Calculation of the Stabilization Energies of Oxidatively Damaged Guanine Base Pairs with Guanine
Abstract
:1. Introduction

2. Results and Discussion
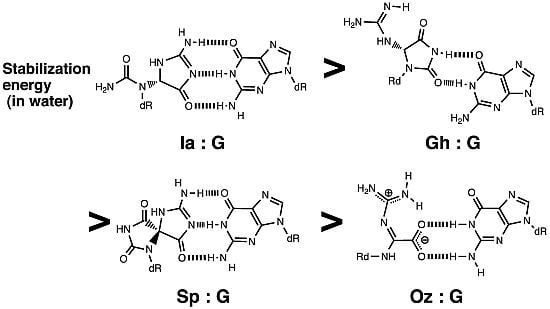
2.1. Optimization and Calculation of the Stabilization Energies of Ia:G Base Pairs
| Base pair | Δ EDFT | Δ ESCRF | Base pair | Δ EDFT | Δ ESCRF |
|---|---|---|---|---|---|
| Ia1:G | 29.5 | 24.1 | Gh7:G | 19.9 | 19.5 |
| Ia2:G | 28.7 | 19.3 | Gh8:G | 19.8 | 16.9 |
| Ia3:G | 29.5 | 17.0 | Gh9:G | 21.0 | 19.1 |
| Ia4:G | 29.5 | 23.5 | Gh10:G | 20.9 | 17.3 |
| Ia5:G | 29.5 | 24.0 | Gh11:G | 20.6 | 20.6 |
| Ia6:G | 28.7 | 19.7 | Gh12:G | 20.4 | 19.6 |
| Ia7:G | 29.5 | 18.1 | Gh13:G | 20.3 | 16.6 |
| Ia8:G | 29.5 | 22.6 | Gh14:G | 20.5 | 17.1 |
| Gh1:G | 21.0 | 18.9 | Gh15:G | 20.8 | 18.5 |
| Gh2:G | 20.9 | 16.7 | Gh16:G | 21.1 | 19.0 |
| Gh3:G | 20.5 | 21.4 | Sp1:G | 28.2 | 18.8 |
| Gh4:G | 20.4 | 19.5 | Sp2:G | 28.2 | 19.9 |
| Gh5:G | 20.4 | 18.2 | Oz:G | 20.7 | 16.3 |
| Gh6:G | 20.5 | 16.8 |

2.2. Optimization and Calculation of the Stabilization Energies of Gh:G Base Pairs

2.3. Optimization and Calculation of the Stabilization Energies of Sp:G Base Pairs

2.4. Comparison of the Above Results with the Calculated Stabilization Energy of Oz:G Base Pairs

3. Experimental

4. Conclusions
Acknowledgments
- Sample Availability: Not available.
References
- Maehira, F.; Miyagi, I.; Asato, T.; Eguchi, Y.; Takei, H.; Nakatsuki, K.; Fukuoka, M.; Zaha, F. Alterations of protein kinase C, 8-hydroxydeoxyguanosine, and K-ras oncogene in rat lungs exposed to passive smoking. Clin. Chim. Acta 1999, 289, 133–144. [Google Scholar] [CrossRef]
- Shibutani, S.; Takeshita, M.; Grollman, A.P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 1991, 349, 431–434. [Google Scholar]
- Cadet, J.; Berger, M.; Buchko, G.W.; Joshi, P.C.; Raoul, S.; Ravanat, J.-L. 2,2-Diamino-4-[(3,5-di-O-acetyl-2-deoxy-β-D-erythro-pentofuranosyl)amino]-5-(2H)-oxazolone: A novel and predominant radical oxidation product of 3',5'-di-O-acetyl-2'-deoxyguanosine. J. Am. Chem. Soc. 1994, 116, 7403–7404. [Google Scholar]
- Kino, K.; Saito, I.; Sugiyama, H. Product analysis of GG-specific photooxidation of DNA via electron transfer: 2-Aminoimidazolone as a major guanine oxidation product. J. Am. Chem. Soc. 1998, 120, 7373–7374. [Google Scholar] [CrossRef]
- Kino, K.; Sugiyama, H. Possible cause of G•C->C•G transversion mutation by guanine oxidation product, imidazolone. Chem. Biol. 2001, 8, 369–378. [Google Scholar] [CrossRef]
- Kino, K.; Sugasawa, K.; Mizuno, T.; Bando, T.; Sugiyama, H.; Akita, M.; Miyazawa, H.; Hanaoka, F. Eukaryotic DNA polymerases α, β and ε incorporate guanine opposite 2,2,4-triamino-5(2H)-oxazolone. ChemBioChem 2009, 10, 2613–2616. [Google Scholar] [CrossRef]
- Duarte, V.; Gasparutto, D.; Jaquinod, M.; Cadet, J. In vitro DNA synthesis opposite oxazolone and repair of this DNA damage using modified oligonucleotides. Nucleic Acids Rec. 2000, 28, 1555–1563. [Google Scholar] [CrossRef]
- Luo, W.; Muller, J.G.; Rachlin, E.M.; Burrows, C.J. Characterization of spiroiminodihydantoin as a product of one-electron oxidation of 8-oxo-7,8-dihydroguanosine. Org. Lett. 2000, 2, 613–616. [Google Scholar] [CrossRef]
- Kino, K.; Morikawa, M.; Kobayashi, T.; Komori, R.; Sei, Y.; Miyazawa, H. The oxidation of 8-oxo-7,8-dihydroguanine by iodine. Bioorg. Med. Chem. Lett. 2010, 20, 3818–3820. [Google Scholar] [CrossRef]
- Munk, B.H.; Burrows, C.J.; Schlegel, H.B. An exploration of mechanisms for the transformation of 8-oxoguanine to guanidinohydantoin and spiroiminodihydantoin by density functional theory. J. Am. Chem. Soc. 2008, 130, 5245–5256. [Google Scholar]
- Ye, Y.; Munk, B.H.; Muller, J.G.; Gogbill, A.; Burrows, C.J.; Schlegel, H.B. Mechanistic aspects of the formation of guanidinohydantoin from spiroiminodihydantoin under acidic conditions. Chem. Res. Toxicol. 2009, 22, 526–535. [Google Scholar] [CrossRef]
- Kornyushyna, O.; Berges, A.M.; Muller, J.G.; Burrows, C.J. In vitro nucleotide misinsertion opposite the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin and DNA synthesis past the lesions using Escherichia coli DNA polymerase I (Klenow fragment). Biochemistry 2002, 41, 15304–15314. [Google Scholar] [CrossRef]
- Luo, W.; Muller, J.G.; Rachlin, E.M.; Burrows, C.J. Characterization of hydantoin products from one-electron oxidation of 8-oxo-7,8-dihydroguanosine in a nucleoside model. Chem. Res. Toxicol. 2001, 14, 927–938. [Google Scholar] [CrossRef]
- Kino, K.; Sugiyama, H. UVR-induced G-C to C-G transversions from oxidative DNA damage. Mutat. Res. 2005, 571, 33–42. [Google Scholar] [CrossRef]
- Ding, S.; Jia, L.; Durandin, A.; Crean, C.; Kolbanovskiy, A.; Shafirovich, V.; Broyde, S.; Geacintov, N.E. Absolute configurations of spiroiminodihydantoin and allantoin stereoisomers: comparison of computed and measured electronic circular dichroism spectra. Chem. Res. Toxicol. 2009, 22, 1189–1193. [Google Scholar] [CrossRef]
- Wong, M.W.; Frisch, M.J.; Wiberg, K.B. Solvent effects. 1. The mediation of electrostatic effects by solvents. J. Am. Chem. Soc. 1991, 113, 4776–4782. [Google Scholar] [CrossRef]
- Beckman, J.; Wang, M.; Blaha, G.; Wang, J.; Konigsberg, W.H. Substitution of Ala for Tyr567 in RB69 DNA polymerase allows dAMP and dGMP to be inserted opposite guanidinohydantoin. Biochemistry 2010, 49, 8554–8563. [Google Scholar] [CrossRef]
- Verdolino, V.; Cammi, R.; Munk, B.H.; Schlegel, H.B. Calculation of pKa values of nucleobases and the guanine oxidation products guanidinohydantoin and spiroiminodihydantoin using density functional theory and a polarizable continuum model. J. Phys. Chem. B 2008, 112, 16860–16873. [Google Scholar]
- Aller, P.; Ye, Y.; Wallace, S.S.; Burrows, C.J.; Doublié, S. Crystal structure of a replicative DNA polymerase bound to the oxidized guanine lesion guanidinohydantoin. Biochemistry 2010, 49, 2502–2509. [Google Scholar]
- Chinyengetere, F.; Jamieson, E.R. Impact of the oxidized guanine lesion spiroiminodihydantoin on the conformation and thermodynamic stability of a 15-mer DNA duplex. Biochemistry 2008, 47, 2584–2591. [Google Scholar] [CrossRef]
- Yanson, I.K.; Teplitsky, A.B.; Sukhodub, L.F. Experimental studies of molecular interactions between nitrogen bases of nucleic acids. Biopolymers 1979, 18, 1149–1170. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Montgomery, J.A.; Vreven, T., Jr.; Kudin, K.N.; Burant, J.C.; et al. Gaussian 03,Revision C.02; Gaussian, Inc.: Wallingford, CT, USA, 2004. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Suzuki, M.; Kino, K.; Morikawa, M.; Kobayashi, T.; Komori, R.; Miyazawa, H. Calculation of the Stabilization Energies of Oxidatively Damaged Guanine Base Pairs with Guanine. Molecules 2012, 17, 6705-6715. https://doi.org/10.3390/molecules17066705
Suzuki M, Kino K, Morikawa M, Kobayashi T, Komori R, Miyazawa H. Calculation of the Stabilization Energies of Oxidatively Damaged Guanine Base Pairs with Guanine. Molecules. 2012; 17(6):6705-6715. https://doi.org/10.3390/molecules17066705
Chicago/Turabian StyleSuzuki, Masayo, Katsuhito Kino, Masayuki Morikawa, Takanobu Kobayashi, Rie Komori, and Hiroshi Miyazawa. 2012. "Calculation of the Stabilization Energies of Oxidatively Damaged Guanine Base Pairs with Guanine" Molecules 17, no. 6: 6705-6715. https://doi.org/10.3390/molecules17066705