In Vitro and in Vivo Studies of the Inhibitory Effects of Emodin Isolated from Polygonum cuspidatum on Coxsakievirus B4
Abstract
:1. Introduction
2. Results and Discussion
2.1. In Vitro Experiments
2.1.1. Cytotoxicity of Emodin on HEp-2 Cells

| Compound | CC50 (μM) | EC50 (μM) | Selectivity index | ||
|---|---|---|---|---|---|
| MTT | MTT | PRA | MTT | PRA | |
| Emodin | 61.35 ± 4.40 | 14.10 ± 0.74 | 12.06 ± 1.85 | 4.35 ± 0.6 | 5.09 ± 0.3 |
| Ribavirin | 2645.14 ± 54.05 | 629.99 ± 1.64 | 460.91 ± 1.23 | 4.20 ± 0.7 | 5.74 ± 0.1 |
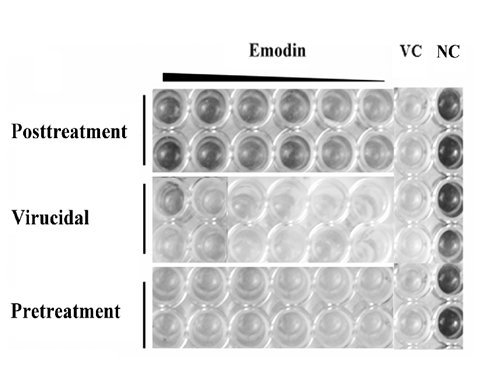
2.1.2. Inhibitory Effect of Emodin on CVB4 Infection

2.1.3. Emodin Mitigates CVB4-Induced Apoptosis in Vitro

2.2. in Vivo Experiments
2.2.1. Emodin Increases the Survival Rate of CVB4 Infected Mice
2.2.2. Emodin Alleviates the Myocardial Lesions Induced by CVB4 Infection


| Group | Pathologic score (means ± S.D.) | |||
|---|---|---|---|---|
| 7 day | 14 day | |||
| Inflammatory infiltration | Necrosis | Inflammatory Infiltration | Necrosis | |
| NC | 0.93 ± 0.67 | 0.90 ± 0.58 | 0.85 ± 0.64 | 0.80 ± 0.48 |
| VC | 3.04 ± 0.29 | 3.01 ± 0.45 | 3.18 ± 0.44 | 2.94 ± 0.39 |
| Ribavirin 10 mg/kg/d | 1.19 ± 0.17 ** | 1.13 ± 0.25 ** | 1.12 ± 0.46 ** | 1.05 ± 0.25 ** |
| Emodin 30 mg/kg/d | 1.15 ± 0.24 ** | 1.07 ± 0.31 ** | 1.07 ± 0.33 ** | 0.99 ± 0.25 ** |
| Emodin 15 mg/kg/d | 1.49 ± 0.22 ** | 1.39 ± 0.39 ** | 1.41 ± 0.20 ** | 1.33 ± 0.30 ** |
| Emodin 7.5 mg/kg/d | 2.08 ± 0.12 * | 2.01 ± 0.16 * | 2.00 ± 0.22 | 1.98 ± 0.23 * |
2.2.3. Emodin Can Alter Transcription and Protein Levels of Apoptosis-Related Genes in Vivo


3. Experimental
3.1. Extraction and Isolation of Emodin
3.2. Experiments in Vitro
3.2.1. Cells and Virus Stocks
3.2.2. Evaluation of Cytotoxicity
3.2.3. Drug Treatment after Infection
3.2.4. Virucidal Assay
3.2.5. Drug Treatment before Infection
3.2.6. Plaque Reduction Assay
3.2.7. Time of Addition Assay
3.2.8. Quantification of CVB4 RNA Level
3.2.9. Detection of Apoptosis by Flow Cytometry
3.3. Experiments in Vivo
3.3.1. Experimental Design and Tissue Sampling
3.3.2. Apoptosis-Related Genes Isolation and Real-Time RT-PCR
3.3.3. Immunoblot Analysis
3.4. Statistical Analysis
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Crowell, R.L.; Landau, B.J. A short history and introductory background on the coxsackieviruses of group B. Curr. Top. Microbiol. Immunol. 1997, 233, 1–11. [Google Scholar]
- Modlin, J.F.; Rotart, H.A. Group B coxsackie disease in children. Curr. Top. Microbiol. Immunol. 1997, 233, 54–80. [Google Scholar]
- Rotbart, H.A. Treatment of picornavirus infections. Antiviral Res. 2002, 53, 83–98. [Google Scholar] [CrossRef]
- Pallansch, M.A. Coxsackievirus B epidemiology and public health concerns. Curr. Top. Microbiol. Immunol. 1997, 233, 13–30. [Google Scholar]
- Carrasco, L. Picornavirus inhibitors. Pharmacol. Ther. 1994, 64, 215–290. [Google Scholar] [CrossRef]
- Heim, A.; Grumbach, I.; Pring-Akerblom, P.; Stille-Siegener, M.; Müller, G.; Kandolf, R.; Figulla, H.R. Inhibition of coxsackievirus B3 carrier state infection of cultured human myocardial fibroblasts by ribavirin and human natural interferon-alpha. Antiviral Res. 1997, 34, 101–111. [Google Scholar] [CrossRef]
- Kishimoto, C.; Crumpacker, C.S.; Abelmann, W.H. Ribavirin treatment of murine coxsackievirus B3 myocarditis with analyses of lymphocyte subsets. J. Am. Coll. Cardio. 1988, 12, 1334–1341. [Google Scholar] [CrossRef]
- Feigelstock, D.A.; Mihalik, K.B.; Feinstone, S.M. Selection of hepatitis C virus resistant to ribavirin. Virol. J. 2011, 8, 402. [Google Scholar] [CrossRef]
- Nomura, H.; Tanimoto, H.; Kajiwara, E.; Shimono, J.; Maruyama, T.; Yamashita, N.; Nagano, M.; Higashi, M.; Mukai, T.; Matsui, Y.; et al. Factors contributing to ribavirin-induced anemia. J. Gastroenterol. Hepatol. 2004, 19, 1312–1317. [Google Scholar] [CrossRef]
- De Clercq, E. Antiviral drugs in current clinical use. J. Clin. Virol. 2004, 30, 115–133. [Google Scholar] [CrossRef]
- Heo, S.K.; Yun, H.J.; Park, W.H.; Park, S.D. Emodin inhibits TNF-alpha-induced human aortic smooth muscle cell proliferation via caspase and mitochondrial-dependent apoptosis. J. Cell. Biochem. 2008, 105, 70–80. [Google Scholar] [CrossRef]
- Kuo, Y.C.; Tsai, W.J.; Meng, H.C.; Chen, W.P.; Yang, L.Y.; Lin, C.Y. Immune reponses in human mesangial cells regulated by emodin from Polygonum hypoleucum Ohwi. Life Sci. 2001, 68, 1271–1286. [Google Scholar] [CrossRef]
- Alves, D.S.; Pérez-Fons, L.; Estepa, A.; Micol, V. Membrane-related effects underlying the biological activity of the anthraquinones emodin and barbaloin. Biochem. Pharmacol. 2004, 68, 549–561. [Google Scholar] [CrossRef]
- Hsiang, C.Y.; Ho, T.Y. Emodin is a novel alkaline nuclease inhibitor that sup-presses herpes simplex virus type 1 yields in cell cultures. Br. J. Pharmacol. 2008, 155, 227–235. [Google Scholar] [CrossRef]
- Shuangsuo, D.; Zhengguo, Z.; Yunru, C.; Xin, Z.; Baofeng, W.; Lichao, Y.; Yan’an, C. Inhibition of the replication of hepatitis B virus in vitro by emodin. Med. Sci. Monit. 2006, 12, BR302–BR306. [Google Scholar]
- Schwarz, S.; Wang, K.; Yu, W.; Sun, B.; Schwarz, W. Emodin inhibits current through SARS-associated coronavirus 3a protein. Antiviral Res. 2011, 90, 64–69. [Google Scholar] [CrossRef]
- Dang, S.; Zhang, X.; Song, P.; Bian, J.; Cheng, Y. Inhibition on hepatitis B virus by emodin and astragalus polysaccharides in vitro. J. Xi’an Jiaotong Univ. Med. Sci. 2007, 28, 521–525. [Google Scholar]
- Andersen, D.O.; Weber, N.D.; Wood, S.G.; Hughes, B.G.; Murray, B.K.; North, J.A. In vitro virucidal activity of selected anthraquinones and anthraquinone derivatives. Antiviral Res. 1991, 16, 185–196. [Google Scholar] [CrossRef]
- Sydiskis, R.J.; Owen, D.G.; Lohr, J.L.; Rosler, K.H.; Blomster, R.N. Inactivation of enveloped viruses by anthraquinones extracted from plants. Antimicrob. Agents Chemother. 1991, 35, 2463–2466. [Google Scholar] [CrossRef]
- Adrian, F.R. A left superior vena cava draining the blood from a closed coronary sinus. J. Anat. 1938, 73, 195–197. [Google Scholar]
- Shi, L.; Xiong, H.; He, J.; Deng, H.; Li, Q.; Zhong, Q.; Hou, W.; Cheng, L.; Xiao, H.; Yang, Z. Antiviral activity of arbidol against influenza A virus, respiratory syncytial virus, rhinovirus, coxsackie virus and adenovirus in vitro and in vivo. Arch. Virol. 2007, 152, 1447–1455. [Google Scholar] [CrossRef]
- Xiong, H.R.; Luo, J.; Hou, W.; Xiao, H.; Yang, Z.Q. The effect of emodin, an anthraquinone derivative extracted from the roots of Rheum tanguticum, against herpes simplex virus in vitro and in vivo. J. Ethnopharmacol. 2011, 133, 718–723. [Google Scholar] [CrossRef]
- Ling, J.X.; Wei, F.; Li, N.; Li, J.L.; Chen, L.J.; Liu, Y.Y. Amelioration of influenza virus-induced reactive oxygen species formation by epigallocatechin gallate derived from green tea. Acta Pharmacol. Sin. 2012, 33, 1533–1541. [Google Scholar] [CrossRef]
- Brilot, F.; Jaïdane, H.; Geenen, V.; Hober, D. Coxsackievirus B4 Infection of Murine Foetal Thymus Organ Cultures. J. Med. Virol. 2008, 80, 659–666. [Google Scholar] [CrossRef]
- Wei, F.; Li, J.L.; Ling, J.X.; Chen, L.J.; Li, N.; Liu, Y.Y. Establishment of SYBR green-based qPCR assay for rapid evaluation and quantification for anti-Hantaan virus compounds in vitro. Virus Genes 2013, 46, 54–62. [Google Scholar] [CrossRef]
- Kluck, R.M.; Bossy-Wetzel, E.; Green, D.R.; Newmeyer, D.D. The release of cytochrome c from mitochondria: A primary site for Bcl-2 regulation of apoptosis. Science 1997, 275, 1132–1136. [Google Scholar] [CrossRef]
- Battistutta, R.; Sarno, S.; de Moliner, E.; Papinutto, E.; Zanotti, G.; Pinna, L.A. The replacement of ATP by the competitive inhibitor emodin induces conformational modifications in the catalytic site of protein kinase CK2. J. Biol. Chem. 2000, 275, 29618–29622. [Google Scholar]
- Ho, T.Y.; Wu, S.L.; Chen, J.C.; Li, C.C.; Hsiang, C.Y. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antiviral Res. 2007, 74, 92–101. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds of emodin and ribavirin are available from the authors.
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Liu, Z.; Wei, F.; Chen, L.-J.; Xiong, H.-R.; Liu, Y.-Y.; Luo, F.; Hou, W.; Xiao, H.; Yang, Z.-Q. In Vitro and in Vivo Studies of the Inhibitory Effects of Emodin Isolated from Polygonum cuspidatum on Coxsakievirus B4 . Molecules 2013, 18, 11842-11858. https://doi.org/10.3390/molecules181011842
Liu Z, Wei F, Chen L-J, Xiong H-R, Liu Y-Y, Luo F, Hou W, Xiao H, Yang Z-Q. In Vitro and in Vivo Studies of the Inhibitory Effects of Emodin Isolated from Polygonum cuspidatum on Coxsakievirus B4 . Molecules. 2013; 18(10):11842-11858. https://doi.org/10.3390/molecules181011842
Chicago/Turabian StyleLiu, Zhao, Fei Wei, Liang-Jun Chen, Hai-Rong Xiong, Yuan-Yuan Liu, Fan Luo, Wei Hou, Hong Xiao, and Zhan-Qiu Yang. 2013. "In Vitro and in Vivo Studies of the Inhibitory Effects of Emodin Isolated from Polygonum cuspidatum on Coxsakievirus B4 " Molecules 18, no. 10: 11842-11858. https://doi.org/10.3390/molecules181011842