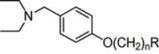
Hologram QSAR Models of 4-[(Diethylamino)methyl]-phenol Inhibitors of Acetyl/Butyrylcholinesterase Enzymes as Potential Anti-Alzheimer Agents
Abstract
:1. Introduction
2. Results and Discussion
2.1. HQSAR Analysis
| # | n | R | pIC50 a | # | n | R | pIC50 [16] | |||
|---|---|---|---|---|---|---|---|---|---|---|
| AChE | BChE | AChE | BChE | |||||||
| 1 | 2 |  | 4.86 | 4.99 | 19 | 6 | 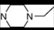 | 5.79 | 5.66 | |
| 2 | 2 | 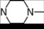 | 4.94 | 4.69 | 20 | 6 |  | 6.02 | 5.77 | |
| 3 | 4 |  | 5.47 | 5.85 | 21 | 6 | 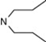 | 5.67 | 5.87 | |
| 4 | 4 |  | 5.17 | 6.14 | 22* | 6 |  | 6.17 | 6.64 | |
| 5 | 4 | 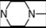 | 5.25 | 5.01 | 23 | 8 |  | 7.08 | 7.82 | |
| 6 * | 4 |  | 4.97 | 5.56 | 24 | 8 |  | 7.04 | 8.13 | |
| 7 | 4 |  | 4.86 | 4.93 | 25 | 8 |  | 6.68 | 7.96 | |
| 8 | 4 |  | 4.99 | 5.60 | 26 | 8 |  | 7.11 | 7.85 | |
| 9 | 5 |  | 6.30 | 5.81 | 27* | 8 |  | 6.85 | 7.31 | |
| 10 | 5 | 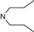 | 5.37 | 6.17 | 28 | 8 |  | 6.77 | 8.04 | |
| 11 * | 5 |  | 5.61 | 5.03 | 29 | 8 | 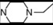 | 6.85 | 7.77 | |
| 12 | 5 |  | 5.75 | 5.51 | 30 * | 10 |  | 6.52 | 7.17 | |
| 13 | 5 |  | 5.08 | 5.35 | 31 | 10 | 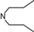 | 6.22 | 7.11 | |
| 14 | 5 | 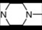 | 5.25 | 5.75 | 32 | 10 |  | 6.27 | 7.48 | |
| 15 | 5 | 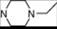 | 5.61 | 5.72 | 33* | 10 | 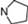 | 5.24 | 6.72 | |
| 16 | 6 | 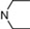 | 6.36 | 5.89 | 34 | 10 |  | 6.51 | 7.57 | |
| 17 | 6 | 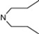 | 6.05 | 5.86 | 35 | 10 | 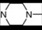 | 6,47 | 7.58 | |
| 18 | 6 | 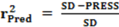 | 5.74 | 5.42 | 36 | 10 |  | 6.59 | 7.82 | |
| AChE Statistical Indexes a | BChE Statistical Indexes a | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FD b | q2 | r2 | SE | SEcv | NC | HL | q2 | r2 | SE | SEcv | NC | HL | |
| A/B | 0.660 | 0.833 | 0.328 | 0.468 | 6 | 59 | 0.706 | 0.780 | 0.545 | 0.630 | 2 | 61 | |
| A/C | 0.642 | 0.833 | 0.328 | 0.480 | 6 | 307 | 0.696 | 0.777 | 0.549 | 0.640 | 2 | 199 | |
| A/H | 0.635 | 0.785 | 0.364 | 0.475 | 5 | 53 | 0.704 | 0.754 | 0.566 | 0.621 | 1 | 97 | |
| A/DA | 0.655 | 0.826 | 0.321 | 0.452 | 4 | 71 | 0.656 | 0.761 | 0.568 | 0.682 | 2 | 401 | |
| B/C | 0.674 | 0.787 | 0.348 | 0.431 | 3 | 307 | 0.636 | 0.753 | 0.588 | 0.715 | 3 | 307 | |
| B/H | 0.657 | 0.732 | 0.383 | 0.434 | 2 | 59 | 0.656 | 0.733 | 0.600 | 0.682 | 2 | 307 | |
| C/H | 0.672 | 0.783 | 0.358 | 0.456 | 3 | 307 | 0.637 | 0.750 | 0.592 | 0.713 | 3 | 53 | |
| C/DA | 0.638 | 0.811 | 0.335 | 0.463 | 4 | 97 | 0.676 | 0.792 | 0.530 | 0.661 | 2 | 59 | |
| A/B/C | 0.643 | 0.830 | 0.324 | 0.469 | 5 | 401 | 0.690 | 0.777 | 0.548 | 0.647 | 2 | 199 | |
| A/C/H | 0.624 | 0.782 | 0.367 | 0.482 | 5 | 257 | 0.694 | 0.744 | 0.578 | 0.631 | 1 | 61 | |
| A/B/C/H | 0.619 | 0.781 | 0.368 | 0.485 | 5 | 257 | 0.693 | 0.743 | 0.578 | 0.632 | 1 | 61 | |
| AChE Statistical Indexes a | BChE Statistical Indexes a | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FS | q2 | r2 | SE | SEcv | NC | HL | q2 | r2 | SE | SEcv | NC | HL | |
| 2–5 | 0.598 | 0.764 | 0.390 | 0.509 | 6 | 353 | 0.703 | 0.768 | 0.560 | 0.633 | 2 | 53 | |
| 3–6 | 0.605 | 0.758 | 0.379 | 0.484 | 4 | 307 | 0.705 | 0.775 | 0.557 | 0.631 | 2 | 53 | |
| 4–7 | 0.674 | 0.787 | 0.348 | 0.431 | 3 | 307 | 0.706 | 0.780 | 0.545 | 0.630 | 2 | 61 | |
| 5–8 | 0.695 | 0.810 | 0.329 | 0.417 | 3 | 61 | 0.862 | 0.957 | 0.272 | 0.489 | 8 | 401 | |
| 6–9 | 0.698 | 0.815 | 0.324 | 0.415 | 3 | 97 | 0.880 | 0.959 | 0.268 | 0.456 | 8 | 61 | |
| 7–10 | 0.693 | 0.813 | 0.326 | 0.418 | 3 | 59 | 0.894 | 0.966 | 0.244 | 0.430 | 8 | 199 | |
| 8–11 | 0.782 | 0.940 | 0.202 | 0.383 | 7 | 307 | 0.893 | 0.961 | 0.250 | 0.412 | 6 | 151 | |
| 9–12 | 0.787 | 0.965 | 0.156 | 0.388 | 8 | 151 | 0.904 | 0.952 | 0.265 | 0.373 | 4 | 151 | |

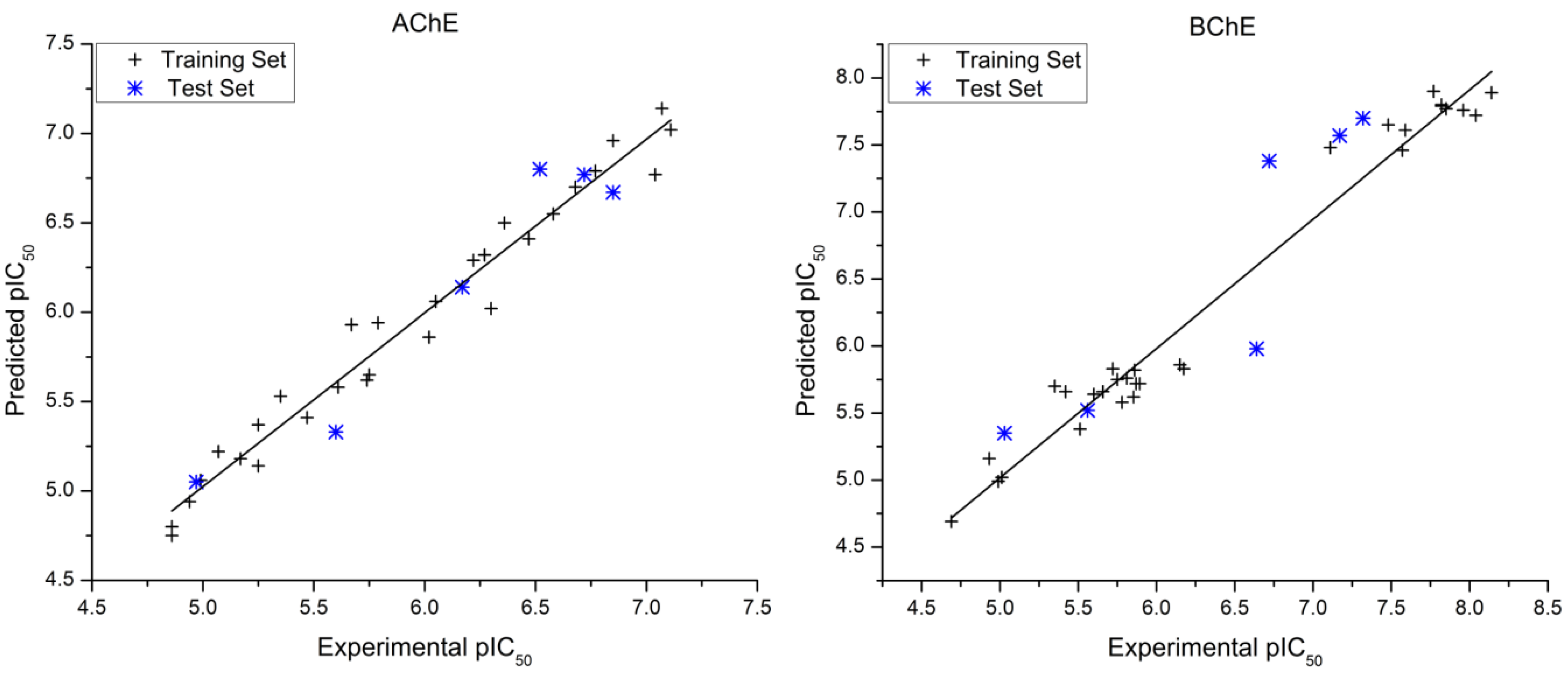
| # | AChE pIC50 | Residual | BChE pIC50 | Residual | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Exp. a | Pred. | Exp. [16] | Pred. | |||||||
| 1 * | 4.86 | 4.8 | 0.06 | 4.99 | 4.99 | 0.00 | ||||
| 2 | 4.94 | 4.94 | 0.00 | 4.69 | 4.69 | 0.00 | ||||
| 3 | 5.47 | 5.41 | 0.06 | 5.85 | 5.62 | 0.23 | ||||
| 4 | 5.17 | 5.18 | −0.01 | 6.15 | 5.86 | 0.29 | ||||
| 5 | 5.25 | 5.37 | −0.12 | 5.01 | 5.02 | −0.01 | ||||
| 6 * | 4.97 | 5.05 | −0.08 | 5.56 | 5.52 | 0.04 | ||||
| 7 | 4.86 | 4.75 | 0.11 | 4.93 | 5.16 | −0.23 | ||||
| 8 | 4.99 | 5.06 | −0.07 | 5.60 | 5.64 | −0.04 | ||||
| 9 | 6.3 | 6.02 | 0.28 | 5.81 | 5.76 | 0.05 | ||||
| 10 | 5.35 | 5.53 | −0.18 | 6.17 | 5.83 | 0.34 | ||||
| 11* | 5.60 | 5.33 | 0.27 | 5.03 | 5.35 | −0.32 | ||||
| 12 | 5.75 | 5.65 | 0.10 | 5.51 | 5.38 | 0.13 | ||||
| 13 | 5.07 | 5.22 | −0.15 | 5.35 | 5.70 | −0.35 | ||||
| 14 | 5.25 | 5.14 | 0.11 | 5.75 | 5.75 | 0.00 | ||||
| 15 | 5.61 | 5.58 | 0.03 | 5.72 | 5.83 | −0.11 | ||||
| 16 | 6.36 | 6.5 | −0.14 | 5.89 | 5.72 | 0.17 | ||||
| 17 | 6.05 | 6.06 | −0.01 | 5.86 | 5.82 | 0.04 | ||||
| 18 | 5.74 | 5.62 | 0.12 | 5.42 | 5.66 | −0.24 | ||||
| 19 | 5.79 | 5.94 | −0.15 | 5.66 | 5.66 | 0.00 | ||||
| 20 | 6.02 | 5.86 | 0.16 | 5.78 | 5.58 | 0.20 | ||||
| 21 | 5.67 | 5.93 | −0.26 | 5.87 | 5.72 | 0.15 | ||||
| 22 * | 6.17 | 6.14 | 0.03 | 6.64 | 5.98 | 0.66 | ||||
| 23 | 7.07 | 7.14 | −0.07 | 7.82 | 7.80 | 0.02 | ||||
| 24 | 7.04 | 6,77 | 0.27 | 8.14 | 7.89 | 0.25 | ||||
| 25 | 6.68 | 6.7 | −0.02 | 7.96 | 7.76 | 0.20 | ||||
| 26 | 7.11 | 7.02 | 0.09 | 7.85 | 7.77 | 0.08 | ||||
| 27 * | 6.85 | 6.67 | 0.18 | 7.32 | 7.70 | −0.38 | ||||
| 28 | 6.77 | 6.79 | −0.02 | 8.04 | 7.72 | 0.32 | ||||
| 29 | 6.85 | 6.96 | −0.11 | 7.77 | 7.90 | −0.13 | ||||
| 30 * | 6.52 | 6.80 | −0.28 | 7.17 | 7.57 | −0.40 | ||||
| 31 | 6.22 | 6.29 | −0.07 | 7.11 | 7.48 | −0.37 | ||||
| 32 | 6.27 | 6.32 | −0.05 | 7.48 | 7.65 | −0.17 | ||||
| 33 * | 6.72 | 6.77 | −0.05 | 6.72 | 7.38 | −0.66 | ||||
| 34 | 6.47 | 6.41 | 0.06 | 7.57 | 7.46 | 0.11 | ||||
| 35 | 6.58 | 6.55 | 0.03 | 7.59 | 7.61 | −0.02 | ||||
| 36 | 4.86 | 4.8 | 0.06 | 7.82 | 7.79 | 0.03 | ||||

3. Experimental
3.1. Data Set and Molecular Modeling
4. Conclusions
Acknowledgments
References
- Mangialasche, F.; Solomon, A.; Winblad, B.; Mecocci, P.; Kivipelto, M. Alzheimer’s disease: Clinical trials and drug development. Lancet Neurol. 2010, 9, 702–716. [Google Scholar] [CrossRef]
- Wimo, A.; Winblad, B.; Jönsson, L. The worldwide societal costs of dementia: Estimates for 2009. Alzheimers Dement. 2010, 6, 98–103. [Google Scholar]
- Winblad, B.; Fioravanti, M.; Dolezal, T.; Logina, I.; Milanov, I.G.; Popescu, D.C.; Solomon, A. Therapeutic Use of Nicergoline. Clin. Drug Investig. 2008, 28, 533–552. [Google Scholar] [CrossRef]
- Benedikz, E.; Kloskowska, E.; Winblad, B. The rat as an animal model of Alzheimer’s disease. J. Cell. Mol. Med. 2009, 13, 1034–1042. [Google Scholar] [CrossRef]
- Robichaud, A.J. Approaches to Palliative Therapies for Alzheimers Disease. Curr. Top. Med. Chem. 2006, 6, 553–568. [Google Scholar] [CrossRef]
- Gualtieri, F.; Dei, S.; Manetti, D.; Novella Romanelli, M.; Scapecchi, S.; Teodori, E. The medicinal chemistry of Alzheimer’s and Alzheimer-like diseases with emphasis on the cholinergic hypothesis. Farmaco Prat. 1995, 50, 489–503. [Google Scholar]
- Schliebs, R.; Arendt, T. The significance of the cholinergic system in the brain during aging and in Alzheimer’s disease. J. Neural. Transm. 2006, 113, 1625–1644. [Google Scholar] [CrossRef]
- Scarpini, E.; Schelterns, P.; Feldman, H. Treatment of Alzheimer’s disease; current status and new perspectives. Lancet Neurol. 2003, 2, 539–547. [Google Scholar] [CrossRef]
- Francis, P.T.; Palmer, A.M.; Snape, M.; Wilcock, G.K. The cholinergic hypothesis of Alzheimer’s disease: A review of progress. J. Neurol. Neurosurg. Psychiatry 1999, 66, 137–147. [Google Scholar]
- Giacobini, E. Cholinesterase inhibitors: New roles and therapeutic alternatives. Pharmacol. Res. 2004, 50, 433–440. [Google Scholar] [CrossRef]
- Heritage, T.W.; Lowis, D.R. Molecular Hologram QSAR. In Rational Drug Design: Novel Methodology and Practical Applications; Parrill, A.L., Reddy, M.R., Eds.; American Chemical Society: Washington, DC, USA, 1999; Volume 719, Chapter 14, pp. 212–225. [Google Scholar]
- Myint, K.Z.; Xie, X.-Q. Recent Advances in Fragment-Based QSAR and Multi-Dimensional QSAR Methods. Int. J. Mol. Sci. 2010, 11, 3846–3866. [Google Scholar] [CrossRef]
- Liu, P.; Long, W. Current Mathematical Methods Used in QSAR/QSPR Studies. Int. J. Mol. Sci. 2009, 10, 1978–1998. [Google Scholar] [CrossRef]
- Castilho, M.S.; Postigo, M.P.; de Paula, C.B.V.; Montanari, C.A.; Oliva, G.; Andricopulo, A.D. Two- and three-dimensional quantitative structure-activity relationships for a series of purine nucleoside phosphorylase inhibitors. Bioorg. Med. Chem. 2006, 14, 516–527. [Google Scholar]
- HQSARTM; Tripos: St. Louis, MO, USA. 2010.
- Yu, L.; Cao, R.; Yi, W.; Yan, Q.; Chen, Z.; Ma, L.; Peng, W.; Song, H. Synthesis of 4-[(diethylamino)methyl]-phenol derivatives as novel cholinesterase inhibitors with selectivity towards butyrylcholinesterase. Bioorg. Med. Chem. Lett. 2010, 20, 3254–3258. [Google Scholar] [CrossRef]
- Rodrigues, C.R.; Flaherty, T.M.; Springer, C.; McKerrow, J.H.; Cohen, F.E. CoMFA and HQSAR of acylhydrazide cruzain inhibitors. Bioorg. Med. Chem. Lett. 2002, 12, 1537–1541. [Google Scholar]
- Freitas, R.F.; Oprea, T.I.; Montanari, C.A. 2D QSAR and similarity studies on cruzain inhibitors aimed at improving selectivity over cathepsin L. Bioorg. Med. Chem. Lett. 2008, 16, 838–853. [Google Scholar] [CrossRef]
- Cramer, R.D.; Patterson, D.E.; Bunce, J.D. Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J. Am. Chem. Soc. 1998, 110, 5959–5967. [Google Scholar]
- Jaiswal, D.; Karthikeyan, C.; Trivedi, P. Rationalization of Physicochemical Properties of Alkanoic Acid Derivatives towards Histone Deacetylase Inhibition. Internet Electron. J. Mol. Des. 2006, 5, 13–26. [Google Scholar]
- Camps, P.; Formosa, X.; Galdeano, C.; Muñoz-Torrero, D.; Ramírez, L.; Gómez, E.; Isambert, N.; Lavilla, R.; Badia, A.; Clos, M.V.; et al. Pyrano[3,2-c]quinoline-6-Chlorotacrine Hybrids as a Novel Family of Acetylcholinesterase- and β-Amyloid-Directed Anti-Alzheimer Compounds. J. Med. Chem. 2009, 52, 5365–5379. [Google Scholar]
- Tang, H.; Wei, Y.-B.; Zhang, C.; Ning, F.-X.; Qiao, W.; Huang, S.-L.; Ma, L.; Huang, Z.-S.; Gu, L.-Q. Synthesis, biological evaluation and molecular modeling of oxoisoaporphine and oxoaporphine derivatives as new dual inhibitors of acetylcholinesterase/butyrylcholinesterase. Eur. J. Med. Chem. 2009, 44, 2523–2532. [Google Scholar]
- Nascimento, É.C.M.; Martins, J.B.L.; dos Santos, M.L.; Gargano, R. Theoretical study of classical acetylcholinesterase inhibitors. Chem. Phys. Lett. 2008, 458, 285–289. [Google Scholar] [CrossRef]
- Gupta, S.; Fallarero, A.; Järvinen, P.; Karlsson, D.; Johnson, M.S.; Vuorela, P.M.; Mohan, C.G. Discovery of dual binding site acetylcholinesterase inhibitors identified by pharmacophore modeling and sequential virtual screening techniques. Bioorg. Med. Chem. Lett. 2011, 21, 1105–1112. [Google Scholar]
- SPARTAN’10; Wavefunction, Inc.: Irvine, CA, USA. 2011.
- SYBYL 8.0; Tripos International: St. Louis, MO, USA. 2010.
- Doddareddy, M.R.; Lee, Y.J.; Cho, Y.S.; Choi, K.I.; Koh, H.Y.; Pae, A.N. Hologram quantitative structure activity relationship studies on 5-HT6 antagonists. Bioorg. Med. Chem. 2004, 12, 3815–3824. [Google Scholar] [CrossRef]
- Sample Availability: Not Available.
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
De Souza, S.D.; De Souza, A.M.T.; De Sousa, A.C.C.; Sodero, A.C.R.; Cabral, L.M.; Albuquerque, M.G.; Castro, H.C.; Rodrigues, C.R. Hologram QSAR Models of 4-[(Diethylamino)methyl]-phenol Inhibitors of Acetyl/Butyrylcholinesterase Enzymes as Potential Anti-Alzheimer Agents. Molecules 2012, 17, 9529-9539. https://doi.org/10.3390/molecules17089529
De Souza SD, De Souza AMT, De Sousa ACC, Sodero ACR, Cabral LM, Albuquerque MG, Castro HC, Rodrigues CR. Hologram QSAR Models of 4-[(Diethylamino)methyl]-phenol Inhibitors of Acetyl/Butyrylcholinesterase Enzymes as Potential Anti-Alzheimer Agents. Molecules. 2012; 17(8):9529-9539. https://doi.org/10.3390/molecules17089529
Chicago/Turabian StyleDe Souza, Simone Decembrino, Alessandra Mendonça Teles De Souza, Ana Carolina Corrêa De Sousa, Ana Carolina Rennó Sodero, Lúcio Mendes Cabral, Magaly Girão Albuquerque, Helena Carla Castro, and Carlos Rangel Rodrigues. 2012. "Hologram QSAR Models of 4-[(Diethylamino)methyl]-phenol Inhibitors of Acetyl/Butyrylcholinesterase Enzymes as Potential Anti-Alzheimer Agents" Molecules 17, no. 8: 9529-9539. https://doi.org/10.3390/molecules17089529