Chemical Genetics — A Versatile Method to Combine Science and Higher Level Teaching in Molecular Genetics
Abstract
:1. Introduction
2. “Chemical Genetics” as Tool to Study Secondary Septum Formation in U. maydis
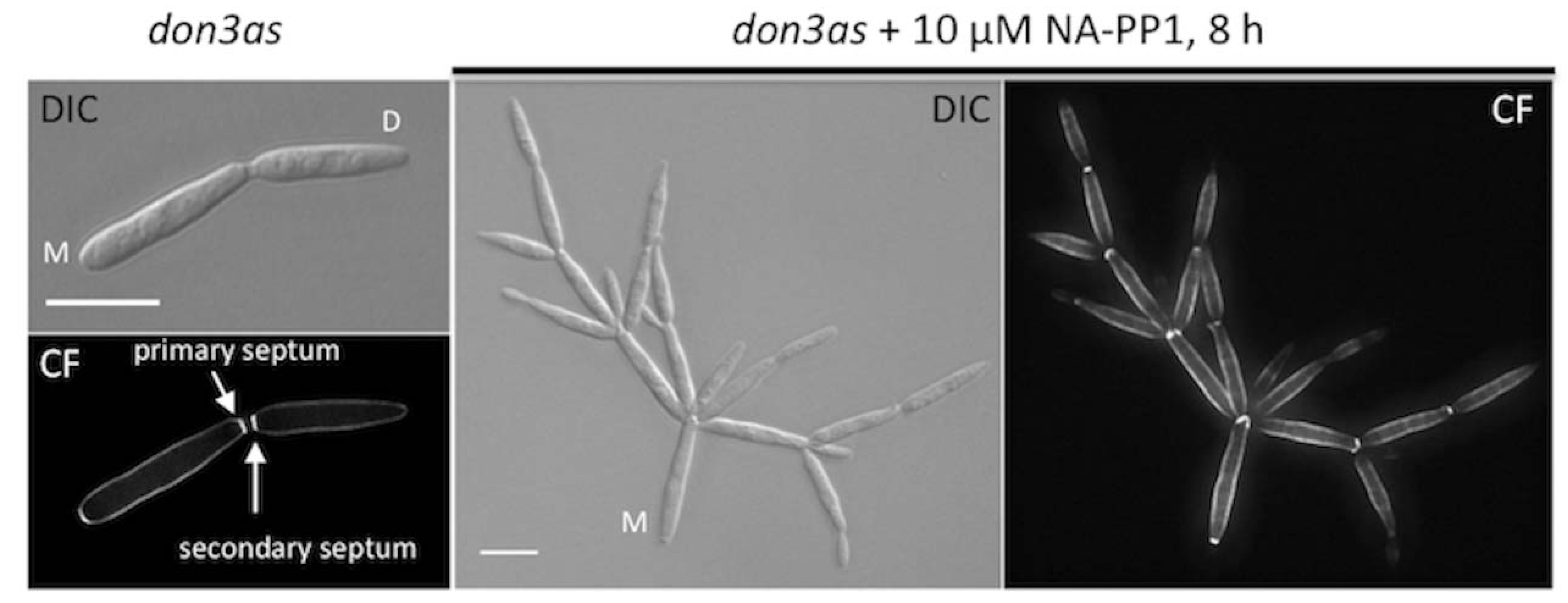
3. Chemical Genetics as Appropriate Technique for Higher Education Systems
4. Structural Organisation of the Course “Chemical Genetics”
| Timeline of the course | |
|---|---|
| Day 1 | Full-time seminar Content: What are kinases? What are their functions? What is the concept behind “Chemical genetics”? Problem: Identification of the gatekeeper amino acid in a given kinase or in a kinase of the students choice. Aim: Design of Oligonucleotide-primers for the site-directed mutagenesis of the gatekeeper codon in the kinase of choice and for the left and right borders for homologous recombination in U. maydis Formation of 12 groups of two students Presentation of a list of original and review literature Collection and Order of the Primers |
| Day 2 | Time for the students to prepare a colloquium |
| Day 3 | Colloquia of about 30 min for each group Preparation of buffers and media Inoculation of strains |
| Day 4 | PCR for the site-directed mutagenesis of the kinase gene of choice (1) The Don3 kinase in Ustilago maydis (2) |
| Day 5 | Cloning of the mutagenized DNA and transformation in E. coli (1) Inhibition and release of different analog-sensitive kinase mutants of the last years courses (3) |
| Day 6 | Inoculation of E. coli transformants and virtual cloning (1) Microscopy of inhibited and released don3as cells surface labelled with Biotin (3) |
| Day 7 | Miniprep, restriction analysis, order for sequences (1) |
| Day 8 | Image processing using e.g., ImageJ open source software (2) |
| Day 9 | Preparation of final talks (1–3), sequence analysis (1) |
| Day 10 | Full-time seminar with each group presenting their results Final discussion |
| U. maydis gene number | Name | Gatekeeper | Observation | Kinase family | Literature |
|---|---|---|---|---|---|
| um05543 | Don3 | M157A | cell separation defect | Germinal centre Kinase | [11] |
| um10145 | Cla4 | M629A | polarity defect | p21-activated kinase | [15] |
| um04956 | Ukc1 | M398A | morphological defect | NDR-like kinase | [16] |
| um05698 | Nrc2 | M454G | no | Phototropin-like kinase | [17] |
| um10119 | Ipl1 | M280A | no | PKA-like kinase | [18] |
| um03841 | Ire1 | L1082A | no | PKc-like kinase | [19] |
| um04902 | Kin28 | L94A | no | Cdk7-like kinase | [20] |
| um02741 | Cbr1 | M798A | no | NDR-like kinase | [21] |
| um11199 | Cdc28 | M267A | wrong gatekeeper | Cdc2-like kinase | [22] |
| um11396 | Nak1 | M769A | no | Germinal centre kinase | [23] |
| um11396 | Nak1 | M769G | morphological defect without inhibitor | Germinal centre kinase | [23] |
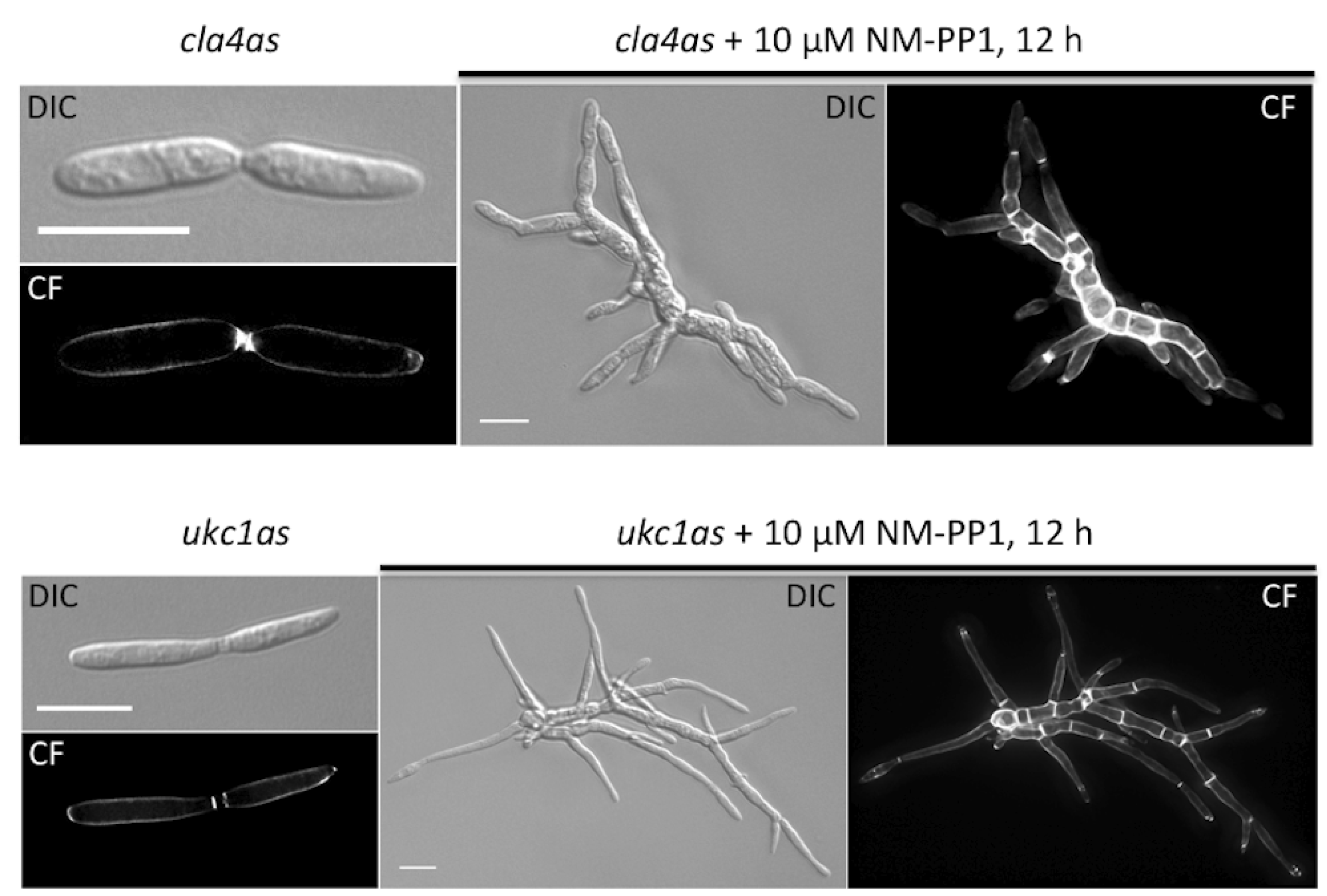
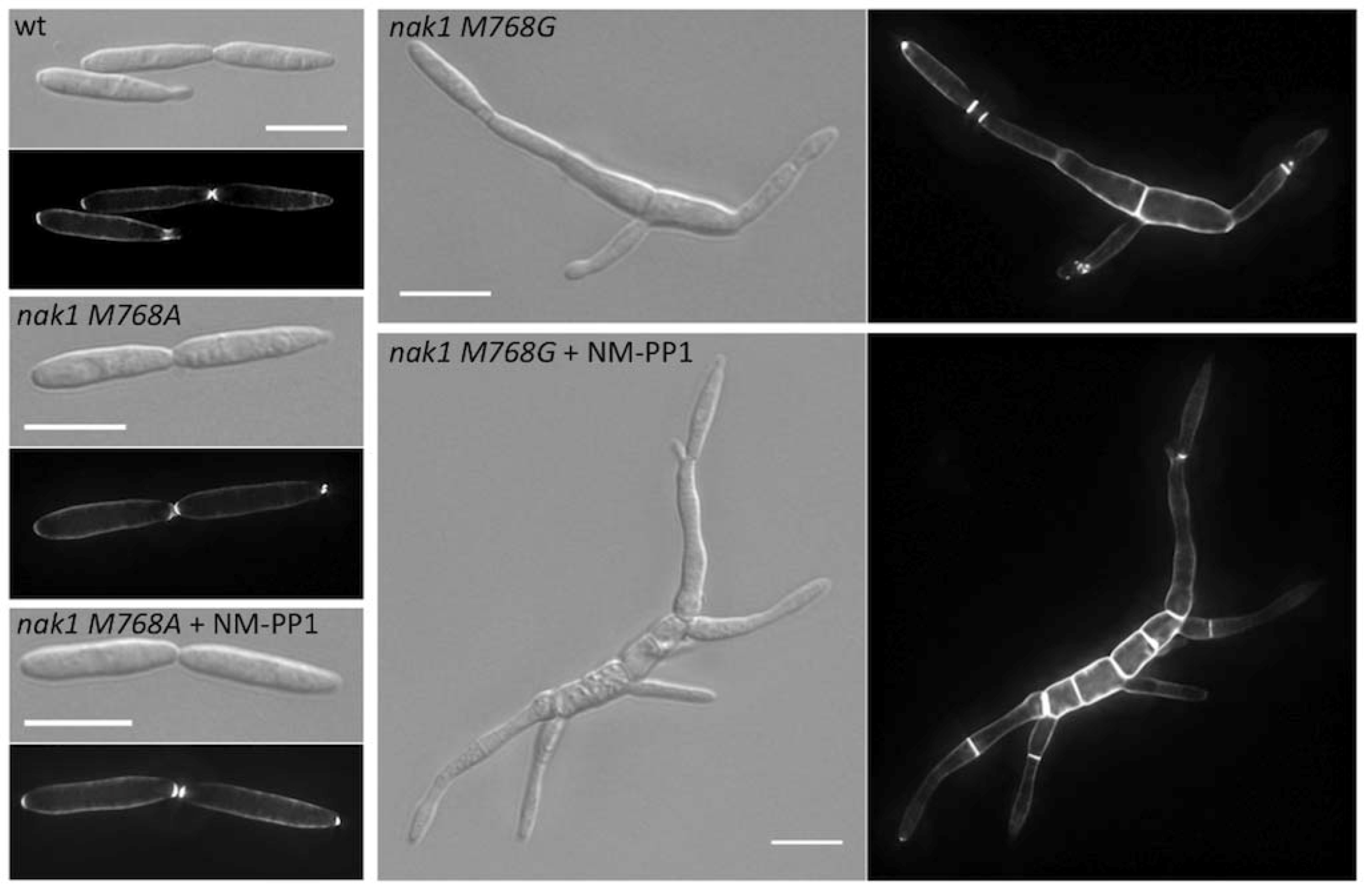
5. Final Remarks and Future Objectives
6. Experimental
6.1. Genetic Methods
6.2. Inhibition
6.3. Microscopy
7. Conclusion
Supplementary Materials
Acknowledgements
Conflict of Interest
References
- Joint Declaration of the European Ministers of Education. Available online: http://www.ehea.info/Uploads/Declarations/BOLOGNA_DECLARATION1.pdf (accessed on 29 September 2012).
- von Humboldt, W. Über die innere und äußere Organisation der höheren wischenschaftlichen Anstalten in Berlin (in German). In von Humboldt, W.: Gesammelte Schriften; Preußische Akademie der Wissenschaften: Berlin, Germany, 1903; pp. 250–261. [Google Scholar]
- Wildt, J. On the way from teaching to learning by competences as learning outcomes. In Higher Education Management and Development in Central, Southern and Eastern Europe; Pausits, A., Pellert, A., Eds.; Waxmann: Münster, Germany, 2007; pp. 115–123. [Google Scholar]
- Wildt, J. Auf dem Weg zu einer Didaktik der Lehrerbildung? (in german). Beitr. zur Lehrerbild. 2005, 23, 183–190. [Google Scholar]
- Barrows, H.S. Problem-based learning in medicine and beyond: A brief overview. In New Directions for Teaching and Learning, no. 68. Bringing Problem-Based Learning to Higher Education: Theory And Practice; Wilkerson, L., Gijselaers, W.H., Eds.; Jossey-Bass: San Francisco, CA, USA, 1996; pp. 3–13. [Google Scholar]
- Böhmer, C.; Böhmer, M.; Bölker, M.; Sandrock, B. Cdc42 and the Ste20-like kinase Don3 act independently in triggering cytokinesis in Ustilago maydis. J. Cell Sci. 2008, 121, 143–148. [Google Scholar] [CrossRef]
- Böhmer, C.; Ripp, C.; Bölker, M. The germinal centre kinase Don3 triggers the dynamic rearrangement of higher-order septin structures during cytokinesis in Ustilago maydis. Mol. Microbiol. 2009, 74, 1484–1496. [Google Scholar] [CrossRef]
- Bishop, A.C.; Ubersax, J.A.; Petsch, D.T.; Matheos, D.P.; Gray, N.S.; Blethrow, J.; Shimizu, E.; Tsien, J.Z.; Schultz, P.G.; Rose, M.D.; et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 2000, 6802, 395–401. [Google Scholar]
- Banuett, F. Ustilago maydis, the delightful blight. Trends Genet. 1992, 8, 174–180. [Google Scholar]
- Kämper, J.; Kahmann, R.; Bölker, M.; Ma, L.J.; Brefort, T.; Saville, B.J.; Banuett, F.; Kronstad, J.W.; Gold, S.E.; Müller, O.; et al. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 2006, 7115, 97–101. [Google Scholar]
- Weinzierl, G.; Leveleki, L.; Hassel, A.; Kost, G.; Wanner, G.; Bölker, M. Regulation of cell separation in the dimorphic fungus Ustilago maydis. Mol. Microbiol. 2002, 45, 219–231. [Google Scholar] [CrossRef]
- Master Studies at the Department of Biology at the Philipps-University of Marburg, Germany. Available online: http://www.uni-marburg.de/fb17/studium/studiengaenge/master (accessed on 29 September 2012).
- Brachmann, A.; König, J.; Julius, C.; Feldbrügge, M. A reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 2004, 272, 216–226. [Google Scholar]
- The suicide vector pJET1.2. Available online: http://www.fermentas.com/en/support/technical-reference/phage-plasmid-dna/pjet12/ (accessed on 29 September 2012).
- Leveleki, L.; Mahlert, M.; Sandrock, B.; Bölker, M. The PAK family kinase Cla4 is required for budding and morphogenesis in Ustilago maydis. Mol. Microbiol. 2004, 54, 396–406. [Google Scholar] [CrossRef]
- Dürrenberger, F.; Kronstad, J. The ukc1 gene encodes a protein kinase involved in morphogenesis, pathogenicity and pigment formation in Ustilago maydis. Mol. Gen. Genet. 1999, 261, 281–289. [Google Scholar] [CrossRef]
- Kothe, G.O.; Free, S.J. The isolation and chracterization of nrc-1 and nrc-2, two genes encoding protein kinases that control growth and development in Neurospora crassa. Genetics 1998, 149, 117–130. [Google Scholar]
- Biggins, S.; Severin, F.F.; Bhalla, N.; Sassonn, I.; Hyman, A.A.; Murray, A.W. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999, 13, 562–544. [Google Scholar]
- Welihinda, A.A.; Kaufman, R.J. The unfolded protein response pathway in Saccharomyces cerevisiae. Oligomerization and trans-phosphorylation of Ire1p (Ern1p) are required for kinase activation. J. Biol. Chem 1996, 271, 18181–18187. [Google Scholar]
- Rodriguez, C.R.; Cho, E.J.; Keogh, M.C.; Moore, C.L.; Greenleaf, A.L.; Buratowski, S. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol. Cell. Biol. 2000, 20, 104–112. [Google Scholar] [CrossRef]
- Racki, W.J.; Bécam, A.M.; Nasr, F.; Herbert, C.J. Cbk1p, a protein similar to the human myotonic dystrophy kinase, is essential for normal morphogenesis in Saccharomyces cerevisia. EMBO J. 2000, 19, 4524–4532. [Google Scholar] [CrossRef]
- Nasmyth, K.A.; Reed, S.I. Isolation of genes by complementation in yeast: Molecular cloning of a cell-cycle gene. Proc. Natl. Acad. Sci. USA 1980, 77, 2119–2123. [Google Scholar] [CrossRef]
- Sartorel, E.; Pérez-Martin, J. The distinct wiring between cell cycle regulation and the widely conserved Morphogenesis-Related (MOR) pathway in the fungus Ustilago maydis determines the morphological outcome. J. Cell Sci. 2012. [Google Scholar] [CrossRef]
- MIPS Ustilago Maydis DataBase MUMDB Homepage. Available online: http://mips.helmholtz-muenchen.de/genre/proj/ustilago (accessed on 29 September 2012).
- NCBI Basic Local Alignment Search Tool BLAST Homepage. Available online: http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi (accessed on 29 September 2012).
- Frieser, S.H.; Hlubek, A.; Sandrock, B.; Bölker, M. Cla4 kinase triggers destruction of the Rac1-GEF Cdc24 during polarized growth in Ustilago maydis. Mol. Biol. Cell 2011, 22, 3253–3262. [Google Scholar] [CrossRef]
- Journal of Cell Science Homepage. Guidelines for image manipulation. Available online: http://jcs.biologists.org/site/journal/editorial_policies.xhtml#text (accessed on 29 September 2012).
- Zhang, C.; Kenski, D.M.; Paulson, J.L.; Bonshtien, A.; Sessa, G.; Cross, J.V.; Templeton, D.J.; Shokat, K.M. A second-site suppressor strategy for chemical genetic analysis of diverse protein kinases. Nat. Methods 2005, 6, 435–441. [Google Scholar] [CrossRef]
- Plasmid Editor Ape Homepage. Available online: http://biologylabs.utah.edu/jorgensen/wayned/ape/ (accessed on 29 September 2012).
- Image Processing Software ImageJ Homepage. Available online: http://rsbweb.nih.gov/ij/ (accessed on 29 September 2012).
- Presentation Software Apache Open Office Homepage. Available online: http://www.openoffice.org/ (accessed on 29 September 2012).
- Schulz, B.; Banuett, F.; Dahl, M.; Schlesinger, R.; Schäfer, W.; Martin, T.; Herskowitz, I.; Kahmann, R. The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 1990, 60, 295–306. [Google Scholar] [CrossRef]
- Freitag, J.; Lanver, D.; Böhmer, C.; Schink, K.O.; Bölker, M.; Sandrock, B. Septation of infectious hyphae is critical for appressoria formation and virulence in the smut fungus Ustilago maydis. PloS Pathog. 2011, 7, e1002044. [Google Scholar] [CrossRef]
- Homepage of the Deutsche Forschungsgemeinschaft. Available online: http://www.dfg.de/index.jsp/ (accessed on 29 September 2012).
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Sandrock, B. Chemical Genetics — A Versatile Method to Combine Science and Higher Level Teaching in Molecular Genetics. Molecules 2012, 17, 11920-11930. https://doi.org/10.3390/molecules171011920
Sandrock B. Chemical Genetics — A Versatile Method to Combine Science and Higher Level Teaching in Molecular Genetics. Molecules. 2012; 17(10):11920-11930. https://doi.org/10.3390/molecules171011920
Chicago/Turabian StyleSandrock, Björn. 2012. "Chemical Genetics — A Versatile Method to Combine Science and Higher Level Teaching in Molecular Genetics" Molecules 17, no. 10: 11920-11930. https://doi.org/10.3390/molecules171011920




