Chemical Arsenal for the Study of O-GlcNAc
Abstract
:1. Introduction

2. Chemical Tools for O-GlcNAc Proteomics
2.1. O-GlcNAc Enrichment Strategies
2.1.1. Conventional antibody/lectin-based enrichment approaches
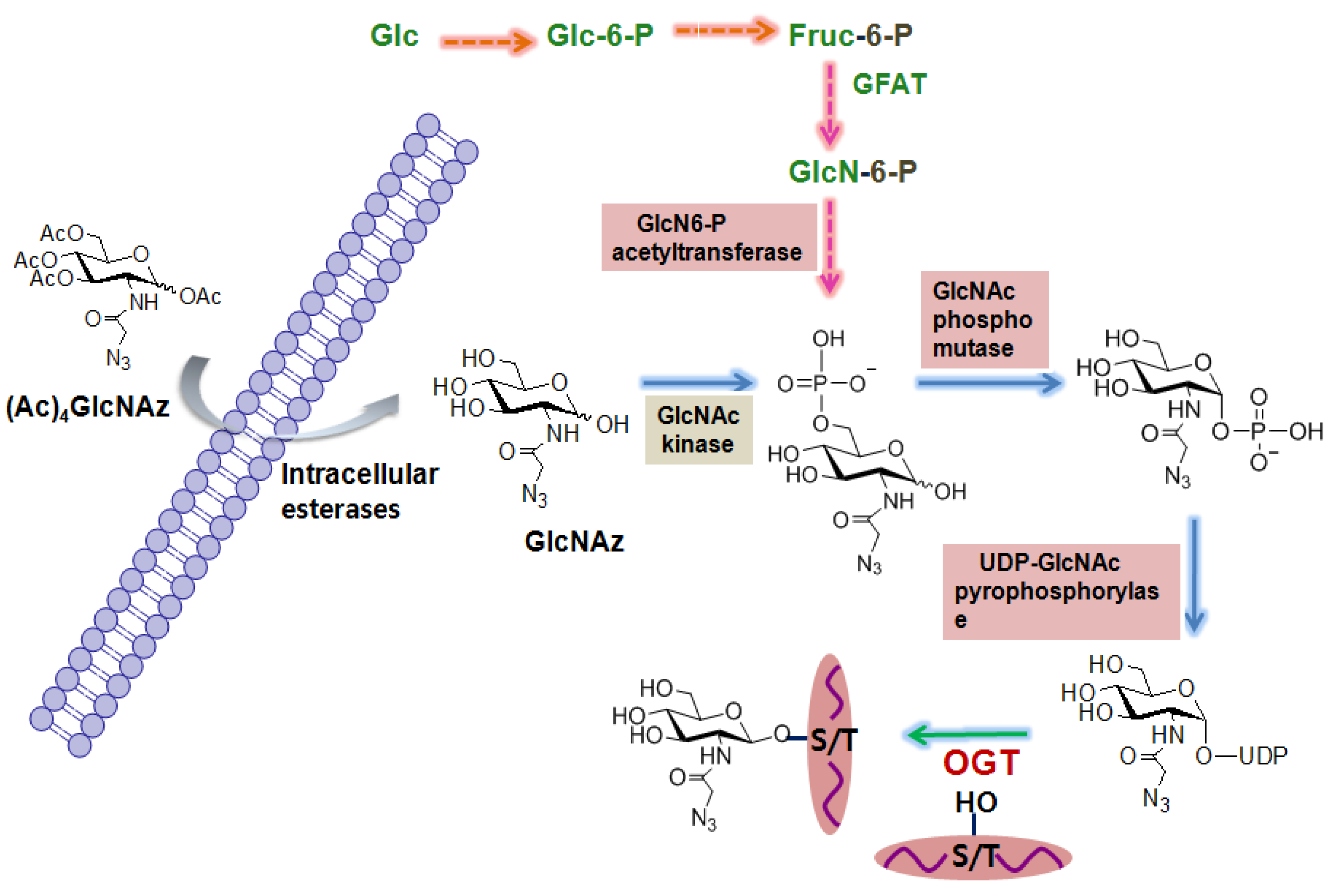
2.1.2. Metabolic labeling of O-GlcNAc-based enrichment approaches

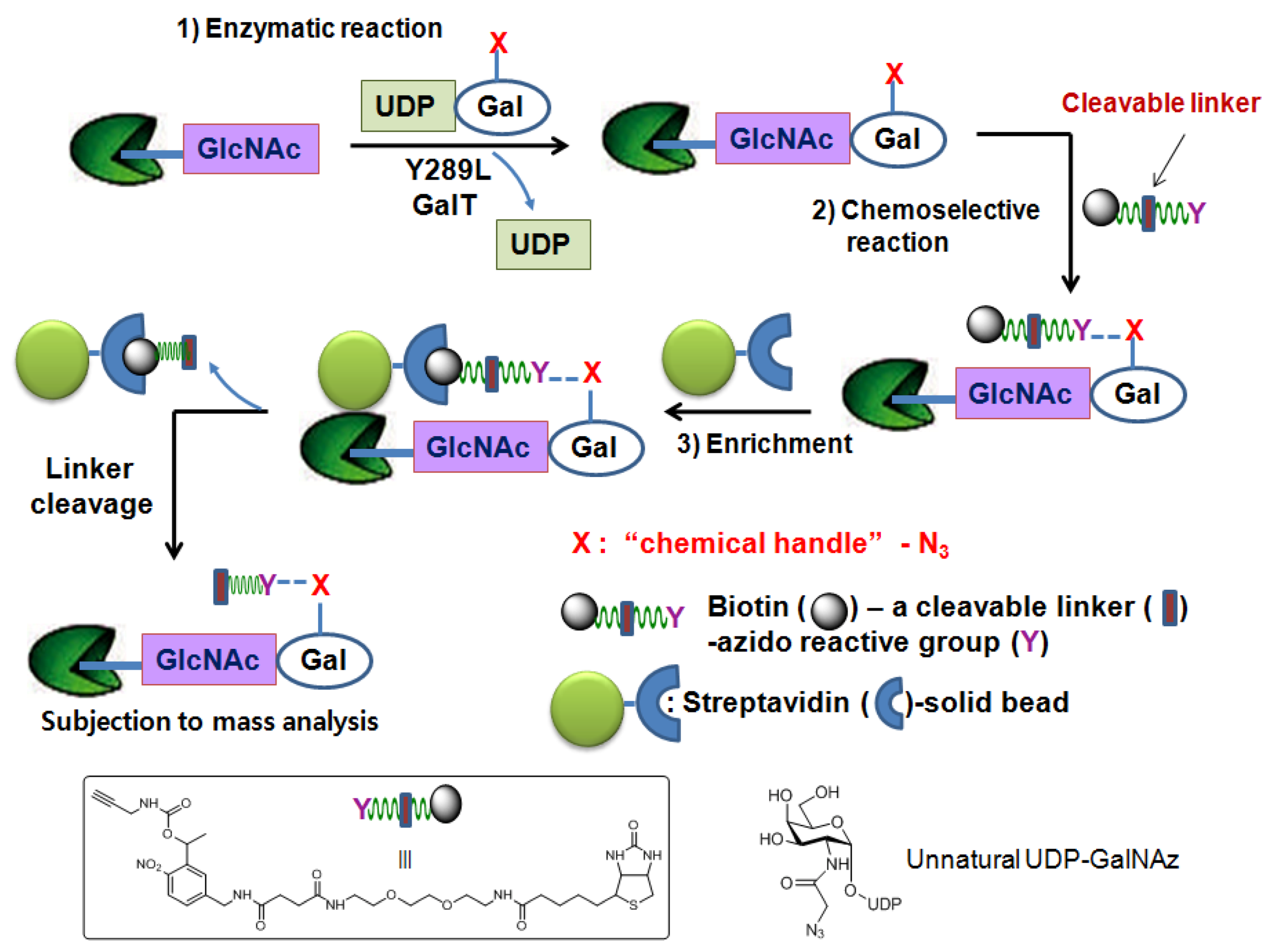
2.1.3. Chemoenzymatic tagging of O-GlcNAc-based enrichment approach

2.1.4. β-Elimination followed by Michael Addition Chemistry-based enrichment approach
2.2. New MS Fragmentation Techniques
2.3. Quantitative Proteomics of O-GlcNAc
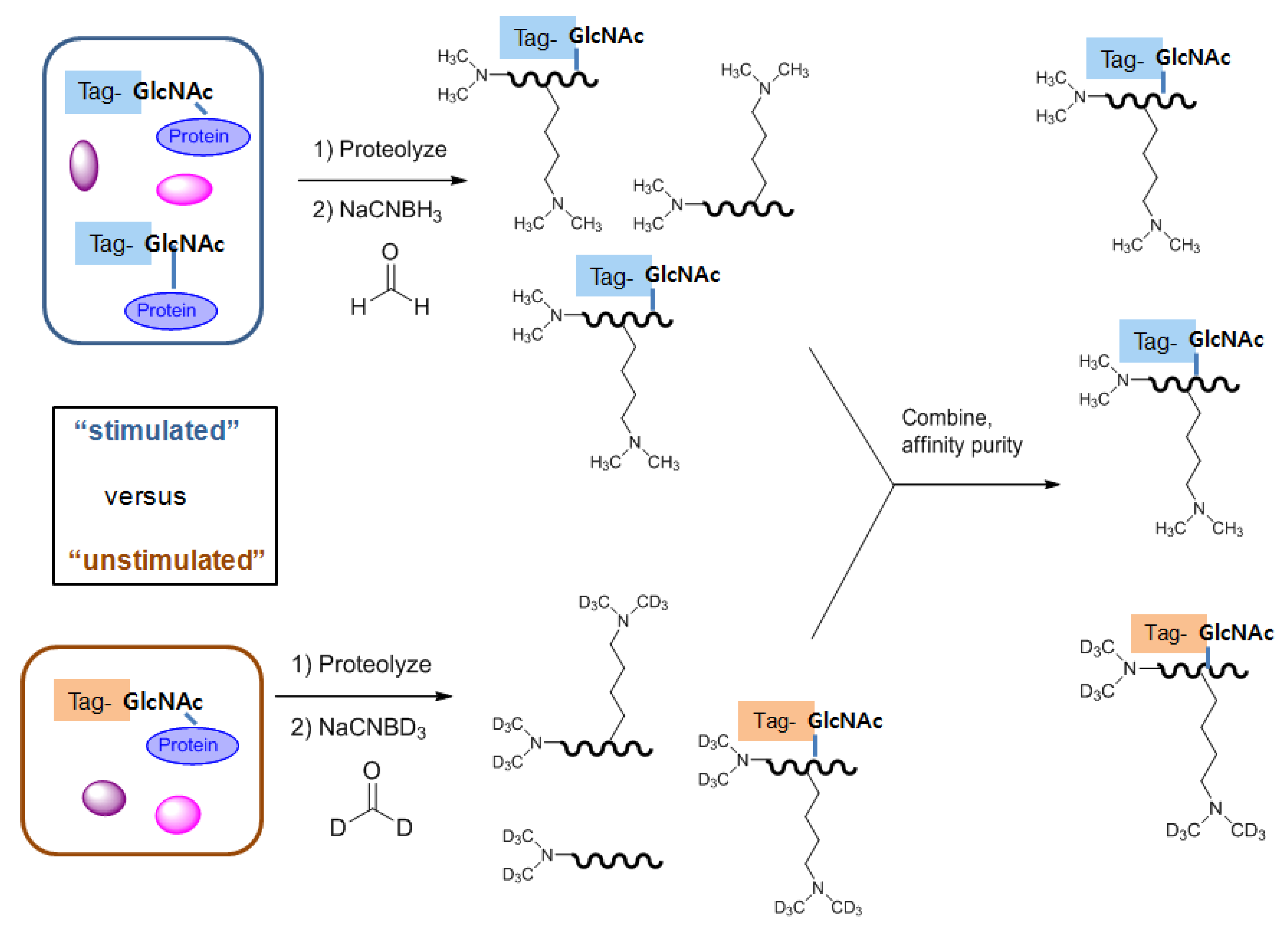
2.3.1. Identification and quantification of O-GlcNAc glycosylation in vivo
2.3.2. Identification and quantification of O-GlcNAc glycosylation in vitro
2.4. Significances of the Chemical Tools Designed for O-GlcNAc Proteomic Studies and Future Directions
3. O-GlcNAc Regulatory Enzymes
3.1. OGA
3.1.1. Brief background
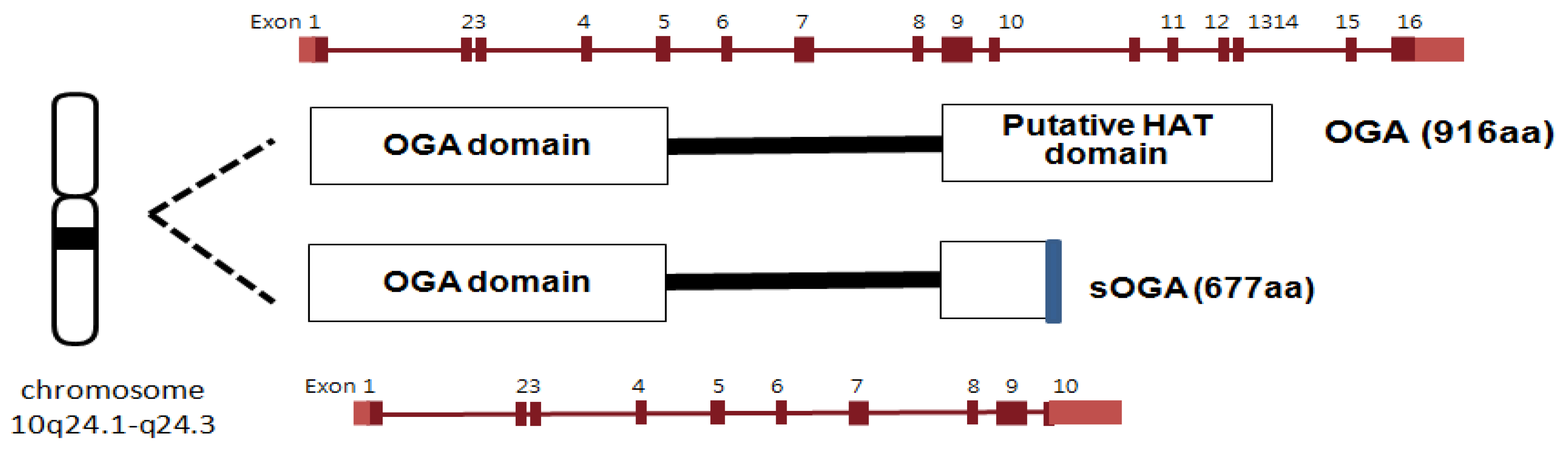
3.1.2. OGA activity assay methods and mechanistic studies of OGA



3.2. OGT
3.2.1. Brief background

3.2.2. OGT activity assay methods and the structural insights on OGT

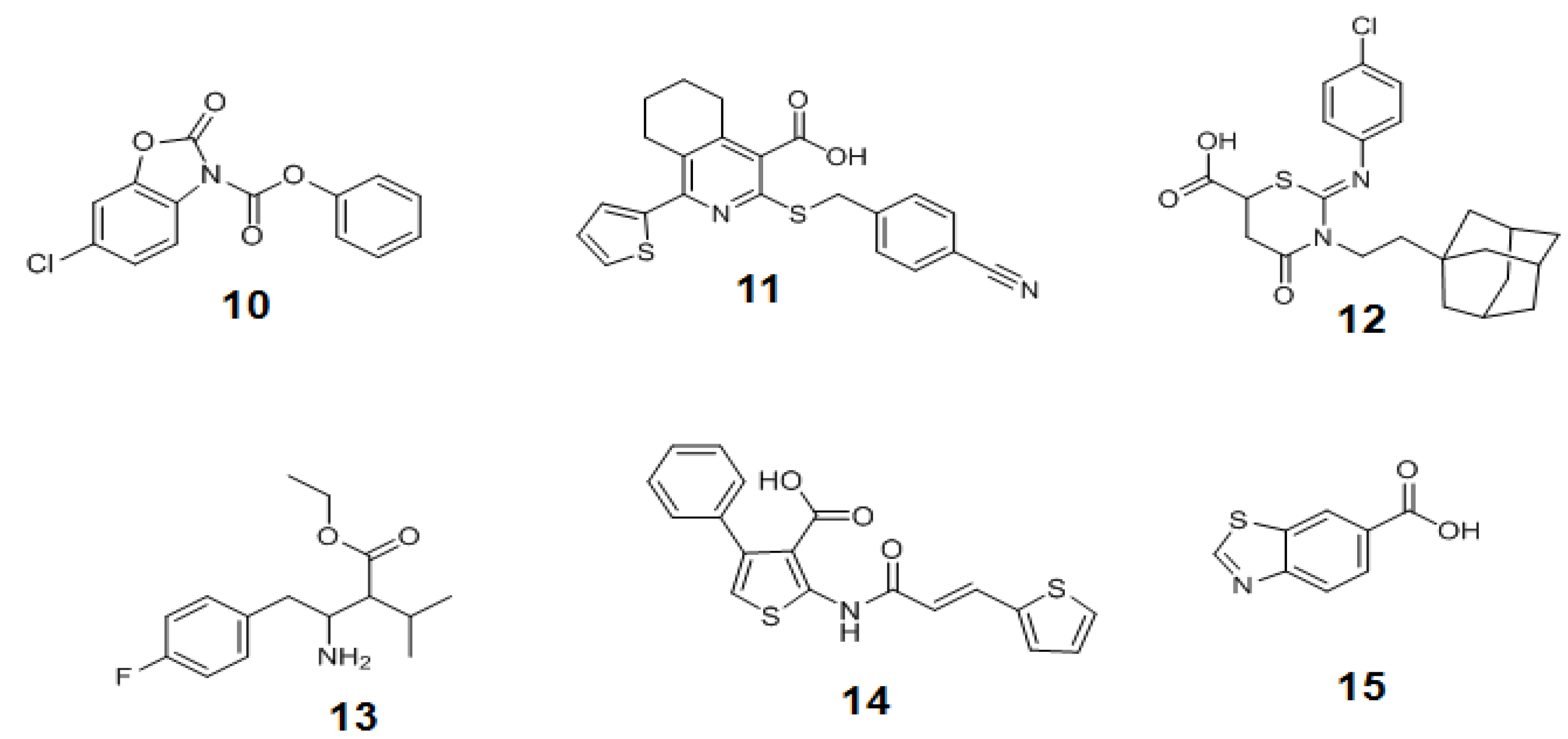

3.3. Significances of the Chemical Tools Designed for Modulating the O-GlcNAc Enzymes and Future Directions
4. Conclusions
Acknowledgements
References
- Torres, C.R.; Hart, G.W. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surface of intact lymphocytes. Evidence for O-linked GlcNAc. J. Biol. Chem. 1984, 259, 3308–3317. [Google Scholar]
- Roquemore, E.P.; Chevrier, M.R.; Cotter, R.J.; Hart, G.W. Dynamic O-GlcNAcylation of the small heat shock protein alpha B-crystallin. Biochemistry 1996, 35, 3578–3586. [Google Scholar] [CrossRef]
- Chou, C.F.; Smith, A.J.; Omary, M.B. Characterization and dynamics of O-linked glycosylation of human cytokeratin 8 and 18. J. Biol. Chem. 1992, 267, 3901–3906. [Google Scholar]
- Kearse, K.P.; Hart, G.W. Lymphocyte activation induces rapid changes in nuclear and cytoplasmic glycoproteins. Proc. Natl. Acad. Sci. USA 1991, 88, 1701–1705. [Google Scholar] [CrossRef]
- Walgren, J.L.; Vincent, T.S.; Schey, K.L.; Buse, M.G. High glucose and insulin promote O-GlcNAc modification of proteins, including alpha-tubulin. Am. J. Physiol. Endocrinol. Metab. 2003, 284, E424–E434. [Google Scholar]
- Gandy, J.C.; Rountree, A.E.; Bijur, G.N. Akt1 is dynamically modified with O-GlcNAc following treatments with PUGNAc and insulin-like growth factor-1. FEBS Lett. 2006, 580, 3051–3058. [Google Scholar] [CrossRef]
- Zachara, N.E.; O’Donnell, N.; Cheung, W.D.; Mercer, J.J.; Marth, J.D.; Hart, G.W. Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. J. Biol. Chem. 2004, 279, 30133–30142. [Google Scholar]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef]
- Haltiwanger, R.S.; Holt, G.D.; Hart, G.W. Enzymatic addition of O-GlcNAc to nuclear and cytoplasmic proteins. Identification of a uridine diphospho-N-acetylglucosamine: peptide β-N-acetylglucosaminyltransferase. J. Biol. Chem. 1990, 265, 2563–2568. [Google Scholar]
- Kreppel, L.K.; Blomberg, M.A.; Hart, G.W. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 1997, 272, 9308–9315. [Google Scholar]
- Lubas, W.A.; Frank, D.W.; Krause, M.; Hanover, J.A. O-Linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J. Biol. Chem. 1997, 272, 9316–9324. [Google Scholar] [CrossRef]
- Dong, D.L.; Hart, G.W. Purification and characterization of an O-GlcNAc selective Nacetyl-β-D-glucosaminidase from rat spleen cytosol. J. Biol. Chem. 1994, 269, 19321–19330. [Google Scholar]
- Gao, Y.; Wells, L.; Comer, F.I.; Parker, G.J.; Hart, G.W. Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic b-Nacetylglucosaminidase from human brain. J. Biol. Chem. 2001, 276, 9838–9845. [Google Scholar]
- Housley, M.P.; Rodgers, J.T.; Udeshi, N.D.; Kelly, T.J.; Shabanowitz, J.; Hunt, D.F.; Puigserver, P.; Hart, G.W. O-GlcNAc regulates FoxO activation in response to glucose. J. Biol. Chem. 2008, 283, 16283–16292. [Google Scholar]
- Kang, E.S.; Han, D.; Park, J.; Kwak, T.K.; Oh, M.A.; Lee, S.A.; Choi, S.; Park, Z.Y.; Kim, Y.; Lee, J.W. O-GlcNAc modulation at Akt1 Ser473 correlates with apoptosis of murine pancreatic beta cells. Exp. Cell Res. 2008, 314, 2238–2248. [Google Scholar]
- Wang, Z.; Gucek, M.; Hart, G.W. Cross-talk between GlcNAcylation and phosphorylation: Site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc. Natl. Acad. Sci. USA 2008, 105, 13793–13798. [Google Scholar] [CrossRef]
- Sanchez, M.; Picard, N.; Sauve, K.; Tremblay, A. Challenging estrogen receptor beta with phosphorylation. Trends Endocrinol. Metab. 2010, 21, 104–110. [Google Scholar] [CrossRef]
- Cheng, X.; Hart, G.W. Alternative O-glycosylation/Ophosphorylation of serine-16 in murine estrogen receptor beta: posttranslational regulation of turnover and transactivation activity. J. Biol. Chem. 2001, 276, 10570–10575. [Google Scholar] [CrossRef]
- Chen, Y.X.; Du, J.T.; Zhou, L.X.; Liu, X.H.; Zhao, Y.F.; Nakanishi, H.; Li, Y.M. Alternative O-GlcNAcylation/O-phosphorylation of Ser16 induce different conformational disturbances to the N-terminus of murine estrogen receptor beta. Chem. Biol. 2006, 13, 937–944. [Google Scholar] [CrossRef]
- Musicki, B.; Kramer, M.F.; Becker, R.E.; Burnett, A.L. Inactivation of phosphorylated endothelial nitric oxide synthase (Ser-1177) by O-GlcNAc in diabetes-associated erectile dysfunction. Proc. Natl. Acad. Sci. USA 2005, 102, 11870–11875. [Google Scholar]
- Yang, W.H.; Kim, J.E.; Nam, H.W.; Ju, J.W.; Kim, H.S.; Kim, Y.S.; Cho, J.W. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat. Cell Biol. 2006, 8, 1074–1083. [Google Scholar] [CrossRef]
- Dias, W.B.; Cheung, W.D.; Wang, Z.; Hart, G.W. Regulation of calcium/calmodulin-dependent kinase IV by O-GlcNAc modification. J. Biol. Chem. 2009, 284, 21327–21337. [Google Scholar]
- Gual, P.; Le Marchand-Brustel, Y.; Tanti, J.F. Positive and negative regulation of insulin signaling through IRS-1 phosphorylation. Biochimie 2005, 87, 99–109. [Google Scholar] [CrossRef]
- Yang, X.; Ongusaha, P.P.; Miles, P.D.; Havstad, J.C.; Zhang, F.; So, W.V.; Kudlow, J.E.; Michell, R.H.; Olefsky, J.M.; Field, S.J.; Evans, R.M. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 2008, 451, 964–969. [Google Scholar]
- Whelan, S.A.; Lane, M.D.; Hart, G.W. Regulation of the O-linked β-N-acetylglucosamine transferase by insulin signaling. J. Biol. Chem. 2008, 283, 21411–21417. [Google Scholar]
- Love, D.C; Hanover, J.A. The hexosamine signaling pathway: deciphering the “O-GlcNAc code”. Sci. STKE 2005, 312, re13. [Google Scholar]
- Hanover, J.A.; Krause, M.W.; Love, D.C. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim. Biophys. Acta 2010, 1800, 80–95. [Google Scholar]
- Teo, C.F.; Wollaston-Hayden, E.E.; Wells, L. Hexosamine flux, the O-GlcNAc modification, and the development of insulin resistance in adipocytes. Mol. Cell. Endocrinol. 2010, 318, 44–53. [Google Scholar] [CrossRef]
- Hu, P.; Shimoji, S.; Hart, G.W. Site-specific interplay between O-GlcNAcylation and phosphorylation in cellular regulation. FEBS Lett. 2010, 584, 2526–2538. [Google Scholar] [CrossRef]
- Butkinaree, C.; Park, K.; Hart, G.W. O-linked β-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim. Biophys. Acta 2010, 1800, 96–106. [Google Scholar]
- McClain, D.; Lubas, W.A.; Cooksey, R.C.; Hazel, M.; Parker, G.J.; Love, D.C.; Hanover, J.A. Altered glycandependent signaling induces insulin resistance and hyperleptinemia. Proc. Natl. Acad. Sci. USA 2002, 99, 10695–10699. [Google Scholar] [CrossRef]
- Vosseller, K.; Wells, L.; Lane, M.D.; Hart, G.W. Elevated nucleocytoplasmic glycosylation by O-GlcNAc results in insulin resistance associated with defects in Akt activation in 3T3-L1 adipocytes. Proc. Natl. Acad. Sci. USA 2002, 99, 5313–5318. [Google Scholar] [CrossRef]
- Chou, T.Y.; Dang, C.V.; Hart, G.W. Glycosylation of the c-Myc transactivation domain. Proc. Natl. Acad. Sci. USA 1995, 92, 4417–4421. [Google Scholar] [CrossRef]
- Liu, F.; Iqbal, K.; Grundke-Iqbal, I.; Hart, G.W.; Gong, C.X. O-GlcNAcylation regulates phosphorylation of tau: a mechanism involved in Alzheimer's disease. Proc. Natl. Acad. Sci. USA 2004, 101, 10804–10809. [Google Scholar]
- Lazarus, B.D.; Love, D.C.; Hanover, J.A. O-GlcNAc cycling: Implications for neurodegenerative disorders. Biochem. Cell Biol. 2009, 41, 2134–2146. [Google Scholar] [CrossRef]
- Love, D.C.; Krause, M.W.; Hanover, J.A. O-GlcNAc cycling: Emerging roles in development and epigenetics. Semin. Cell Dev. Biol. 2010, 21, 646–654. [Google Scholar]
- Kim, E.J.; Perreira, M.; Thomas, C.J.; Hanover, J.A. An O-GlcNAcase-specific inhibitor and substrate engineered by the extension of the N-acetyl moiety. J. Am. Chem. Soc. 2006, 128, 4234–4235. [Google Scholar]
- Macauley, M.S.; Whitworth, G.E.; Debowski, A.W.; Chin, D.; Vocadlo, D.J. O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors. J. Biol. Chem. 2005, 280, 25313–25322. [Google Scholar]
- Cetinbas, N.; Macauley, M.S.; Stubbs, K.A.; Drapala, R.; Vocadlo, D.J. Identification of Asp174 and Asp175 as the key catalytic residues of human O-GlcNAcase by functional analysis of site-directed mutants. Biochemistry 2006, 45, 3835–3844. [Google Scholar]
- Dorfmueller, H.C.; Borodkin, V.S.; Schimpl, M.; Shepherd, S.M.; Shpiro, N.A.; van Aalten, D.M. GlcNAcstatin: a picomolar, selective O-GlcNAcase inhibitor that modulates intracellular O-glcNAcylation levels. J. Am. Chem. Soc. 2006, 128, 16484–16485. [Google Scholar]
- Kim, E.J.; Kang, D.O.; Love, D.C.; Hanover, J.A. Enzymatic characterization of O-GlcNAcase isoforms using a fluorogenic GlcNAc substrate. Carbohydr. Res. 2006, 341, 971–982. [Google Scholar] [CrossRef]
- Copeland, R.J.; Bullen, J.W.; Hart, G.W. Cross-talk between GlcNAcylation and phosphorylation: roles in insulin resistance and glucose toxicity. Am. J. Physiol. Endocrinol. Metab. 2008, 295, E17–E28. [Google Scholar] [CrossRef]
- Macauley, M.S.; Vocadlo, D.J. Increasing O-GlcNAc levels: An overview of small-molecule inhibitors of O-GlcNAcase. Biochim. Biophys. Acta 2010, 1800, 107–121. [Google Scholar]
- Gloster, T.M.; Vocadlo, D.J. Mechanism, structure, and inhibition of O-GlcNAc processing enzymes. Curr. Signal. Transd. Ther. 2010, 5, 74–91. [Google Scholar] [CrossRef]
- Sullivan, S.; Wong, T.W. A manual sequencing method for identification of phosphorylated amino acids in phosphopeptides. Anal. Biochem. 1991, 197, 65–68. [Google Scholar] [CrossRef]
- Dong, D.L.; Xu, Z.S.; Hart, G.W.; Cleveland, D.W. Cytoplasmic O-GlcNAc modification of the head domain and the KSP repeat motif of the neurofilament protein neurofilament-H. J. Biol. Chem. 1996, 271, 20845–20852. [Google Scholar]
- Dong, D.L.; Xu, Z.S.; Chevrier, M.R.; Cotter, R.J.; Cleveland, D.W.; Hart, G.W. Glycosylation of mammalian neurofilaments. Localization of multiple O-linked N-acetylglucosamine moieties on neurofilament polypeptides L and M. J. Biol. Chem. 1993, 268, 16679–16687. [Google Scholar]
- Jiang, M.S.; Hart, G.W. A subpopulation of estrogen receptors are modified by O-linked N-acetylglucosamine. J. Biol. Chem. 1997, 272, 2421–2428. [Google Scholar]
- Cheng, X.; Hart, G.W. Glycosylation of the murine estrogen receptor-alpha. J. Steroid. Biochem. Mol. Biol. 2000, 75, 147–158. [Google Scholar] [CrossRef]
- Cole, R.N.; Hart, G.W. Glycosylation sites flank phosphorylation sites on synapsin I: O-linked N-acetylglucosamine residues are localized within domains mediating synapsin I interactions. J. Neurochem. 1999, 73, 418–428. [Google Scholar]
- Wells, L.; Vosseller, K.; Cole, R.N.; Cronshaw, J.M.; Matunis, M.J.; Hart, G.W. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol. Cell. Proteomics 2002, 1, 791–804. [Google Scholar] [CrossRef]
- Vosseller, K.; Trinidad, J.C.; Chalkley, R.J.; Specht, C.G.; Thalhammer, A.; Lynn, A.J.; Snedecor, J.O.; Guan, S.; Medzihradszky, K.F.; Maltby, D.A.; Schoepfer, R.; Burlingame, A.L. O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol. Cell. Proteomics 2006, 5, 923–934. [Google Scholar] [CrossRef]
- Wang, Z.; Pandey, A.; Hart, G.W. Dynamic interplay between O-linked N-acetylglucosaminylation and glycogen synthase kinase-3-dependent phosphorylation. Mol. Cell. Proteomics 2007, 6, 1365–1379. [Google Scholar] [CrossRef]
- Snow, C. M.; Senior, A.; Gerace, L. Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J. Cell Biol. 1987, 104, 1143–1156. [Google Scholar] [CrossRef]
- Comer, F.I.; Vosseller, K.; Wells, L.; Accavitti, M.A.; Hart, G.W. Characterization of a mouse monoclonal antibody specific for O-linked N-acetylglucosamine. Anal. Biochem. 2001, 293, 169–177. [Google Scholar]
- Chalkley, R.J.; Burlingame, A.L. Identification of GlcNAcylation sites of peptides and alpha-crystallin using Q-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 2001, 12, 1106–1113. [Google Scholar] [CrossRef]
- Haynes, P.A.; Aebersold, R. Simultaneous detection and identification of O-GlcNAc-modified glycoproteins using liquid chromatography-tandem mass spectrometry. Anal. Chem. 2000, 72, 5402–5410. [Google Scholar] [CrossRef]
- Khidekel, N.; Arndt, S; Lamarre-Vincent, N.; Lippert, A.; Poulin-Kerstien, K.G.; Ramakrishnan, B.; Qasba, P.K.; Hsieh-Wilson, L.C. A chemoenzymatic approach toward the rapid and sensitive detection of O-GlcNAc posttranslational modifications. J. Am. Chem. Soc. 2003, 125, 16162–16163. [Google Scholar]
- Roquemore, E.P.; Chou, T.Y.; Hart, G.W. Detection of O-linked N-acetylglucosamine (O-GlcNAc) on cytoplasmic and nuclear proteins. Methods Enzymol. 1994, 230, 443–460. [Google Scholar]
- Kayser, H.; Zeitler, R.; Kannicht, C.; Grunow, D.; Nuck, R.; Reutter, W. Biosynthesis of a nonphysiological sialic acid in different rat organs, using N-propanoyl-D-hexosamines as precursors. J. Biol. Chem. 1992, 267, 16934–16938. [Google Scholar]
- Kayser, H.; Geilen, C.C.; Paul, C.; Zeitler, R.; Reutter, W. New amino sugar analogues are incorporated at different rates into glycoproteins of mouse organs. Experientia 1993, 49, 885–887. [Google Scholar] [CrossRef]
- Mahal, L.K.; Yarema, K.J.; Bertozzi, C.R. Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis. Science 1997, 276, 1125–1128. [Google Scholar] [CrossRef]
- Hang, H.C.; Bertozzi, C.R. Ketone isosteres of 2-N-Acetamidosugars as substrates for metabolic cell surface engineering. J. Am. Chem. Soc. 2001, 123, 1242–1243. [Google Scholar]
- Saxon, E.; Bertozzi, C.R. Cell surface engineering by a modified staudinger reaction. Science 2000, 287, 2007–2010. [Google Scholar] [CrossRef]
- Hang, H.C.; Yu, C.; Kato, D.L.; Bertozzi, C.R. A metabolic labeling approach towards proteomic analysis of mucin-type O-linked glycosylation. Proc. Natl. Acad. Sci. USA 2003, 100, 14846–14851. [Google Scholar] [CrossRef]
- Vocadlo, D.J.; Hang, H.C.; Kim, E.J.; Hanover, J.A.; Bertozzi, C.R. A chemical approach for identifying O-GlcNAc modified proteins in cells. Proc. Natl. Acad. Sci. USA 2003, 100, 9116–9121. [Google Scholar] [CrossRef]
- Rabuka, D.; Hubbard, S.C.; Laughlin, S.T.; Argade, S.P.; Bertozzi, C.R. A chemical reporter strategy to probe Glycoprotein fucosylation. J. Am. Chem. Soc. 2006, 128, 12078–12079. [Google Scholar]
- Sampathkumar, S.G.; Jones, M.B.; Yarema, K.J. Metabolic expression of thiol-derivatized sialic acids on the cell surface and their quantitative estimation by flow cytometry. Nat. Protoc. 2006, 1, 1840–1851. [Google Scholar] [CrossRef]
- Laughlin, S.T.; Agard, N.J.; Baskin, J.M.; Carrico, I.S.; Chang, P.V.; Ganguli, A.S.; Hangauer, M. J.; Lo, A.; Prescher, J.A.; Bertozzi, C.R. Metabolic labeling of Glycans with azido sugars for visualization and glycoproteomics. Methods Enzymol. 2006, 415, 230–250. [Google Scholar] [CrossRef] [PubMed]
- Laughlin, S.T.; Bertozzi, C.R. Metabolic labeling of glycans with azido sugars and subsequent glycan-profiling and visualization via Staudinger ligation. Nat. Protoc. 2007, 2, 2930–2944. [Google Scholar] [CrossRef]
- Prescher, J.A.; Bertozzi, C.R. Chemistry in living systems. Nat. Chem. Biol. 2005, 1, 13–21. [Google Scholar]
- Rostovtsev, V.V.; Green, L.G.; Fokin, V.V.; Sharpless, K.B. A stepwise Huisgen cycloaddition process: Copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. 2002, 41, 2596–2599. [Google Scholar] [CrossRef]
- Speers, A.E.; Adam, G.C.; Cravatt, B.F. Activity-based protein profiling in vivo using a copper(I)-catalyzed azide-alkyne [3 + 2] cycloaddition. J. Am. Chem. Soc. 2003, 125, 4686–4687. [Google Scholar]
- Agard, N.J.; Prescher, J.A.; Bertozzi, C.R. A strain-promoted [3 + 2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 2004, 126, 15046–15047. [Google Scholar]
- Poloukhtine, A.A.; Mbua, N.E.; Wolfert, M.A.; Boons, G.J.; Popik, V.V. Selective labeling of living cells by a Photo-Triggered Click reaction. J. Am. Chem. Soc. 2009, 131, 15769–15776. [Google Scholar]
- Agard, N.J.; Bertozzi, C.R. Chemical approaches to perturb, profile, and perceive glycans. Acc. Chem. Res. 2009, 42, 788–797. [Google Scholar] [CrossRef]
- Sprung, R.; Nandi, A.; Chen, Y.; Kim, S.C.; Barma, D.; Falck, J.R.; Zhao, Y.M. Tagging-via-substrate strategy for probing O-GlcNAc modified proteins. J. Proteome Res. 2005, 4, 950–957. [Google Scholar] [CrossRef]
- Laughlin, S.T.; Baskin, J.M.; Amacher, S.L.; Bertozzi, C.R. In vivo imaging of membrane-associated glycans in developing zebrafish. Science 2008, 320, 664–667. [Google Scholar]
- Nandi, A.; Sprung, R.; Barma, D.K.; Zhao, Y.; Kim, S.C.; Falck, J.R. Global identification of O-GlcNAc-modified proteins. Anal. Chem. 2006, 78, 452–458. [Google Scholar] [CrossRef]
- Ramakrishnan, B.; Qasba, P.K. Structure-based design of beta 1,4-galactosyltransferase I (beta 4Gal-T1) with equally efficient Nacetylgalactosaminyltransferase activity: point mutation broadens beta 4Gal-T1 donor specificity. J. Biol. Chem. 2002, 277, 20833–20839. [Google Scholar] [CrossRef]
- Khidekel, N.; Arndt, S.; Lamarre-Vincent, N.; Lippert, A.; Poulin-Kerstien, K.G.; Ramakrishnan, B.; Qasba, P.K.; Hsieh-Wilson, L.C. A chemoenzymatic approach toward the rapid and sensitive detection of O-GlcNAc posttranslational modifications. J. Am. Chem.Soc. 2003, 125, 16162–16163. [Google Scholar]
- Wang, Z.; Park, K.; Comer, F.; Hsieh-Wilson, L.C.; Saudek, C.D.; Hart, G.W. Site-specific GlcNAcylation of human erythrocyte proteins: potential biomarker(s) for diabetes. Diabetes 2009, 58, 309–317. [Google Scholar]
- Khidekel, N.; Ficarro, S.B.; Peters, E.C.; Hsieh-Wilson, L.C. Exploring the O-GlcNAc proteome: direct identification of O-GlcNAc-modified proteins from the brain. Proc. Natl. Acad. Sci. USA 2004, 101, 13132–13137. [Google Scholar]
- Tai, H.C.; Khidekel, N.; Ficarro, S.B.; Peters, E.C.; Hsieh-Wilson, L.C. Parallel identification of O-GlcNAc-modified proteins from cell lysates. J. Am. Chem. Soc. 2004, 126, 10500–10501. [Google Scholar]
- Wang, Z.; Udeshi, N.D.; O’Malley, M.; Shabanowitz, J.; Hunt, D.F.; Hart, G.W. Enrichment and site mapping of O-linked N-acetylglucosamine by a combination of chemical/enzymatic tagging, photochemical cleavage, and electron transfer dissociation mass spectrometry. Mol. Cell. Proteomics 2010, 9, 153–160. [Google Scholar] [CrossRef]
- Greis, K.D.; Hayes, B.K.; Comer, F.I.; Kirk, M.; Barnes, S.; Lowary, T.L.; Hart, G.W. Selective detection and site-analysis of O-GlcNAc-modified glycopeptides by b-elimination and tandem electrospray mass spectrometry. Anal. Biochem. 1996, 234, 38–49. [Google Scholar]
- McLachlin, D.T.; Chait, B.T. Improved â-elimination-based affinity purification strategy for enrichment of phosphopeptides. Anal. Chem. 2003, 75, 6826–6836. [Google Scholar] [CrossRef]
- Whelan, S.A.; Hart, G.W. Identification of O-GlcNAc sites on proteins. Methods Enzymol. 2006, 415, 113–133. [Google Scholar] [CrossRef]
- Reinhold, V.N.; Reinhold, B.B.; Costello, C.E. Carbohydrate molecular weight profiling, sequence, linkage, and branching data: ES-MS and CID. Anal. Chem. 1995, 67, 1772–1784. [Google Scholar]
- Zubarev, R.A.; Kelleher, N.L.; McLafferty, F.W. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J. Am. Chem. Soc. 1998, 120, 2. [Google Scholar]
- Woodling, K.A.; Eyler, J.R.; Tsybin, Y.O.; Nilsson, C.L.; Marshall, A.G.; Edison, A.S.; Al-Naggar, I.M.; Bubb, M.R. Identification of single and double sites of phosphorylation by ECD FT-ICR/MS in peptides related to the phosphorylation site domain of the myristoylated alanine-rich C kinase protein. J. Am. Soc. Mass. Spectr. 2007, 18, 2137–2145. [Google Scholar]
- Creese, A.J.; Cooper, H.J. The Effect of phosphorylation on the electron capture dissociation of peptide ions. J. Am. Soc. Mass. Spectr. 2008, 19, 1263–1274. [Google Scholar] [CrossRef]
- Syka, J.E.; Coon, J.J.; Schroeder, M.J.; Shabanowitz, J.; Hunt, D.F. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. USA 2004, 101, 9528–9533. [Google Scholar] [CrossRef]
- Mikesh, L.M.; Ueberheide, B.; Chi, A.; Coon, J.J.; Syka, J.E.; Shabanowitz, J.; Hunt, D.F. The utility of ETD mass spectrometry in proteomic analysis. Biochim. Biophys. Acta 2006, 1764, 1811–1822. [Google Scholar]
- Klein, A.L.; Berkaw, M.N.; Buse, M.G.; Ball, L.E. O-linked Nacetylglucosamine modification of insulin receptor substrate-1 occurs in close proximity to multiple SH2 domain binding motifs. Mol. Cell. Proteomics 2009, 8, 2733–2745. [Google Scholar] [CrossRef]
- Housley, M.P.; Udeshi, N.D.; Rodgers, J.T.; Shabanowitz, J.; Puigserver, P.; Hunt, D.F.; Hart, G.W. A PGC-1alpha-O-GlcNAc transferase complex regulates FoxO transcription factor activity in response to glucose. J. Biol. Chem. 2009, 284, 5148–5157. [Google Scholar]
- Wang, Z.; Udeshi, N.D.; Slawson, C.; Compton, P.D.; Sakabe, K.; Cheung, W.D.; Shabanowitz, J.; Hunt, D.F.; Hart, G.W. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci. Signal. 2010, 3, ra2. [Google Scholar] [CrossRef]
- Orlando, R.; Lim, J.M.; Atwood, J.A., III; Angel, P.M.; Fang, M.; Aoki, K.; Alvarez-Manilla, G.; Moremen, K.W.; York, W.S.; Tiemeyer, M.; Pierce, M.; Dalton, S.; Wells, L. IDAWG: Metabolic incorporation of stable isotope labels for quantitative glycomics of cultured cells. J. Proteome Res. 2009, 8, 3816–3823. [Google Scholar]
- Zhu, H.; Pan, S.; Gu, S.; Bradbury, E.M.; Chen, X. Amino acid residue specific stable isotope labeling for quantitative proteomics. Rapid Commun. Mass Sp. 2002, 16, 2115–2123. [Google Scholar] [CrossRef]
- Amanchy, R.; Kalume, D.E.; Pandey, A. Stable isotope labeling with amino acids in cell culture (SILAC) for studying dynamics of protein abundance and posttranslational modifications. Sci. STKE 2005, 267, pl2. [Google Scholar]
- Harsha, H.C.; Molina, H.; Pandey, A. Quantitative proteomics using stable isotope labeling with amino acids in cell culture. Nat. Protoc. 2008, 3, 505–516. [Google Scholar] [CrossRef]
- Thompson, A.; Schäfer, J.; Kuhn, K.; Kienle, S.; Schwarz, J.; Schmidt, G.; Neumann, T.; Johnstone, R.; Mohammed, A.K.; Hamon, C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 2003, 75, 1895–1904. [Google Scholar] [CrossRef]
- Viner, R.I.; Zhang, T.; Second, T.; Zabrouskov, V. Quantification of post-translationally modified peptides of bovine alpha-crystallin using tandem mass tags and electron transfer dissociation. J. Proteomics 2009, 72, 874–885. [Google Scholar] [CrossRef]
- Ross, P.L; Huang, Y.N.; Marchese, J.N.; Williamson, B.; Parker, K.; Hattan, S.; Khainovski, N.; Pillai, S.; Dey, S.; Daniels, S.; Purkayastha, S.; Juhasz, P.; Martin, S.; Bartlet-Jones, M.; He, F.; Jacobson, A.; Pappin, D.J. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 2004, 3, 1154–1169. [Google Scholar]
- Hsu, J.L.; Huang, S.Y.; Chow, N.H.; Chen, S.H. Stable-isotope dimethyl labeling for quantitative proteomics. Anal. Chem. 2003, 75, 6843–6852. [Google Scholar]
- Khidekel, N.; Ficarro, S.B.; Clark, P.M.; Bryan, M.C.; Swaney, D.L.; Rexach, J.E.; Sun, Y.E.; Coon, J.J.; Peters, E.C.; Hsieh-Wilson, L.C. Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat. Chem. Biol. 2007, 3, 339–348. [Google Scholar]
- Majumdar, G.; Harrington, A.; Hungerford, J.; Martinez-Hernandez, A.; Gerling, I.C.; Raghow, R.; Solomon, S. Insulin dynamically regulates calmodulin gene expression by sequential O-Glycosylation and phosphorylation of sp1 and its subcellular compartmentalization in liver cells. J. Biol. Chem. 2006, 281, 3642–3650. [Google Scholar]
- Knight, Z.A.; Shokat, K.M. Chemical genetics: where genetics and pharmacology meet. Cell 2007, 128, 425–430. [Google Scholar] [CrossRef]
- Henrissat, B.; Davies, G. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 1997, 7, 637–644. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The carbohydrate-active enzymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Campbell, J.A.; Davies, G.J.; Bulone, V.; Henrissat, B. A classification of nucleotidediphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 1997, 326, 929–939. [Google Scholar]
- Coutinho, P.; Deleury, E.; Davies, G.J.; Henrissat, B. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 2003, 328, 307–317. [Google Scholar] [CrossRef]
- Farook, V.S.; Bogardus, C.; Prochazka, M. Analysis of MGEA5 on 10q24.1-q24.3 encoding the b-O-linked N-acetylglucosaminidase as a candidate gene for type 2 diabetes mellitus in Pima Indians. Mol. Genet. Metab. 2002, 77, 189–193. [Google Scholar]
- Heckel, D.; Comtesse, N.; Brass, N.; Blin, N.; Zang, K.D.; Meese, E. Novel immunogenic antigen homologous to hyaluronidase in meningioma. Hum. Mol. Genet. 1998, 7, 1859–1872. [Google Scholar] [CrossRef]
- Lehman, D.M.; Fu, D.J.; Freeman, A.B.; Hunt, K.J.; Leach, R.J.; Johnson-Pais, T.; Hamlington, J.; Dyer, T.D.; Arya, R.; Abboud, H.; Goring, H.H.; Duggirala, R.; Blangero, J.; Konrad, R.J.; Stern, M.P. A single nucleotide polymorphism in MGEA5 encoding O-GlcNAc-selective N-acetyl-β-D-glucosaminidase is associated with type 2 diabetes in Mexican Americans. Diabetes 2005, 54, 1214–1221. [Google Scholar]
- Comtesse, N.; Maldener, E.; Meese, E. Identification of a nuclear variant of MGEA5, a cytoplasmic hyaluronidase and a β-N-acetylglucosaminidase. Biochem. Biophys. Res. Commun. 2001, 283, 634–640. [Google Scholar] [CrossRef]
- Toleman, C.; Paterson, A.J.; Shin, R.; Kudlow, J.E. Streptozotocin inhibits O-GlcNAcase via the production of a transition state analog. Biochem. Biophys. Res. Commun. 2006, 340, 526–534. [Google Scholar] [CrossRef]
- Toleman, C.; Paterson, A.J.; Whisenhunt, T.R.; Kudlow, J.E. Characterization of the histone acetyltransferase (HAT) domain of a bifunctional protein with activable O-GlcNAcase and HAT activities. J. Biol. Chem. 2004, 279, 53665–53673. [Google Scholar]
- Butkinaree, C.; Cheung, W.D.; Park, S.; Park, K.; Barber, M.; Hart, G.W. Characterization of β-N-acetylglucosaminidase cleavage by caspase-3 during apoptosis. J. Biol. Chem. 2008, 283, 23557–23566. [Google Scholar]
- Wells, L.; Gao, Y.; Mahoney, J.A.; Vosseller, K.; Chen, C.; Rosen, A.; Hart, G.W. Dynamic O-glycosylation of nuclear and cytosolic proteins: further characterization of the nucleocytoplasmic β-N-acetylglucosaminidase, O-GlcNAcase. J. Biol.Chem. 2002, 277, 1755–1761. [Google Scholar]
- Glasgow, L.R.; Raulson, J.A.; Hill, R.L. Systematic purification of five glycosidases from Streptococcus pneumonia. J. Biol. Chem. 1977, 252, 8615–8623. [Google Scholar] [PubMed]
- Rao, F.V.; Dorfmueller, H.C.; Villa, F.; Allwood, M.; Eggleston, I.M.; van Aalten, D.M. Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis. EMBO J. 2006, 25, 1569–1578. [Google Scholar]
- Ficko-Blean, E.; Boraston, A.B. Cloning, recombinant production, crystallization and preliminary X-ray diffraction studies of a family 84 glycoside hydrolase from Clostridium perfringens. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2005, 61, 834–836. [Google Scholar] [CrossRef]
- Dennis, R.J.; Taylor, E.J.; Macauley, M.S.; Stubbs, K.A.; Turkenburg, J.P.; Hart, S.J.; Black, G.N.; Vocadlo, D.J.; Davies, G.J. Structure and mechanism of a bacterial β-glucosaminidase having O-GlcNAcase activity. Nat. Struct. Mol. Biol. 2006, 13, 365–371. [Google Scholar]
- Miller, D.J.; Gong, X.; Shur, B.D. Sperm require β-N-acetylglucosaminidase to penetrate through the egg zona pellucid. Development 1993, 118, 1279–1289. [Google Scholar]
- Knapp, S.; Vocadlo, D.; Gao, Z.N.; Kirk, B.; Lou, J.P.; Withers, S.G. NAG-thiazoline, an N-acetyl-β-hexosaminidase inhibitor that implicates acetamido participation. J. Am. Chem. Soc. 1996, 118, 6804–6805. [Google Scholar]
- Yuzwa, S.A.; Macauley, M.S.; Heinonen, J.E.; Shan, X.; Dennis, R.J.; He, Y.; Whitworth, G.E.; Stubbs, K.A.; McEachern, E.J.; Davies, G.J.; Vocadlo, D.J. A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat. Chem. Biol. 2008, 4, 483–490. [Google Scholar] [CrossRef]
- Dorfmueller, H.C.; Borodkin, V.S.; Schimpl, M.; van Aalten, D.M. GlcNAcstatins are nanomolar inhibitors of human O-GlcNAcase inducing cellular hyper-O-GlcNAcylation. Biochem. J. 2009, 420, 221–227. [Google Scholar] [CrossRef]
- Aoyagi, T.; Suda, H.; Uotani, K.; Kojima, F.; Aoyama, T.; Horiguchi, K.; Hamada, M.; Takeuchi, T. Nagstatin, a new inhibitor of N-acetyl-β-D-glucosaminidase, produced by Streptomyces amakusaensis MG846-fF3. Taxonomy, production, isolation, physico-chemical properties and biological activities. J. Antibiot.(Tokyo) 1992, 45, 1404–1408. [Google Scholar]
- Terinek, M.; Vasella, A. Synthesis of N-acetylglucosamine-derived nagstatin analogues and their evaluation as glycosidase inhibitors. Helv. Chim. Acta 2005, 88, 10–22. [Google Scholar] [CrossRef]
- Shanmugasundaram, B.; Debowski, A.W.; Dennis, R.J.; Davies, G.J.; Vocadlo, D.J.; Vasella, A. Inhibition of O-GlcNAcase by a gluco-configured nagstatin and a PUGNAc-imidazole hybrid inhibitor. Chem. Commun. 2006, 4372–4374. [Google Scholar]
- Lubas, W.A.; Hanover, J.A. Functional expression of O-linked GlcNAc transferase. Domain structure and substrate specificity. J. Biol. Chem. 2000, 275, 10983–10988. [Google Scholar]
- Shafi, R.; Iyer, S.P.; Ellies, L.G.; O'Donnell, N.; Marek, K.W.; Chui, D.; Hart, G.W.; Marth, J.D. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. USA 2000, 97, 5735–5739. [Google Scholar]
- Hanover, J.A.; Yu, S.; Lubas, W.B.; Shin, S.H.; Ragano-Caracciola, M.; Kochran, J.; Love, D.C. Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene. Arch. Biochem. Biophys. 2003, 409, 287–297. [Google Scholar] [CrossRef]
- O’Donnell, N.; Zachara, N.E.; Hart, G.W.; Marth, J.D. Ogtdependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol. Cell Biol. 2004, 24, 1680–1690. [Google Scholar] [CrossRef]
- Hanover, J.A.; Forsythe, M.E.; Hennessey, P.T.; Brodigan, T.M.; Love, D.C.; Ashwell, G.; Krause, M.A. Caenorhabditis elegans model of insulin resistance: altered macronutrient storage and dauer formation in an OGT-1 knockout. Proc. Natl. Acad. Sci. USA 2005, 102, 11266–11271. [Google Scholar]
- Haltiwanger, R.S.; Blomberg, M.A.; Hart, G.W. Glycosylation of nuclear and cytoplasmic proteins. Purification and characterization of a uridine diphospho-N-acetylglucosamine:polypeptide b-N-acetylglucosaminyltransferase. J. Biol. Chem. 1992, 267, 9005–9013. [Google Scholar]
- Love, D.C.; Kochan, J.; Cathey, R.L.; Shin, S.H.; Hanover, J.A. Mitochondrial and nucleocytoplasmic targeting of O-linked GlcNAc transferase. J. Cell Sci. 2003, 116, 647–654. [Google Scholar] [CrossRef]
- Sikorski, R.S.; Boguski, M.S.; Goebl, M.; Hieter, P. A repeating amino acid motif in CDC23 defines a new family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell 1990, 26, 307–317. [Google Scholar]
- Blatch, G.L.; Lassle, M. The tetratricopeptide repeat: A structural motif mediating protein-protein interactions. Bioessays 1999, 21, 932–939. [Google Scholar] [CrossRef]
- Wrabl, J.O.; Grishin, N.V. Homology between O-linked GlcNAc transferases and proteins of the glycogen phosphorylase superfamily. J. Mol. Biol. 2001, 314, 365–374. [Google Scholar] [CrossRef]
- Lazarus, B.D.; Love, D.C.; Hanover, J.A. Recombinant O-GlcNAc transferase isoforms: identification of O-GlcNAcase, yes tyrosine kinase, and tau as isoform-specific substrates. Glycobiology 2006, 16, 415–421. [Google Scholar] [CrossRef]
- Comer, F.I.; Hart, G.W. Reciprocity between O-GlcNAc and O-phosphate on the carboxyl terminal domain of RNA polymerase II. Biochemistry 2001, 40, 7845–7852. [Google Scholar] [CrossRef]
- Zhang, F.; Su, K.; Yang, X.; Bowe, D.B.; Paterson, A.J.; Kudlow, J.E. O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell 2003, 115, 715–725. [Google Scholar] [CrossRef]
- Zhang, F.; Hu, Y.; Huang, P.; Toleman, C.A.; Paterson, A.J.; Kudlow, J.E. Proteasome function is regulated by cyclic AMP-dependent protein kinase through phosphorylation of Rpt6. J. Biol. Chem. 2007, 282, 22460–22471. [Google Scholar]
- Zachara, N.E.; O’Donnell, N.; Cheung, W.D.; Mercer, J.J.; Marth, J.D.; Hart, G.W. Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. A survival response of mammalian cells. A survival response of mammalian cells. J. Biol. Chem. 2004, 279, 30133–30142. [Google Scholar]
- Du, X.L.; Edelstein, D.; Dimmeler, S.; Ju, Q.; Sui, C.; Brownlee, M. Hyperglycemia inhibits endothelial nitric oxide synthase activity by posttranslational modification at the Akt site. J. Clin. Invest. 2001, 108, 1341–1348. [Google Scholar]
- Fletcher, B.S.; Dragstedt, C.; Notterpek, L.; Nolan, G.P. Functional cloning of SPIN-2, a nuclear anti-apoptotic protein with roles in cell cycle progression. Leukemia 2002, 16, 1507–1518. [Google Scholar] [CrossRef]
- Gross, B.J.; Kraybill, B.C.; Walker, S. Discovery of O-GlcNAc transferase inhibitors. J. Am. Chem. Soc. 2005, 127, 14588–14589. [Google Scholar] [CrossRef]
- Kreppel, L.K.; Hart, G.W. Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J. Biol. Chem. 1999, 274, 32015–32022. [Google Scholar] [CrossRef]
- Jinek, M.; Rehwinkel, J.; Lazarus, B.D.; Izaurralde, E.; Hanover, J.A.; Conti, E. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat. Struct. Mol. Biol. 2004, 11, 1001–1007. [Google Scholar]
- Conti, E.; Uy, M.; Leighton, L.; Blobel, G.; Kuriyan, J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell 1998, 94, 193–204. [Google Scholar] [CrossRef]
- Huber, A.H.; Weis, W.I. The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell 2001, 105, 391–402. [Google Scholar] [CrossRef]
- Wongkongkatep, J.; Miyahara, Y.; Ojida, A. Label-free, real-time Glycosyltransferase assay based on a fluorescent artificial chemosensor. Angew. Chem. Int. Ed. 2006, 45, 665–668. [Google Scholar] [CrossRef]
- Helm, J.S.; Hu, Y.; Chen, L.; Gross, B.; Walker, S. Identification of active-site inhibitors of MurG using a generalizable, high-throughput glycosyltransferase screen. J. Am. Chem. Soc. 2003, 125, 11168–11169. [Google Scholar] [CrossRef]
- Gross, B.J.; Kraybill, B.C.; Walker, S. Discovery of O-GlcNAc transferase inhibitors. J. Am. Chem. Soc. 2005, 127, 14588–14589. [Google Scholar]
- Gross, B.J.; Swoboda, J.G.; Walker, S. A strategy to discover inhibition of O-linked glycosylation. J. Am. Chem. Soc. 2008, 130, 440–441. [Google Scholar] [CrossRef]
- Leavy, T.M.; Bertozzi, C.R. A high-throughput assay for O-GlcNAc transferase detects primary sequence preferences in peptide substrates. Bioorg.Med. Chem. Lett. 2007, 17, 3851–3854. [Google Scholar] [CrossRef]
- Martinez-Fleites, C.; Macauley, M.S.; He, Y.; Shen, D.L.; Vocadlo, D.J.; Davies, G.J. Structure of an O-GlcNAc transferase homolog provides insight into intracellular glycosylation. Nat. Struct. Mol. Biol. 2008, 15, 764–765. [Google Scholar] [CrossRef]
- Clarke, A.J.; Hurtado-Guerrero, R.; Pathak, S.; Schüttelkopf, A.W.; Borodkin, V.; Shepherd, S.M.; Ibrahim, A.F.; van Aalten, D.M. Structural insights into mechanism and specificity of O-GlcNAc transferase. EMBO J. 2008, 27, 2780–2788. [Google Scholar] [CrossRef]
- Lairson, L.L.; Henrissat, B.; Davies, G.J.; Withers, S.G. Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 2008, 77, 521–555. [Google Scholar] [CrossRef]
- Herr, R.; Jahnke, H.K.; Argoudelis, A.D. Structure of streptozotocin. J. Am. Chem. Soc. 1967, 89, 4808–4809. [Google Scholar] [CrossRef]
- Mansford, K.R.; Opie, L. Comparison of metabolic abnormalities in diabetes mellitus induced by streptozotocin or by alloxan. Lancet 1968, 1, 670–671. [Google Scholar]
- O'Brien, B.A.; Harmon, B.V.; Cameron, D.P.; Allan, D.J. Beta-cell apoptosis is responsible for the development of IDDM in the multiple low-dose streptozotocin model. J. Pathol. 1996, 178, 176–181. [Google Scholar] [CrossRef]
- Saini, K.S.; Thompson, C.; Winterford, C.M.; Walker, N.I.; Cameron, D.P. Streptozotocin at low doses induces apoptosis and at high doses causes necrosis in a murine pancreatic beta cell line, INS-1. Biochem. Mol. Biol. Int. 1996, 39, 1229–1236. [Google Scholar]
- Arias, E.B.; Kim, J.; Cartee, G.D. Prolonged incubation in PUGNAc results in increased protein O-linked glycosylation and insulin resistance in rat skeletal muscle. Diabetes 2004, 53, 921–930. [Google Scholar] [CrossRef]
- Yamamoto, H.; Uchigata, Y.; Okamoto, H. Streptozotocin and alloxan induce dna strand breaks and poly(adp-ribose) synthetase in pancreatic-islets. Nature 1981, 294, 284–286. [Google Scholar]
- Kroncke, K.D.; Fehsel, K.; Sommer, A.; Rodriguez, M.L.; Kolb-Bachofen, V. Nitric oxide generation during cellular metabolization of the diabetogenic N-methyl-Nnitroso-urea streptozotozin contributes to islet cell DNA damage. Biol. Chem. Hoppe Seyler 1995, 376, 179–185. [Google Scholar] [CrossRef]
- Turk, J.; Corbett, J.A.; Ramanadham, S.; Bohrer, A.; McDaniel, M.L. Biochemical evidence for nitric oxide formation from streptozotocin in isolated pancreatic islets. Biochem. Biophys. Res. Commun. 1993, 197, 1458–1464. [Google Scholar] [CrossRef]
- Kaneto, H.; Fujii, J.; Seo, H.G.; Suzuki, K.; Matsuoka, T.; Nakamura, M.; Tatsumi, H.; Yamasaki, Y.; Kamada, T.; Taniguchi, N. Apoptotic cell death triggered by nitric oxide in pancreatic beta-cells. Diabetes 1995, 44, 733–738. [Google Scholar]
- Gao, Y.; Parker, G.J.; Hart, G.W. Streptozotocin-induced beta-cell death is independent of its inhibition of O-GlcNAcase in pancreatic Min6 cells. Arch. Biochem. Biophys. 2000, 383, 296–302. [Google Scholar] [CrossRef]
- Okuyama, R.; Yachi, M. Cytosolic O-GlcNAc accumulation is not involved in beta-cell death in HIT-T15 or Min6. Biochem. Biophys. Res. Commun. 2001, 287, 366–371. [Google Scholar] [CrossRef]
- Nojiri, H.; Stroud, M.; Hakomori, S. A specific type of ganglioside as a modulator of insulin-dependent cell growth and insulin receptor tyrosine kinase activity. Possible association of ganglioside-induced inhibition of insulin receptor function and monocytic differentiation induction in HL-60 cells. J. Biol. Chem. 1991, 266, 4531–4537. [Google Scholar]
- Macauley, M.S.; Bubb, A.K.; Martinez-Fleites, C.; Davies, G.J.; Vocadlo, D.J. Elevation of global O-GlcNAc levels in 3 T3-L1 adipocytes by selective inhibition of O-GlcNAcase does not induce insulin resistance. J. Biol. Chem. 2008, 283, 34687–34695. [Google Scholar]
- Macauley, M.S.; He, Y.; Gloster, T.M.; Stubbs, K.A.; Davies, G.J.; Vocadlo, D.J. Inhibition of O-GlcNAcase using a potent and cell-permeable inhibitor does not induce insulin resistance in 3T3-L1 adipocytes. Chem. Biol. 2010, 17, 937–948. [Google Scholar] [CrossRef]
- Macauley, M.S.; Shan, X.; Yuzwa, S.A.; Gloster, T.M., Vocadlo. Elevation of Global O-GlcNAc in rodents using a selective O-GlcNAcase inhibitor does not cause insulin resistance or perturb glucohomeostasis. Chem. Biol. 2010, 17, 949–958. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kim, E.J. Chemical Arsenal for the Study of O-GlcNAc. Molecules 2011, 16, 1987-2022. https://doi.org/10.3390/molecules16031987
Kim EJ. Chemical Arsenal for the Study of O-GlcNAc. Molecules. 2011; 16(3):1987-2022. https://doi.org/10.3390/molecules16031987
Chicago/Turabian StyleKim, Eun J. 2011. "Chemical Arsenal for the Study of O-GlcNAc" Molecules 16, no. 3: 1987-2022. https://doi.org/10.3390/molecules16031987



