Design of Novel 4-Hydroxy-chromene-2-one Derivatives as Antimicrobial Agents
Abstract
:1. Introduction
2. Results and Discussion
2.1. Antimicrobial activity
2.2. SAR
2.3. QSAR
2.4. Molecular docking
2.5. Design of novel coumarin derivatives
3. Experimental
3.1. Synthesis
3.2. Assay for in vitro antimicrobial activity
3.3. Molecular modeling
3.4. Molecular docking
4. Conclusions
Acknowledgements
References and Notes
- Stein, A.C.; Álvarez, S.; Avancini, C.; Zacchinos, S.; von Poser, G. Antifungal activity of some coumarins obtained from species of Pterocaulon (Asteraceae). J. Ethnopharmacol. 2006, 107, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Ojala, T.; Remes, S.; Haansuu, P.; Vuorela, H.; Hiltunen, R.; Haahtela, K.; Vuorela, P. Antimicrobial activity of some coumarin containing herbal plants growing in Finland. J. Ethnopharmacol. 2000, 73, 299–305. [Google Scholar] [CrossRef]
- Mladenović, M.; Vuković, N.; Niciforović, N.; Sukdolak, S.; Solujić, S. Synthesis and molecular descriptor characterization of novel 4-hydroxy-chromene-2-one derivatives as antimicrobial agents. Molecules 2009, 14, 1495–1512. [Google Scholar] [CrossRef] [PubMed]
- Vuković, N.; Sukdolak, S.; Solujić, S.; Milošević, T. Synthesis and antimicrobial evaluation of some novel 2-aminothiayole derivatives of 4-hydroxy-chromene-2-one. Arch. Pharm. Chem. Life Sci. 2008, 341, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Kayser, O.; Koldrziey, H. Antibacterial activity of simple coumarins structural requirements for biological activity. Z. Naturforch. 1999, 54c, 169–174. [Google Scholar] [CrossRef]
- Zloh, M.; Bucar, F.; Gibbons, S. Quantum chemical studies on structure activity relationship of natural product polyacetylenes. Theor. Chem. Acc. 2007, 117, 247–252. [Google Scholar] [CrossRef]
- Okamoto, A.K.; Gaudio, A.C.; dos Santos Marques, A.; Takahata, Y. QSAR study of inibition by coumarins of IQ induced mutation in S. typhimurium TA98. J. Mol. Struc. Theochem. 2005, 725, 231–238. [Google Scholar] [CrossRef]
- Basile, A.; Sorbo, S.; Spadaro, V.; Bruno, M.; Maggio, A.; Faraone, N.; Rosselli, S. Antimicrobial and antioxidant activities of coumarins from the roots of Ferulago campestris (Apiaceae). Molecules 2009, 14, 939–952. [Google Scholar] [CrossRef] [PubMed]
- Widelski, J.; Popova, M.; Graikou, K.; Glowniak, K.; Chinou, I. Coumarins from Angelica lucida L.-Antibacterial activities. Molecules 2009, 14, 2729–2734. [Google Scholar] [CrossRef] [PubMed]
- Debeljak, Ž.; Škrbo, A.; Jasprica, I.; Mornar, A.; Plečko, V.; Banjanac, M.; Medić-Šaric, M. QSAR Study of antimicrobial activity of some 3-nitrocoumarins and related compounds. J. Chem. Inf. Model. 2007, 47, 918–926. [Google Scholar] [CrossRef] [PubMed]
- Hishmit, O. H.; Miky, A.A.J.; Farrag, A.A.; Fadl-Allah, E. M. Synthesis of some coumarin derivatives and their antimicrobial activity. Arch. Pharm. Res. 1989, 12, 181–185. [Google Scholar] [CrossRef]
- Sharma, M.C.; Sahu, N.K.; Kohali, D.V.; Chaturvedi, S.C.; Sharma, S. QSAR, synthesis and biological activity studies of some thiazolidines derivatives. DJNB 2009, 4, 223–232. [Google Scholar]
- Lewis, J.R.; Singh, M.P.O.; Smith, V.C.; Skarzynski, T.; Maxwell, A.; Wonacott, J.A.; Wigley, B.D. The nature of inhibition of DNA gyrase by the coumarins and the cyclothialidines by X-ray crystallography. EMBO J. 1996, 15, 1412–1240. [Google Scholar] [CrossRef] [PubMed]
- Šarić-Medić, M.; Mornar, A.; Črnjević-Badovinac, T.; Josprica, I. Optimization of chromatographic conditions in thin layer chromatography of flavonoids and phenolic acids. Croat. Chem. Acta 2004, 77, 367–370. [Google Scholar]
- Stanchev, S.; Momekov, G.; Jensen, F.; Manolov, I. Synthesis, computational study and cytotoxic activity of new 4-hydroxycoumarin derivatives. Eur. J. Med. Chem. 2008, 43, 694–706. [Google Scholar] [CrossRef] [PubMed]
- Cravotto, G.; Tagliapietra, S.; Cappelo, R.; Palmisano, G.; Curini, M.; Boccalini, M. Long-chain 3-acyl-4-hydroxycoumarins: Structure and antibacterial activity. Arch. Pharm. Chem. Life. Sci. 2006, 339, 129–132. [Google Scholar] [CrossRef] [PubMed]
- Campos, J.; del Carmen Nunez, M.; Rodrguez, V.; Entrena, A.; Hernandez-Alcoceba, R.; Fernandez, F.; Lacal, I.J.; Gallo, M.A.; Espinoza, A. LUMO energy of model compounds of bispyridinium compounds as an index for the inhibition of choline kinase. Eur. J. Med. Chem. 2001, 36, 215–225. [Google Scholar] [CrossRef]
- Kubinyi, H. The Quantitative Analysis of Structure-Activity Relationships. In Burger´s Medicinal Chemstry and Drug Discovery, Principles and Practice, 5th ed.; Wolff, M.E., Ed.; John Wiley & Sons: New York, NY, USA, 1995; Volume 3, pp. 497–571. [Google Scholar]
- NCCLS. Performance Standards for Antimicrobial Susceptibility Testing 14th Int. Suplement M100-S14; Wayne, PA, USA, 2003. [Google Scholar]
- Umesha, S.; Richardson, P.A.; Kong, P.; Hong, C.X. A novel indicator plant to test the hypersensitivity of phytopathogenic bacteria. J. Microbiol. Meth. 2008, 72, 95–97. [Google Scholar] [CrossRef] [PubMed]
- Carter, C.H.; Gaspar, A.J.; Leise, J.M. Resazurin staining of bacterial colonies on membranes filters; Fort Dietrick: Frederick, MD, USA, 1955. [Google Scholar]
- Software Spartan 2002 for Windows; Wavefunction, Inc.: 18401 Von Karman Ave. Ste. 370, Irvine, CA 92612-8542, USA, 2002.
- Mohamadi, F.; Richards, N.; Guida, W.; Liskamp, R.; Lipton, M.; Caulfield, C.; Chang, G.; Hendrikson, T.; Still, W. Macromodel an integrated software system for modeling organic and bioorganic molecules using molecular mechanics. J. Comput. Chem. 1990, 11, 440–467. [Google Scholar] [CrossRef]
- Stewart, J.J.P. MOPAC 2009; Stewart Computational Chemistry: Colorado Springs, CO, USA; http://www.openMOPAC.net.
- Pedretti, A.; Villa, A.; Vistoli, G. VEGA: A versatile program to convert, handle and virtualize molecular structure on Windows-based PCs. J. Mol. Graph. 2002, 21, 47–49. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Montgomery, J.A.; Vreven, T.; Kudin, K.N.; Burant, J.C.; Millam, J.M.; Iyengar, S.S.; Tomasi, J.; Barone, V.; Mennucci, B.; Cossi, M.; Scalmani, G.; Rega, N.; Petersson, G.A.; Nakatsuji, H.; Hada, M.; Ehara, M.; Toyota, K.; Fukuda, R.; Hasegawa, J.; Ishida, M.; Nakajima, T.; Honda, Y.; Kitao, O.; Nakai, H.; Klene, M.; Li, X.; Knox, J.E.; Hratchian, H.P.; Cross, J.B.; Bakken, V.; Adamo, C.; Jaramillo, J.; Gomperts, R.; Stratmann, R.E.; Yazyev, O.; Austin, A.J.; Cammi, R.; Pomelli, C.; Ochterski, J.W.; Ayala, P.Y.; Morokuma, K.; Voth, G.A.; Salvador, P.; Dannenberg, J.J.; Zakrzewski, V.G.; Dapprich, S.; Daniels, A.D.; Strain, M.C.; Farkas, O.; Malick, D.K.; Rabuck, A.D.; Raghavachari, K.; Foresman, J.B.; Ortiz, J.V.; Cui, Q.; Baboul, A.G.; Clifford, S.; Cioslowski, J.; Stefanov, B.B.; Liu, G.; Liashenko, A.; Piskorz, P.; Komaromi, I.; Martin, R.L.; Fox, D.J.; Keith, T.; Al-Laham, M.A.; Peng, C.Y.; Nanayakkara, A.; Challacombe, M.; Gill, P.M.W.; Johnson, B.; Chen, W.; Wong, M.W.; Gonzalez, C.; Pople, J.A. Gaussian 03, Revision C.02. Gaussian, Inc.: Wallingford, CT, USA.
- Sharma, P.; Kumar, A.; Upadhyay, S.; Sahu, V.; Singh, J. Synthesis and QSAR modeling of 2-acetyl-2-ethoxycarbonyl-1-[4(4’-arylazo)-phenyl]-N,N-dimethylaminophenyl aziridines as potential antibacterial agents. Eur. J. Med. Chem. 2009, 44, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Chem3D Ultra 10.0, ChemOffice Ultra, 2006, Cambridge Scientific Software: CambridgeSoft Corporate Headquarters, 100 Cambridge Park Drive Cambridge, MA 02140, USA, 2006.
- Nikolic, K.M. Design and QSAR study of analogs of a-tocopherol with enhanced antiproliferative activity against human breast adenocarcinoma cells. J. Mol. Graph. Model. 2008, 26, 868–873. [Google Scholar] [CrossRef] [PubMed]
- Origin Pro 8, OriginLab Corporation: Northampton, MA, USA, 2009.
- Lafitte, D.; Lamour, V.; Tsvetkov, P.O.; Makarov, A.A.; Klich, M.; Deprez, P.; Moras, D.; Briand, C.; Gilli, R. DNA gyrase interaction with coumarin-based inhibitors: the role of the hydroxybenzoate isopentenyl moiety and the 5’-methyl group of the noviose. Biochemistry 2002, 41, 7217–7223. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.G.; Goodsell, S.D.; Huey, R.; Olson, J.A. Distributed automated docking of flexible ligands to proteins: Parallel applications of AutoDock 2.4. J. Comp. Aid. Mol. Des. 1996, 10, 293–304. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds 1-10c are available from the authors. |

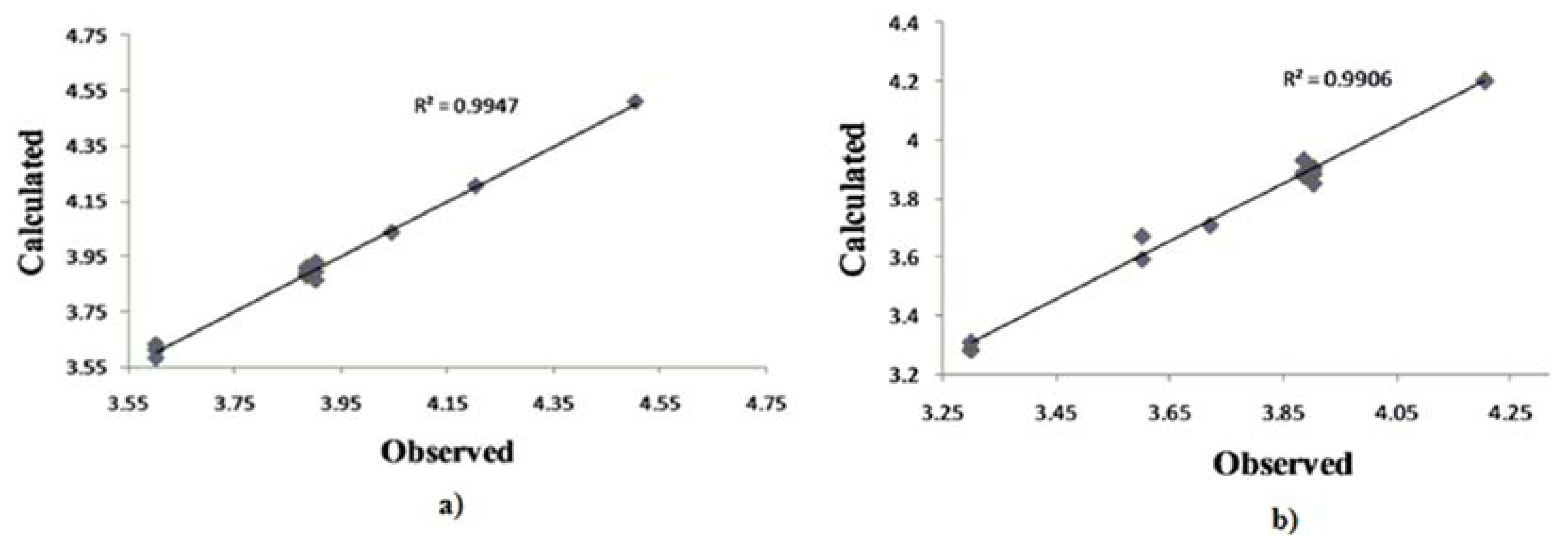

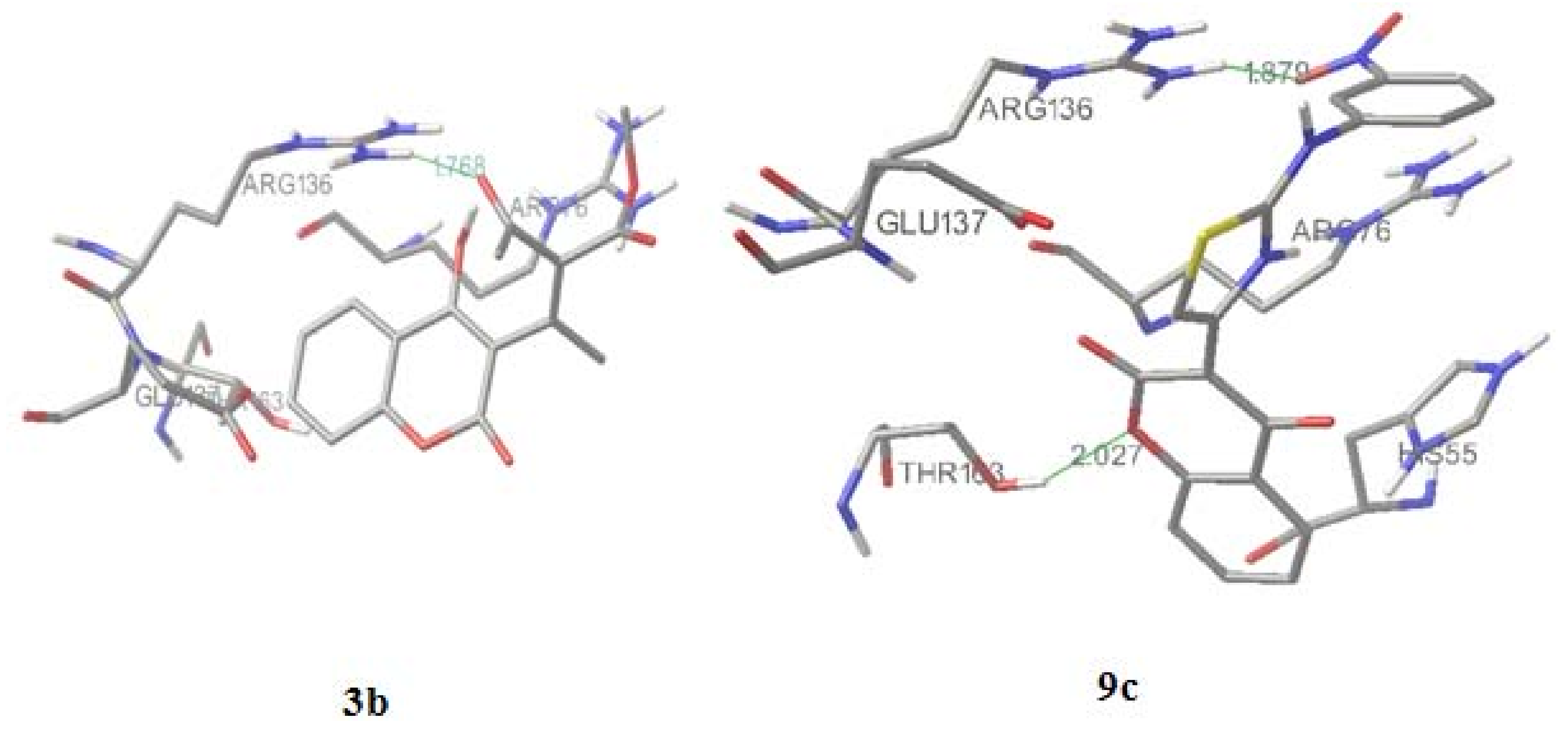

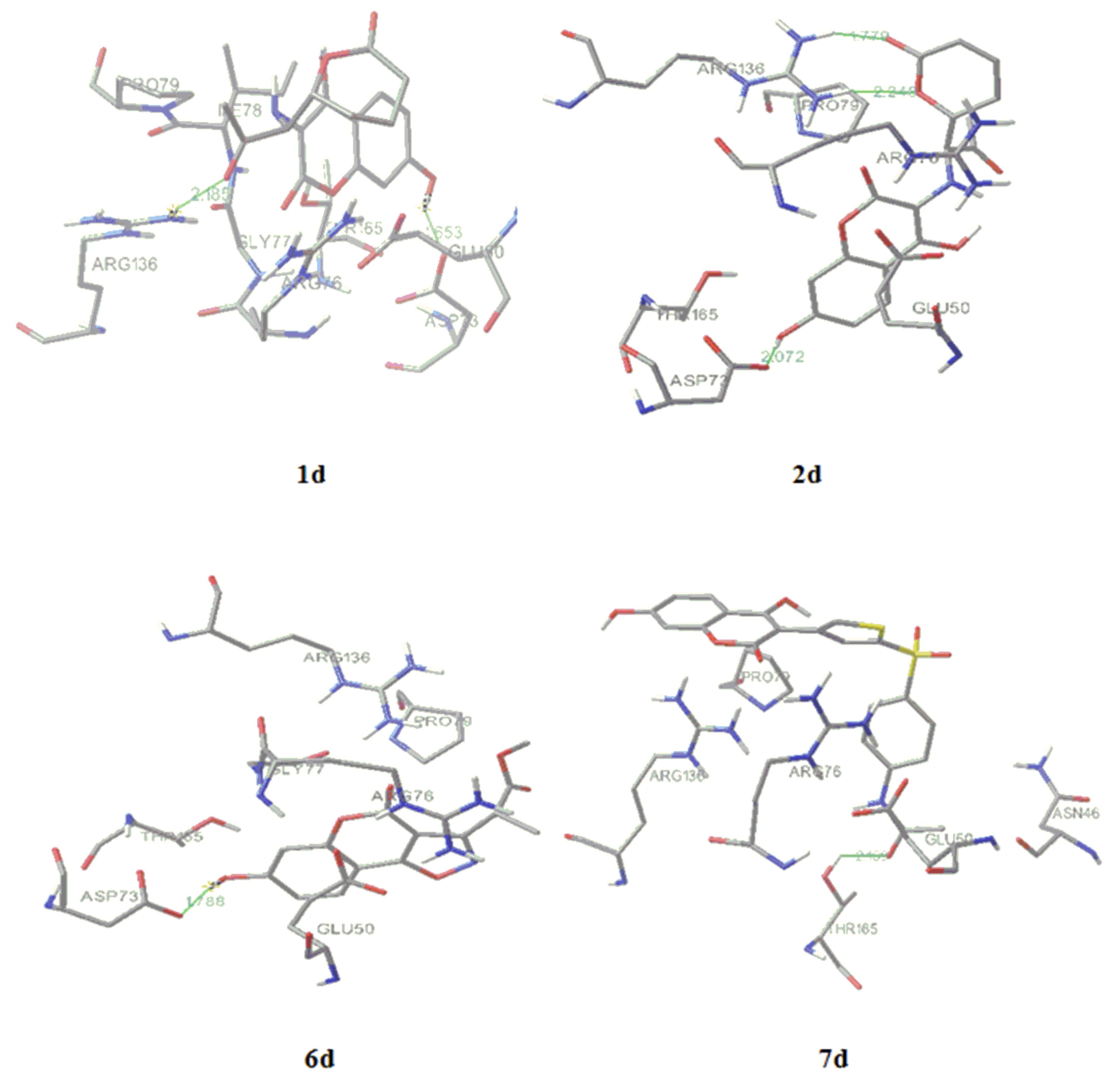
| S. aureus | E. coli | C. albicans | ||||||
|---|---|---|---|---|---|---|---|---|
| Comp. | MICa values of tested compounds (10-6 g/mL) (-log MIC) | |||||||
| Experimental | Calculated | Experimental | Calculated | Experimental | Calculated | |||
| 1 | 90 ± 0.35 (4.046) | 103 (3.984) | 190 ± 0.35 (3.721) | 195 (3.708) | 90 ± 0.05 (4.046) | 92 (4.035) | ||
| 3b | 130 ± 0.22 (3.886) | 156 (3.806) | 130 ± 0.34 (3.886) | 132 (3.878) | 130 ± 0.15 (3.886) | 130 (3.885) | ||
| 4b | 130 ± 0.50 (3.886) | 222 (3.652) | 130 ± 0.25 (3.886) | 129 (3.888) | 250 ± 0.25 (3.602) | 132 (3.878) | ||
| 6b | 130 ± 0.45(3.886) | 116 (3.932) | 250 ± 0.55(3.602) | 255 (3.592) | 130 ± 0.15 (3.886) | 129 (3.889) | ||
| 7b | 130 ± 0.35 (3.886) | 112 (3.947) | 500 ± 0.35 (3.301) | 519 (3.284) | 500 ± 0.35 (3.301) | 124 (3.908) | ||
| 8b | 130 ± 0.26 (3.886) | 136 (3.865) | 130 ± 0.35 (3.886) | 117 (3.932) | 130 ± 0.55 (3.886) | 130 (3.886) | ||
| 2c | 125 ± 0.25 (3.904) | 62.5 (4.206) | 250 ± 0.55 (3.602) | 491 (3.309) | 62.5 ± 0.55 (4.204) | 137 (3.862) | ||
| 3c | 125 ± 0.25 (3.904) | 60 (4.217) | 62.5 ± 0.25 (4.204) | 62.5 (4.204) | 62.5 ± 0.55 (4.204) | 145 (3.893) | ||
| 4c | 62.5 ± 0.29 (4.204) | 62.5 (4.204) | 62.5 ± 0.10 (4.204) | 62.6 (4.203) | 62.5 ± 0.15 (4.204) | 63 (4.205) | ||
| 5c | 62.5 ± 0.24 (4.204) | 33.5 (4.475) | 125 ± 0.50 (3.904) | 124 (3.906) | 31.25 ± 0.10 (4.505) | 63 (4.205) | ||
| 6c | 250 ± 0.35 (3.602) | 66 (4.179) | 125 ± 0.35 (3.904) | 125 (3.902) | 62.5 ± 0.15 (4.204) | 234 (3.630) | ||
| 7c | 250 ± 0.55 (3.602) | 26.2 (4.581) | 125 ± 0.45 (3.904) | 131 (3.884) | 31.25 ± 0.55 (4.505) | 245 (3.610) | ||
| 8c | 250 ± 0.36 (3.602) | 121 (3.914) | 250 ± 0.12 (3.602) | 214 (3.670) | 125 ± 0.15 (3.904) | 262 (3.582) | ||
| 9c | 31.25 ± 0.21 (4.505) | 30 (4.526) | 62.5 ± 0.09 (4.204) | 63.1 (4.200) | 31.25 ± 0.25 (4.505) | 31 (4.508) | ||
| 10c | 125 ± 0.38 (3.903) | 69.5 (4.158) | 125 ± 0.25 (3.904) | 141 (3.851) | 62.5 ± 0.55 (4.204) | 118 (3.928) | ||
| S | 31.25 ± 0.07 | 31.25 ± 0.35 | ||||||
| K | 1.95 ± 0.05 | |||||||
| Comp. | log P | MR | lipole | HOMO | LUMO | CAA | CMA | CSEV | ovality |
|---|---|---|---|---|---|---|---|---|---|
| 1 | -0.529 | 51.062 | 2.161 | -9.989 | -1.490 | 335.170 | 156.104 | 124.411 | 1.368 |
| 3b | -1.318 | 75.494 | 1.622 | -10.015 | -1.562 | 466.977 | 236.476 | 203.327 | 1.590 |
| 4b | -0.035 | 66.062 | 1.646 | -9.958 | -1.537 | 424.305 | 205.095 | 165.182 | 1.495 |
| 6b | -1.679 | 75.277 | 1.905 | -9.977 | -1.453 | 444.049 | 223.344 | 192.322 | 1.561 |
| 7b | -1.765 | 67.538 | 2.645 | -10.040 | -1.646 | 424.466 | 207.570 | 171.656 | 1.482 |
| 8b | -1.709 | 66.570 | 2.507 | -10.037 | -1.694 | 426.269 | 208.634 | 173.406 | 1.512 |
| 2c | 1.216 | 96.238 | 0.557 | -9.042 | -1.449 | 566.431 | 291.664 | 246.339 | 1.645 |
| 3c | 3.129 | 95.320 | 2.244 | -9.003 | -1.954 | 542.789 | 279.873 | 233.719 | 1.604 |
| 4c | 0.702 | 65.468 | 1.415 | -8.999 | -1.621 | 572.035 | 296.286 | 249.537 | 1.654 |
| 5c | 2.904 | 95.722 | 2.513 | -8.900 | -1.135 | 521.823 | 269.577 | 225.940 | 1.608 |
| 6c | 1.921 | 88.488 | 0.281 | -8.923 | -0.936 | 552.855 | 279.213 | 230.128 | 1.680 |
| 7c | 1.856 | 84.807 | 1.112 | -8.890 | -0.950 | 478.322 | 244.811 | 208.372 | 1.588 |
| 8c | 3.380 | 93.722 | 2.552 | -8.913 | -1.072 | 521.339 | 268.518 | 225.738 | 1.604 |
| 9c | 3.129 | 95.320 | 2.101 | -8.972 | -1.759 | 538.849 | 281.111 | 237.198 | 1.614 |
| 10c | 3.603 | 104.966 | 3.169 | -8.866 | -1.231 | 558.547 | 292.497 | 247.999 | 1.641 |
| Partial atomic charges of the compounds | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Functional groups | 1 | 3b | 4b | 6b | 7b | 8b | |||
| 4-OH | -0.615 | -0.646 | -0.637 | -0.648 | -0.614 | -0.625 | |||
| O-lactone | -0.523 | -0.523 | -0.522 | -0.522 | -0.515 | -0.515 | |||
| CO-lactone | -0.474 | -0.491 | -0.493 | -0.491 | -0.473 | -0.464 | |||
| CO | -0.456 | -0.609 | -0.604 | -0.613 | -0.531 | ||||
| 0.221 | 0.235 | 0.225 | |||||||
| CO-carboxyl | -0.743 | -0.747 | -0.745 | -0.456 | -0.488 | ||||
| OH-carboxyl | -0.507 | -0.492 | -0.579 | -0.586 | |||||
| CN | -0.452 | -0.463 | -0.475 | ||||||
| Functional groups | 2c | 3c | 4c | 5c | 6c | 7c | 8c | 9c | 10c |
| 4-OH | -0.638 | -0.644 | -0.204 | -0.617 | -0.675 | -0.671 | -0.646 | -0.637 | -0.648 |
| O-lactone | -0.522 | -0.520 | -0.114 | -0.599 | -0.531 | -0.532 | -0.523 | -0.522 | -0.522 |
| CO-lactone | -0.492 | -0.490 | -0.195 | -0.499 | -0.515 | -0.516 | -0.491 | -0.493 | -0.491 |
| N-thiazole | -0.582 | -0.604 | -0.261 | -0.681 | -0.673 | -0.659 | -0.609 | -0.604 | -0.613 |
| S-thiazole | 0.230 | 0.244 | 0.296 | 0.458 | 0.261 | 0.259 | 0.221 | 0.235 | 0.225 |
| N-amine | -0.750 | -0.744 | -0.262 | -0.753 | -0.610 | -0.419 | -0.743 | -0.747 | -0.745 |
| CO-carboxyl | -0.467 | -0.468 | |||||||
| OH-carboxyl | -0.582 | -0.563 | |||||||
| OH-phenyl | -0.638 | ||||||||
| N-nitro | -0.374 | -0.386 | |||||||
| O-nitro | -0.400b | -0.392b | |||||||
| -0.4016 | -0.386b | ||||||||
| S-SO3H | 0.758 | ||||||||
| O-SO3H | -0.282c | ||||||||
| 0.259c | |||||||||
| OH-SO3 | -0.256 | ||||||||
| aD1 | D2 | D3 | D4 | D5 | D6 | D7 | D8 | D9 | D10 | D11 | D12 | D13 | D14 | bC1 | C2 | C3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D1 | 1.00 | ||||||||||||||||
| D2 | 0.43 | 1.00 | |||||||||||||||
| D3 | 0.35 | 0.99 | 1.00 | ||||||||||||||
| D4 | 0.39 | 0.35 | 0.25 | 1.00 | |||||||||||||
| D5 | 0.77 | 0.54 | 0.46 | 0.45 | 1.00 | ||||||||||||
| D6 | 0.64 | 0.59 | 0.43 | 0.46 | 0.98 | 1.00 | |||||||||||
| D7 | 0.35 | 0.43 | 0.47 | 0.44 | 0.96 | 0.96 | 1.00 | ||||||||||
| D8 | 0.56 | 0.37 | 0.11 | 0.78 | 0.37 | 0.19 | 0.23 | 1.00 | |||||||||
| D9 | 0.12 | 06 | 0.66 | 0.12 | 0.54 | 0.43 | 0.46 | 0 | 1.00 | ||||||||
| D10 | 0.28 | 0.95 | 0.89 | 0.23 | 0.62 | 0.61 | 0.22 | 0 | 0.94 | 1.00 | |||||||
| D11 | 0.11 | 0.72 | 0.68 | 0.27 | 0.63 | 0.64 | 0.58 | 0 | 0.36 | 0.44 | 1.00 | ||||||
| D12 | 0.29 | 0.47 | 0.34 | 0.61 | 0.15 | 0.15 | 0.19 | 0 | 0.18 | 0.19 | 0.36 | 1.00 | |||||
| D13 | 0.36 | 0.44 | 0.95 | 0.25 | 0.31 | 0.37 | 0.31 | 0 | 0.62 | 0.55 | 0.34 | 0.95 | 1.00 | ||||
| D14 | 0.13 | 0.78 | 0.66 | 0.14 | 0.72 | 0.74 | 0.64 | 0.15 | 0.73 | 0.78 | 0.66 | 0.23 | 0.47 | 1.00 | |||
| C1 | 045 | 0.52 | 0.94 | 0 | 0.44 | 0.53 | 0.57 | 0 | 0 | 0.84 | 0.32 | 0.36 | 0.96 | 0.66 | 1.00 | ||
| C2 | 0.77 | 0 | 0 | 0.88 | 0.94 | 0.51 | 0.53 | 0.96 | 0.54 | 0.26 | 0.58 | 0.21 | 0.19 | 0.34 | 0.55 | 1.00 | |
| C3 | 0.75 | 0 | 0 | 0.96 | 0.12 | 0.35 | 0.15 | 0.93 | 0.24 | 0 | 0 | 0 | 0 | 0.1 | 0.52 | 0.75 | 1.00 |
© 2010 by the authors; licensee MDPI, Basel, Switzerland. This article is an Open Access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Mladenović, M.; Vuković, N.; Sukdolak, S.; Solujić, S. Design of Novel 4-Hydroxy-chromene-2-one Derivatives as Antimicrobial Agents. Molecules 2010, 15, 4294-4308. https://doi.org/10.3390/molecules15064294
Mladenović M, Vuković N, Sukdolak S, Solujić S. Design of Novel 4-Hydroxy-chromene-2-one Derivatives as Antimicrobial Agents. Molecules. 2010; 15(6):4294-4308. https://doi.org/10.3390/molecules15064294
Chicago/Turabian StyleMladenović, Milan, Nenad Vuković, Slobodan Sukdolak, and Slavica Solujić. 2010. "Design of Novel 4-Hydroxy-chromene-2-one Derivatives as Antimicrobial Agents" Molecules 15, no. 6: 4294-4308. https://doi.org/10.3390/molecules15064294




