techniques; Elsevier Academic Press: San Diego, USA, 2008; Volume 84, pp. 3–23. [Google Scholar]
- Suh, D.; Chaires, J.B. Criteria for the Mode of Binding of DNA-Binding Agents. Bioorg. Med. Chem. 1995, 3, 723–728. [Google Scholar] [CrossRef]
- Nishimura, T.; Okobira, T.; Kelly, A.M.; Shimada, N.; Takeda, Y.; Sakurai, K. DNA binding of tilorone: H-1 NMR and calorimetric studies of the intercalation. Biochemistry 2007, 46, 8156–8163. [Google Scholar] [CrossRef]
- Assamunt, N.; Denny, W.A.; Leupin, W.; Kearns, D.R. H-1-NMR Study of the Binding of Bis(Acridines) to D(AT)5.D(AT)5.1. Mode of Binding. Biochemistry 1985, 24, 1441–1449. [Google Scholar] [CrossRef]
- Assamunt, N.; Leupin, W.; Denny, W.A.; Kearns, D.R. H-1-NMR Study of the Binding of Bis(Acridines) To D(AT)5.D(AT)5.2. Dynamic Aspects. Biochemistry 1985, 24, 1449–1460. [Google Scholar] [CrossRef]
- Kang, M.; Chouai, A.; Chifotides, H.T.; Dunbar, K.R. 2D NMR spectroscopic evidence for unprecedented interactions of cis-[Rh-2(dap)-(mu-O2CCH3)(2)(eta(1)-O2CCH3)(CH3OH)](O2CCH3) with a DNA oligonucleotide: Combination of intercalative and coordinative binding. Angew. Chem. Int. Ed. Engl. 2006, 45, 6148–6151. [Google Scholar] [CrossRef]
- Lee, J.; Guelev, V.; Sorey, S.; Hoffman, D.W.; Iverson, B.L. NMR structural analysis of a modular threading tetra intercalator bound to DNA. J. Am. Chem. Soc. 2004, 126, 14036–14042. [Google Scholar] [CrossRef]
- Spielmann, H.P. Dynamics of a bis-intercalator DNA complex by H-1-detected natural abundance C-13 NMR spectroscopy. Biochemistry 1998, 37, 16863–16876. [Google Scholar] [CrossRef]
- Kubar, T.; Hanus, M.; Ryjacek, F.; Hobza, P. Binding of cationic and neutral phenanthridine Intercalators to a DNA oligomer is controlled by dispersion energy: Quantum chemical calculations and molecular mechanics simulations. Chem. Eur. J. 2006, 12, 280–290. [Google Scholar] [CrossRef]
- Bondensgaard, K.; Jacobsen, J.P. Solution structure and energy calculation of bis-intercalation of homodimeric thiazole orange dye derivatives in DNA: Effects of modifying the linker. Bioconjug. Chem. 1999, 10, 735–744. [Google Scholar] [CrossRef]
- Cieplak, P.; Rao, S.N.; Grootenhuis, P.D.J.; Kollman, P.A. Free-Energy Calculation on Base Specificity of Drug DNA Interactions Application to Daunomycin and Acridine Intercalation into DNA. Biopolymers 1990, 29, 717–727. [Google Scholar] [CrossRef]
- Chaires, J.B. A thermodynamic signature for drug-DNA binding mode. Arch. Biochem. Biophys. 2006, 453, 26–31. [Google Scholar] [CrossRef]
- Leng, F.F.; Priebe, W.; Chaires, J.B. Ultratight DNA binding of a new bisintercalating anthracycline antibiotic. Biochemistry 1998, 37, 1743–1753. [Google Scholar] [CrossRef]
- Haq, I.; Ladbury, J.E.; Chowdhry, B.Z.; Jenkins, T.C.; Chaires, J.B. Specific binding of Hoechst 33258 to the d(CGCAAATTTGCG)(2) duplex: Calorimetric and spectroscopic studies. J. Mol. Biol. 1997, 271, 244–257. [Google Scholar] [CrossRef]
- Tuite, E.; Sehlstedt, U.; Hagmar, P.; Norden, B.; Takahashi, M. Effects of minor and major groove-binding drugs and intercalators on the DNA association of minor groove-binding proteins RecA and deoxyribonuclease I detected by flow linear dichroism. Eur. J. Biochem. 1997, 243, 482–492. [Google Scholar]
- Lincoln, P.; Norden, B. DNA binding geometries of ruthenium(II) complexes with 1,10-phenanthroline and 2,2 '-bipyridine ligands studied with linear dichroism spectroscopy. Borderline cases of intercalation. J. Phys. Chem. B 1998, 102, 9583–9594. [Google Scholar] [CrossRef]
- Garbett, N.C.; Ragazzon, P.A.; Chaires, J. B. Circular dichroism to determine binding mode and affinity of ligand-DNA interactions. Nat. Protoc. 2007, 2, 3166–3172. [Google Scholar] [CrossRef]
- Lincoln, P.; Broo, A.; Norden, B. Diastereomeric DNA-binding geometries of intercalated ruthenium(II) trischelates probed by linear dichroism: [Ru(phen)(2)DPPZ](2+) and [Ru(phen)(2)BDPPZ](2+). J. Am. Chem. Soc. 1996, 118, 2644–2653. [Google Scholar] [CrossRef]
- Hogan, M.; Dattagupta, N.; Crothers, D.M. Transient Electric Dichroism Studies of the Structure of the DNA Complex with Intercalated Drugs. Biochemistry 1979, 18, 280–288. [Google Scholar] [CrossRef]
- Lippard, S.J.; Bond, P.J.; Wu, K.C.; Bauer, W.R. Stereochemical Requirements for Intercalation of Platinum Complexes into Double-Stranded DNAs. Science 1976, 194, 726–728. [Google Scholar]
- Tsai, C.C.; Jain, S.C.; Sobell, H.M. X-Ray Crystallographic Visualization of Drug Nucleic Acid Intercalative Binding Structure of an Ethidium-Dinucleoside Monophosphate Crystalline Complex, Ethidium - 5-Iodouridylyl(3'-5')Adenosine. Proc. Natl. Acad. Sci. 1975, 72, 628–632. [Google Scholar] [CrossRef]
- Tsai, C.C.; Jain, S.C.; Sobell, H.M. Drug Nucleic Acid Interaction X-Ray Crystallographic Determination of an Ethidium-Dinucleoside Monophosphate Crystalline Complex, Ethidium - 5-Iodouridylyl(3'-5')Adenosine. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 1975, 272, 137–146. [Google Scholar] [CrossRef]
- Ren, J.S.; Chaires, J.B. Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry 1999, 38, 16067–16075. [Google Scholar] [CrossRef]
- Chaires, J.B. Dissecting the free energy of drug binding to DNA. Anti-Cancer Drug Design 1996, 11, 569–580. [Google Scholar]
- Haq, I. Part II: The thermodynamics of drug-bipolymer interaction Thermodynamics of drug-DNA interactions. Arch. Biochem. Biophys. 2002, 403, 1–15. [Google Scholar] [CrossRef]
- Silva, G.L.; Ediz, V.; Yaron, D.; Armitage, B.A. Experimental and computational investigation of unsymmetrical cyanine dyes: Understanding torsionally responsive fluorogenic dyes. J. Am. Chem. Soc. 2007, 129, 5710–5718. [Google Scholar] [CrossRef]
- Mancilha, F.S.; Neto, B.A.D.; Lopes, A.S.; Moreira, P.F.; Quina, F.H.; Goncalves, R.S.; Dupont, J. Are molecular 5,8-pi-extended quinoxaline derivatives good chromophores for photoluminescence applications? Eur. J. Org. Chem. 2006, 4924–4933. [Google Scholar]
- Normand, A.T.; Cavell, K.J. Donor-functionalised N-heterocyclic carbene complexes of group 9 and 10 metals in catalysis: Trends and directions. Eur. J. Inorg. Chem. 2008, 2781–2800. [Google Scholar] [CrossRef]
- Marion, N.; Nolan, S.P. Well-Defined N-Heterocyclic Carbenes-Palladium(II) Precatalysts for Cross-Coupling Reactions. Acc. Chem. Res. 2008, 41, 1440–1449. [Google Scholar] [CrossRef]
- Kakiuchi, F.; Kochi, T. Transition-Metal-Catalyzed Carbon-Carbon Bond Formation via Carbon-Hydrogen Bond Cleavage. Synthesis 2008, 3013–3039. [Google Scholar] [CrossRef]
- Seregin, I.V.; Gevorgyan, V. Direct transition metal-catalyzed functionalization of heteroaromatic compounds. Chem. Soc. Rev. 2007, 36, 1173–1193. [Google Scholar] [CrossRef]
- D'Souza, D.M.; Muller, T.J.J. Multi-component syntheses of heterocycles by transition-metal catalysis. Chem. Soc. Rev. 2007, 36, 1095–1108. [Google Scholar] [CrossRef]
- Felpin, F.X.; Ayad, T.; Mitra, S. Pd/C: An old catalyst for new applications - Its use for the Suzuki-Miyaura reaction. Eur. J. Org. Chem. 2006, 2679–2690. [Google Scholar] [CrossRef]
- Chen, L.; Li, C.J. Catalyzed reactions of alkynes in water. Adv. Synth. Catal. 2006, 348, 1459–1484. [Google Scholar] [CrossRef]
- Marciniec, B. Catalysis by transition metal complexes of alkene silylation - recent progress and mechanistic implications. Coord. Chem. Rev. 2005, 249, 2374–2390. [Google Scholar] [CrossRef]
- Dupont, J.; Consorti, C.S.; Spencer, J. The potential of palladacycles: More than just precatalysts. Chem. Rev. 2005, 105, 2527–2571. [Google Scholar] [CrossRef]
- Granzhan, A.; Ihmels, H. N-Aryl-9-amino-substituted acridizinium derivatives as fluorescent "light-up" probes for DNA and protein detection. Org. Lett. 2005, 7, 5119–5122. [Google Scholar] [CrossRef]
- Granzhan, A.; Ihmels, H.; Viola, G. 9-donor-substituted acridizinium salts: Versatile environment-sensitive fluorophores for the detection of biomacromolecules. J. Am. Chem. Soc. 2007, 129, 1254–1267. [Google Scholar] [CrossRef]
- Neto, B.A.D.; Lapis, A.A.M.; Mancilha, F.S.; Vasconcelos, I.B.; Thum, C.; Basso, L.A.; Santos, D.S.; Dupont, J. New Sensitive Fluorophores for Selective DNA Detection. Org. Lett. 2007, 9, 4001–4004. [Google Scholar]
- Rosa, G.R.; Ebeling, G.; Dupont, J.; Monteiro, A.L. A superior non-symmetrical NCP pincer type palladacycle catalyst precursor for the coupling of aryl boronic acids with aryl chlorides. Synthesis 2003, 2894–2897. [Google Scholar]
- Neto, B.A.D.; Lopes, A.S.A.; Ebeling, G.; Goncalves, R.S.; Costa, V.E.U.; Quina, F.H.; Dupont, J. Photophysical and electrochemical properties of pi-extended molecular 2,1,3-benzothiadiazoles. Tetrahedron 2005, 61, 10975–10982. [Google Scholar] [CrossRef]
- Neto, B.A.D.; Lopes, A.S.; Wust, M.; Costa, V.E.U.; Ebeling, G.; Dupont, J. Reductive sulfur extrusion reaction of 2,1,3-benzothiadiazole compounds: a new methodology using NaBH4/CoCl2 6H2O(cat) as the reducing system. Tetrahedron Lett. 2005, 46, 6843–6846. [Google Scholar]
- Queiroz, M.; Castanheira, E.M.S.; Carvalho, M.S.D.; Abreu, A.S.; Ferreira, P.M.T.; Karadeniz, H.; Erdem, A. New tetracyclic heteroaromatic compounds based on dehydroamino acids: photophysical and electrochemical studies of interaction with DNA. Tetrahedron 2008, 64, 382–391. [Google Scholar] [Green Version]
- Wilson, B.; Fernandez, M.J.; Lorente, A.; Grant, K.B. Syntheses and DNA photocleavage by mono- and bis-phenothiazinium piperazinexylene intercalators. Tetrahedron 2008, 64, 3429–3436. [Google Scholar] [CrossRef]
- Sinan, M.; Panda, M.; Ghosh, A.; Dhara, K.; Fanwick, P.E.; Chattopadhyay, D.J.; Goswami, S. Mild synthesis of a family of planar triazinium cations via proton-assisted cyclization of pyridyl containing azo compounds and studies on DNA intercalation. J. Am. Chem. Soc. 2008, 130, 5185–5193. [Google Scholar]
- Ferguson, L.R.; Denny, W.A. Genotoxicity of non-covalent interactions: DNA intercalators. Mutat. Res. 2007, 623, 14–23. [Google Scholar] [CrossRef]
- Li, H.H.; Aubrecht, J.; Fornace, A.J. Toxicogenomics: Overview and potential applications for the study of non-covalent DNA interacting chemicals. Mutat. Res. 2007, 623, 98–108. [Google Scholar] [CrossRef]
- Andaloussi, M.; Moreau, E.; Masurier, N.; Lacroix, J.; Gaudreault, R.C.; Chezal, J.M.; El Laghdach, A.; Canitrot, D.; Debiton, E.; Teulade, J.C.; Chavignon, O. Novel imidazo[1,2-a]naphthyridinic systems (part 1): Synthesis, antiproliferative and DNA-intercalating activities. Eur. J. Med. Chem. 2008, 43, 2505–2517. [Google Scholar] [CrossRef]
- Wilhelmsson, L.M.; Kingi, N.; Bergman, J. Interactions of Antiviral Indolo[2,3-b]quinoxaline Derivatives with DNA. J. Med. Chem. 2008, 51, 7744–7750. [Google Scholar] [CrossRef]
- da Silva, J.F.M.; Garden, S.J.; Pinto, A.C. The chemistry of isatins: a review from 1975 to 1999. J. Braz. Chem. Soc. 2001, 12, 273–U286. [Google Scholar] [CrossRef]
- Pinto, A.C.; Lapis, A.A.M.; da Silva, B.V.; Bastos, R. S.; Dupont, J.; Neto, B. A. D. Pronounced ionic liquid effect in the synthesis of biologically active isatin-3-oxime derivatives under acid catalysis. Tetrahedron Lett. 2008, 49, 5639–5641. [Google Scholar]
- Tan, J.; Wang, B.C.; Zhu, L.C. DNA binding, cytotoxicity, apoptotic inducing activity, and molecular modeling study of quercetin zinc(II) complex. Bioorg. Med. Chem. 2009, 17, 614–620. [Google Scholar] [CrossRef]
- Shao, H.B.; Chu, L.Y.; Lu, Z.H.; Kang, C.M. Primary antioxidant free radical scavenging and redox signaling pathways in higher plant cells. Int. J. Biol. Sci. 2008, 4, 8–14. [Google Scholar] [CrossRef]
- Martinez, R.; Chacon-Garcia, L. The search of DNA-intercalators as antitumoral drugs: What it worked and what did not work. Current Medicinal Chemistry 2005, 12, 127–151. [Google Scholar] [CrossRef]
- Sample availability: Not available.
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Neto, B.A.D.; Lapis, A.A.M. Recent Developments in the Chemistry of Deoxyribonucleic Acid (DNA) Intercalators: Principles, Design, Synthesis, Applications and Trends. Molecules 2009, 14, 1725-1746. https://doi.org/10.3390/molecules14051725
Neto BAD, Lapis AAM. Recent Developments in the Chemistry of Deoxyribonucleic Acid (DNA) Intercalators: Principles, Design, Synthesis, Applications and Trends. Molecules. 2009; 14(5):1725-1746. https://doi.org/10.3390/molecules14051725
Chicago/Turabian StyleNeto, Brenno A. D., and Alexandre A. M. Lapis. 2009. "Recent Developments in the Chemistry of Deoxyribonucleic Acid (DNA) Intercalators: Principles, Design, Synthesis, Applications and Trends" Molecules 14, no. 5: 1725-1746. https://doi.org/10.3390/molecules14051725