Enzymatic Synthesis of Highly Fluorescent 8-Azapurine Ribosides Using a Purine Nucleoside Phosphorylase Reverse Reaction: Variable Ribosylation Sites
Abstract
:1. Introduction
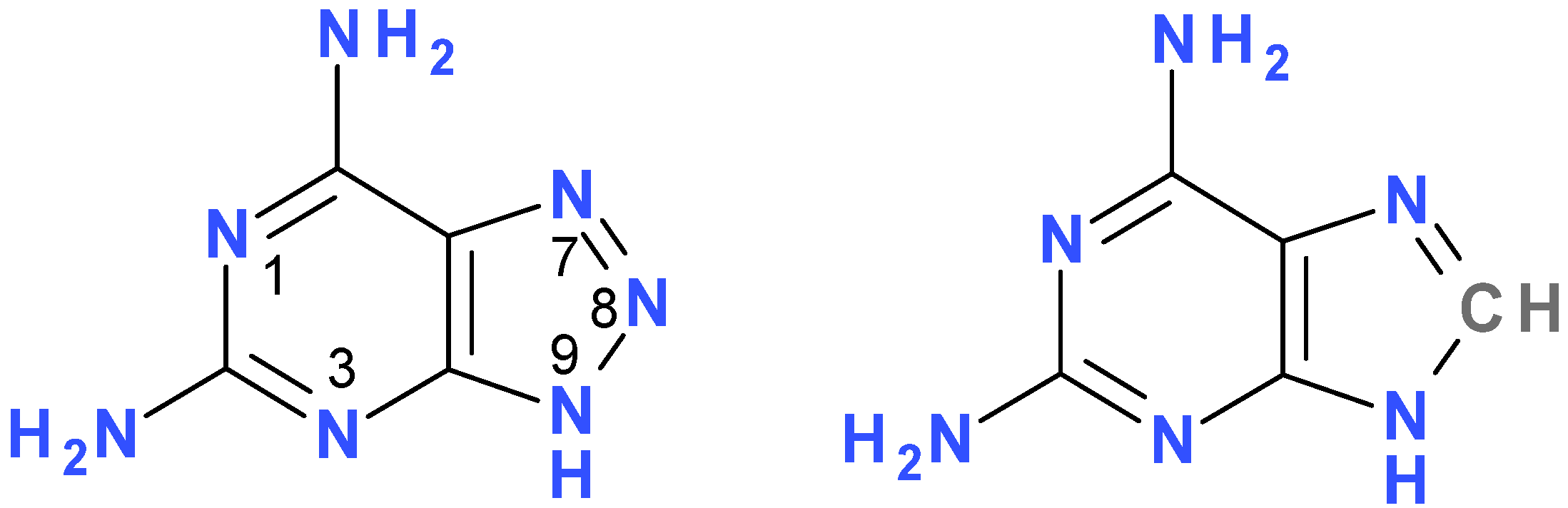
2. Results and Discussion
2.1. Fluorescence of 2,6-diamino-8-azapurine Base and its Alkyl Derivatives
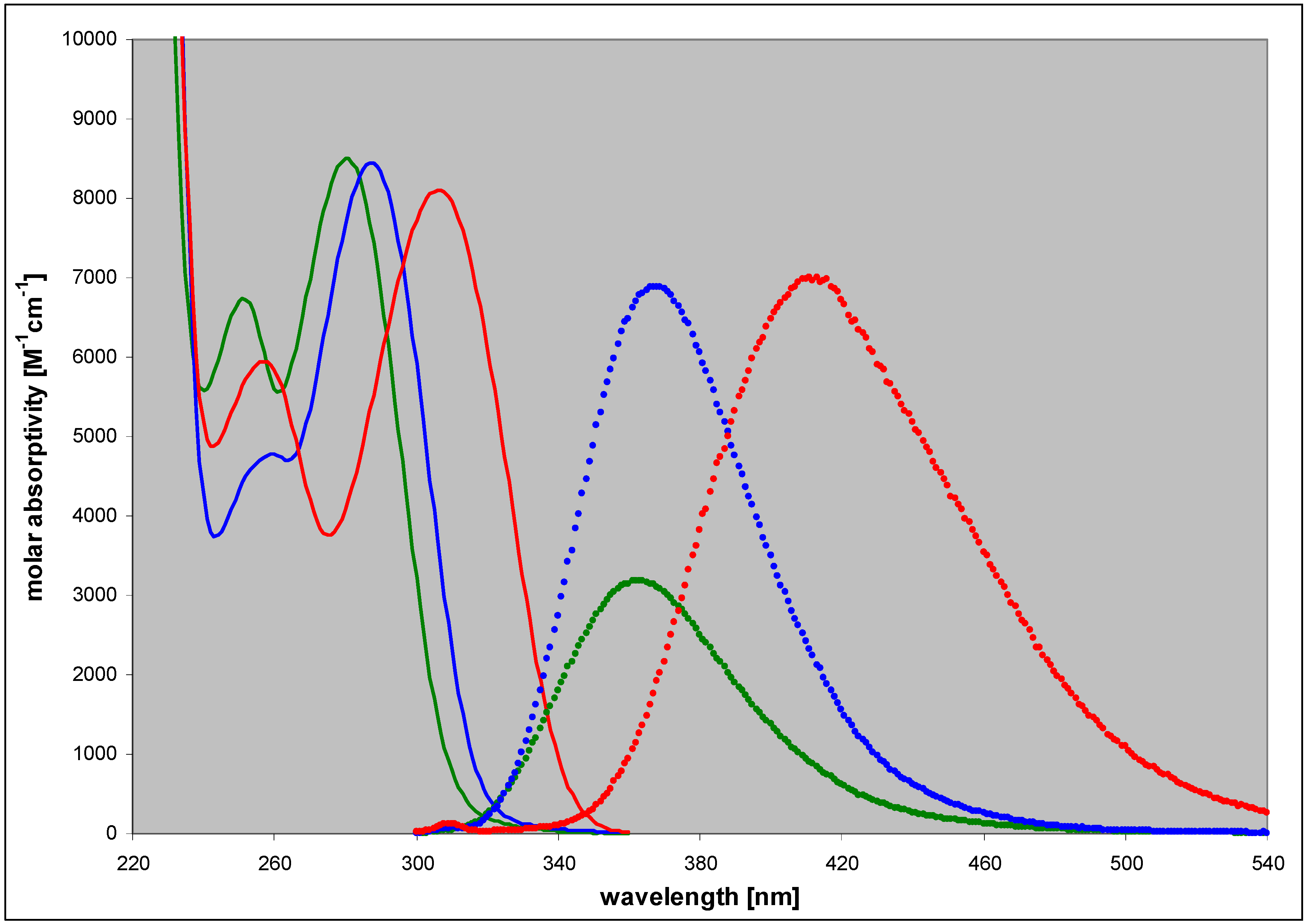
2.2. Enzymatic Ribosylation of DaaPur Using Various Forms of PNP as a Catalyst

| Compound | Relative a kcat | Km [µM] | Enzyme source |
|---|---|---|---|
| 2,6-diamino-8-azapurine | ~0.4% | 54 | calf spleen |
| 2% | 65 | E. coli | |
| 2,6-diaminopurine b | <0.1% | - | calf spleen |
| >120% c | ~7 | E. coli | |
| 8-azaguanine c | 21% | 100 | calf spleen |
| ~4 U/mg | >200 | E. coli |
| Compound | pKa | Form (pH) a | UV absorption b | Fluorescence | ||
|---|---|---|---|---|---|---|
| λmax [nm] | εmax [M−1cm−1] | λmax [nm] | ϕ | |||
| 2,6-diamino-8-azapurine | 3.68; 7.68 | n (6) | 280 | 8500 | 363 | 0.40 |
| ma (10) | 290 | 6400 | 370 | 0.36 | ||
| cat (2.5) | 253 | 9500 | 410 | 0.27 | ||
| N9-phoshonomethoxy-propyl(PMP)- | 3.4 | n (7) | 284 | nd c | 367 | 0.80 |
| N8-methyl- | 4.85 | n (7) | 307 | 8100 | 412 | 0.85 |
| cat (2.5) | 284 | 12000 | 410 | ~0.4 | ||
| N7-methyl- | 4.3 | n (7) | 309 | 6200 | nd | nd |
| cat (2) | 282 | 7900 | nd | nd | ||
| N9-β-D-ribofuranosyl- | ~3.2 | n (7) | 286 | 10000 | 367 | ~0.9 |
| N8-β-D-ribofuranosyl- | 4.9 | n (7) | 313 | 8200 | 430 | 0.41 |
| N7-β-D-ribofuranosyl- | 4.1 | n (7) | 314 | ~5500 | 420 | 0.06 |
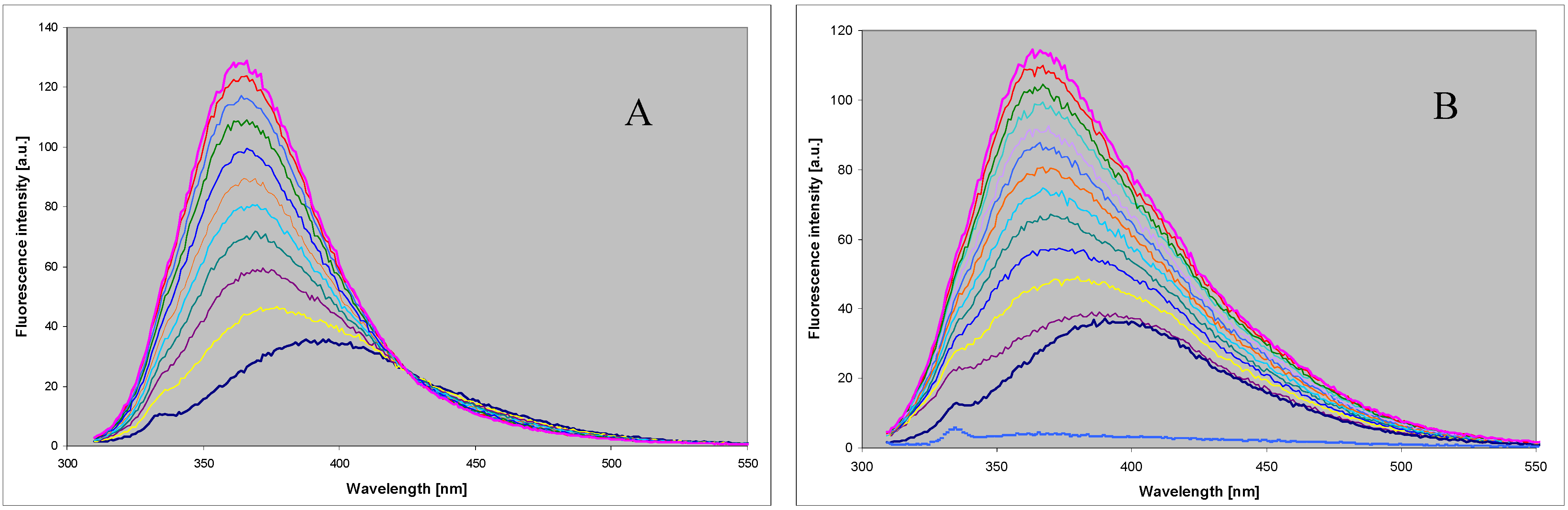
2.3. Spectral and Biochemical Properties of the Ribosylation Products
2.4. Comparison with Enzymatic Ribosylation of 8-azaguanine and Other 8-azapurines
3. Experimental
3.1. General
3.2. Enzymatic Reactions and Separation of the Products
- (1)
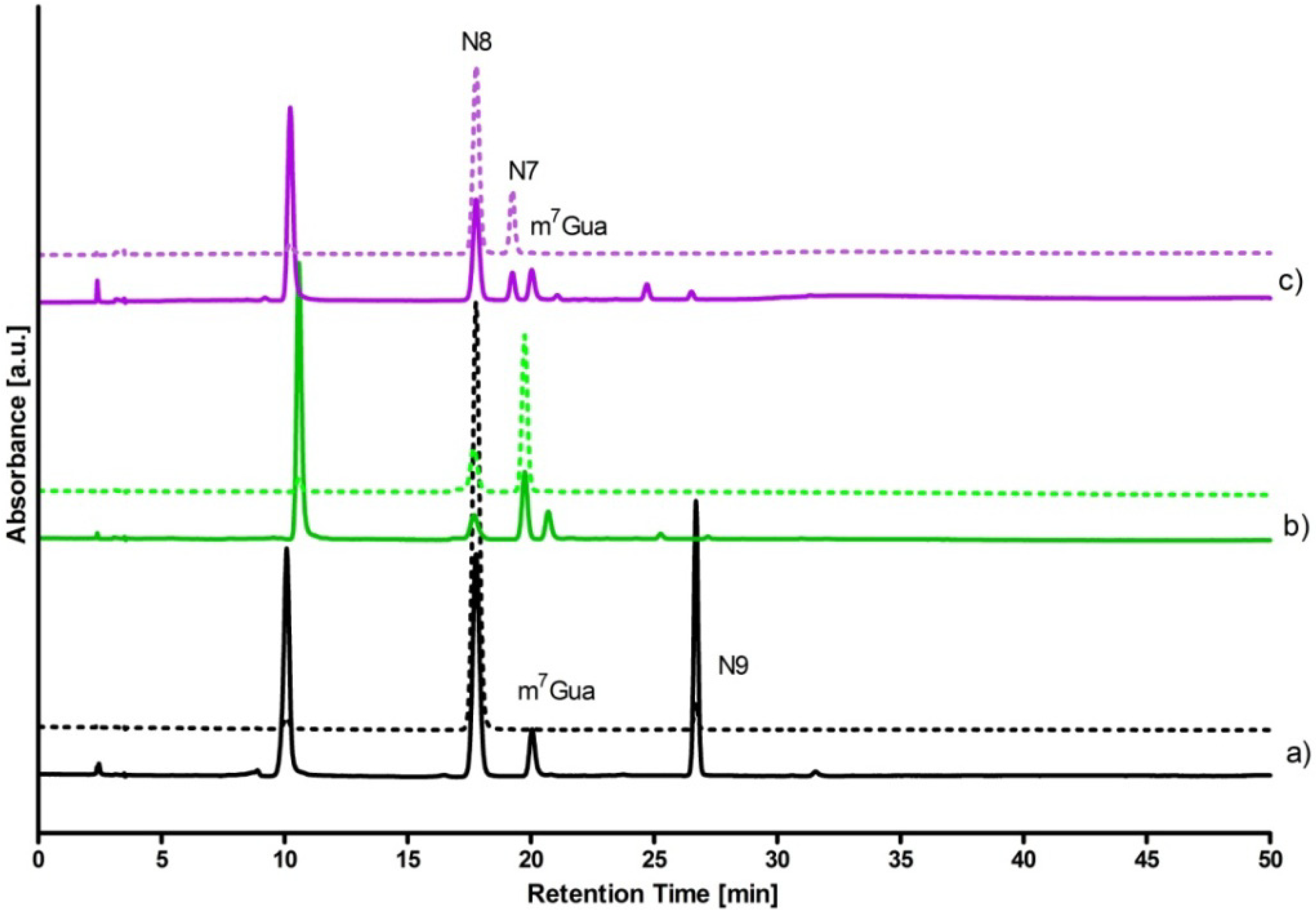
- 5 mM DaaPur, diluted from the ammonium salt solution, in 25 mM HEPES, pH 6.6, reacted with ~7 mM ribose-1-phosphate in 1 mL volume, at 35 °C, for 3 h. Recombinant calf PNP (4 µL of 12.7 mg/mL solution) was used as a catalyst. Reaction progress was monitored fluorimetrically. After 3 h the reaction was stopped by boiling in a microwave oven for ~20 s and the resultant mixture was analyzed by HPLC (see Figure 4). The overall yields of N7-β-d- and N8-β-D-riboside were ca. 25% and 7%, respectively.
- (2)
- To the same starting solution 0.5 mM inorganic phosphate was added. The reaction was allowed to run for 8 h at 32 °C. Analysis of the products (Figure 4) shown ca. 40% yield of the N8-β-d-riboside, with ~10% yield of the N7-β-d-riboside.
- (3)
- Reaction was conducted for ~5 h under the conditions as indicated in (1) above, except that E. coli PNP was used as a catalyst. The resultant mixture contained N8-β-D-riboside (~30%, retention time ~17.5 min) and N9-β-d-riboside (~10%, 29–33 min).
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Krenitsky, T.A.; Koszalka, G.W.; Tuttle, J.V. Purine nucleoside synthesis: An efficient method employing nucleoside phosphorylases. Biochemistry 1981, 20, 3615–3621. [Google Scholar] [CrossRef]
- Mikhailopulo, I.A. Biotechnology of nucleic acid constituents—State of the art and perspectives. Curr. Org. Chem. 2007, 11, 317–335. [Google Scholar] [CrossRef]
- Mikhailopulo, I.A.; Miroshnikov, A.I. Biologically important nucleosides: Modern trends in biotechnology and application. Mendeleev Commun. 2011, 21, 57–68. [Google Scholar] [CrossRef]
- Stepchenko, V.A.; Seela, F.; Esipov, R.S.; Miroshnikov, A.I.; Sokolov, Y.A.; Mikhailopulo, I.A. Enzymatic synthesis of 2-deoxy-β-d-ribonucleosides of 8-azapurines and 8-aza-7-deazapurines. Synlett 2012, 23, 1541–1545. [Google Scholar] [CrossRef]
- Jameson, D.M.; Eccleston, J.F. Fluorescent nucleotide analogs: Synthesis and applications. Methods Enzymol. 1997, 278, 363–390. [Google Scholar] [CrossRef]
- Sinkeldam, R.W.; Greco, N.J.; Tor, Y. Fluorescent analogs of biomolecular building blocks: Design, properties, and applications. Chem. Rev. 2010, 110, 2579–2619. [Google Scholar] [CrossRef]
- Tanpure, A.A.; Pawar, M.G.; Srivatsan, S.G. Fluorescent nucleoside analogs: Probes for investigating nucleic acid structure and function. Israel J. Chem. 2013, 53, 366–378. [Google Scholar] [CrossRef]
- Albert, A. Chemistry of 8-azapurines. Adv. Heterocycl. Chem. 1986, 39, 117–178. [Google Scholar] [CrossRef]
- Wierzchowski, J.; Wielgus-Kutrowska, B.; Shugar, D. Fluorescence emission properties of 8-azapurines and their nucleosides, and application to the kinetics of the reverse synthetic reaction of PNP. Biochim. Biophys. Acta 1996, 1290, 9–17. [Google Scholar]
- Wierzchowski, J.; Bzowska, A.; Stępniak, K.; Shugar, D. Interactions of calf spleen purine nucleoside phosphorylase with 8-azaguanine, and a bisubstrate analogue inhibitor: Implications for the reaction mechanism. Z. Naturforschung 2004, 59c, 713–725. [Google Scholar]
- Da Costa, C.P.; Fedor, M.J.; Scott, L.G. 8-Azaguanine reporter of purine ionization states in structured RNAs. J. Am. Chem. Soc. 2007, 129, 3426–3432. [Google Scholar] [CrossRef]
- Liu, L.; Cottrell, J.W.; Scott, L.G.; Fedor, M.J. Direct measurement of the ionization state of an essential guanine in the hairpin ribozyme. Nat. Chem. Biol. 2009, 5, 351–357. [Google Scholar] [CrossRef]
- Cottrell, J.W.; Scott, L.G.; Fedor, M.J. The pH dependence of hairpin ribozyme catalysis reflects ionization of an active site adenine. J. Biol. Chem. 2011, 286, 17658–17664. [Google Scholar] [CrossRef]
- Seela, F.; Jiang, D.; Budow, S. Triplexes with 8-aza-2′-deoxyisoguanosine replacing protonated dC: Probing third strand stability with a fluorescent nucleobase targeting duplex DNA. ChemBioChem 2010, 11, 1443–1450. [Google Scholar] [CrossRef]
- Wierzchowski, J.; Ogiela, M.; Iwańska, B.; Shugar, D. Selective fluorescent and fluorogenic substrates for purine-nucleoside phosphorylases from various sources, and direct fluorimetric determination of enzyme levels in human and animal blood. Anal. Chim. Acta 2002, 472, 63–74. [Google Scholar] [CrossRef]
- Wierzchowski, J.; Stachelska-Wierzchowska, A. Two fluorogenic substrates for purine-nucleoside phosphorylase, selective for mammalian and bacterial forms of the enzyme. Anal. Biochem. 2013. submitted. [Google Scholar]
- Wierzchowski, J.; Mędza, G.; Szabelski, M.; Stachelska-Wierzchowska, A. Properties of 2,6-diamino-8-azapurine, a highly fluorescent purine analog and its N-alkyl derivatives: Tautomerism and excited-state proton transfer reactions. Photochem. Photobiol. A 2013, 265, 49–57. [Google Scholar] [CrossRef]
- Bzowska, A.; Kulikowska, E.; Shugar, D. Purine nucleoside phosphorylases: Properties, functions, and clinical aspects. Pharmacol. Therapeutics 2000, 88, 349–425. [Google Scholar] [CrossRef]
- Montgomery, J.A.; Shortnacy, A.T.; Secrist, J.A., III. Synthesis and biological evaluation of 2-fluoro-8-azaadenosine and related compounds. J. Med. Chem. 1983, 26, 1483–1489. [Google Scholar] [CrossRef]
- Seela, F.; Lampe, S. 8-Aza-2′-deoxyguanosine and related 1,2,3-triazolo[4,5-d]pyrimidine 2′-deoxyribofuranosides. Helv. Chim. Acta 1993, 72, 2388–2397. [Google Scholar] [CrossRef]
- Ward, D.C.; Reich, E.; Stryer, L. Fluorescence studies of nucleotides and polynucleotides. I. Formycin, 2-aminopurine riboside, 2,6-diaminopurine riboside, and their derivatives. J. Biol. Chem. 1969, 244, 1228–1237. [Google Scholar]
- Ottink, O.M.; Nelissen, F.H.T; Derks, Y.; Wijmenga, S.S.; Heus, H.A. Enzymatic stereospecific preparation of fluorescent S-adenosyl-L-methionine analogs. Anal. Biochem 2010, 396, 280–283. [Google Scholar] [CrossRef]
- Olsen, A.G.; Dahl, O.; Petersen, A.B.; Nielsen, J.N.; Nielsen, P.E. A novel pseudo-complementary PNA G-C base pair. Artif. DNA: PNA XNA 2011, 2, 33–37. [Google Scholar]
- Kath-Schorr, S.; Wilson, T.J.; Li, N.-S.; Lu, J.; Piccirilli, J.A.; Lilley, D.M.J. General acid-base catalysis mediated by nucleobases in the hairpin ribozyme. J. Am. Chem. Soc. 2012, 134, 16717–16724. [Google Scholar] [CrossRef]
- Albert, A.; Taguchi, H. V-triazolo(4,5-d)pyrimidines (8-azapurines). 8. Synthesis, from 1,2,3-triazoles, of 1- and 2-methyl derivatives of 5,7-disubstituted V-triazolo(4,5-d)pyrimidines (7- and 8-methyl 2,6-disubstituted 8-azapurines). J. Chem. Soc. Perkin Trans. I 1972, 4, 449–456. [Google Scholar] [CrossRef]
- Leonard, N.J. Etheno-substituted nucleotides and coenzymes: Fluorescence and biological activity. CRC Crit. Rev. Biochem. 1984, 15, 125–199. [Google Scholar] [CrossRef]
- Kierdaszuk, B.; Wierzchowski, J. 8-Azapurines as substrates for the E. coli purine nucleoside phosphorylase—II (xanthosine phosphorylase): Does the ribosylation always go to the aza-purine N(9)? Collect. Symp. Series 2005, 7, 489–491. [Google Scholar]
- Bzowska, A.; Ananiev, A.V.; Ramzaeva, N.; Alksnis, E.; Maurins, J.A.; Kulikowska, E.; Shugar, D. Purine nucleoside phosphorylase: Inhibition by purine N(7)- and N(9)-acyclonucleosides; and substrate properties of 7-β-D-ribofuranosylguanine and 7-β-D-ribofuranosylhypoxanthine. Biochem. Pharmacol. 1994, 48, 937–947. [Google Scholar] [CrossRef]
- Bzowska, A.; Kulikowska, E.; Poopeiko, N.E.; Shugar, D. Kinetics of phosphorolysis of 3-(β-D-ribofuranosyl)adenine and 3-(β-D-ribofuranosyl)hypoxanthine, non-conventional substrates of purinenucleoside phosphorylase. Eur. J. Biochem. 1996, 239, 229–234. [Google Scholar]
- Roivainen, J.; Elizarova, T.; Lapinjoki, S.; Mikhailopulo, I.A.; Esipov, R.S.; Miroshnikov, A.I. An enzymatic transglycosylation of purine bases. Nucleosides, Nucleotides Nucleic Acids 2007, 26, 905–909. [Google Scholar] [CrossRef]
- Mikleušević, G.; Štefanić, Z.; Narczyk, M.; Wielgus-Kutrowska, B.; Bzowska, A.; Luić, M. Validation of the catalytic mechanism of Escherichia coli purine nucleoside phosphorylase by structural and kinetic studies. Biochimie 2011, 93, 1610–1622. [Google Scholar] [CrossRef]
- Wielgus-Kutrowska, B.; Breer, K.; Hashimoto, M.; Hikishima, S.; Yokomatsu, T.; Narczyk, M.; Dyzma, A.; Girstun, A.; Staroń, K.; Bzowska, A. Trimeric purine nucleoside phosphorylase: Exploring postulated one-third-of-the-sites binding in the transition state. Bioorg. Med. Chem. 2012, 20, 6758–6769. [Google Scholar] [CrossRef]
- Suzuki, K.; Kobayashi, A.; Kaneko, S.; Takehira, K.; Yoshihara, T.; Ishida, H.; Shiina, Y.; Oishi, S.; Tobita, S. Reevaluation of absolute luminescence quantum yields of standard solutions using a spectrometer with an integrating sphere and a back-thinned CCD detector. Phys. Chem. Chem. Phys. 2009, 11, 9850–9860. [Google Scholar] [CrossRef]
- Portsmouth, D.; Hlavaty, J.; Renner, M. Suicide genes for cancer therapy. Mol. Aspects Med. 2007, 28, 4–41. [Google Scholar] [CrossRef]
- Štefanić, Z.; Mikleušević, G.; Narczyk, M.; Wielgus-Kutrowska, B.; Bzowska, A.; Luić, M. Still a long way to fully understanding the molecular mechanism of Escherichia coli Purine Nucleoside Phosphorylase. Croat. Chem. Acta 2013, 86, 117–127. [Google Scholar]
- Zhou, X.; Szeker, K.; Janocha, B.; Böhme, T.; Albrecht, D.; Mikhailopulo, I.A.; Neubauer, P. Recombinant purine nucleoside phosphorylases from thermophiles: Preparation, properties and activity towards purine and pyrimidine nucleosides. FEBS J. 2013, 280, 1475–1490. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds are available from the authors.
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Stachelska-Wierzchowska, A.; Wierzchowski, J.; Wielgus-Kutrowska, B.; Mikleušević, G. Enzymatic Synthesis of Highly Fluorescent 8-Azapurine Ribosides Using a Purine Nucleoside Phosphorylase Reverse Reaction: Variable Ribosylation Sites. Molecules 2013, 18, 12587-12598. https://doi.org/10.3390/molecules181012587
Stachelska-Wierzchowska A, Wierzchowski J, Wielgus-Kutrowska B, Mikleušević G. Enzymatic Synthesis of Highly Fluorescent 8-Azapurine Ribosides Using a Purine Nucleoside Phosphorylase Reverse Reaction: Variable Ribosylation Sites. Molecules. 2013; 18(10):12587-12598. https://doi.org/10.3390/molecules181012587
Chicago/Turabian StyleStachelska-Wierzchowska, Alicja, Jacek Wierzchowski, Beata Wielgus-Kutrowska, and Goran Mikleušević. 2013. "Enzymatic Synthesis of Highly Fluorescent 8-Azapurine Ribosides Using a Purine Nucleoside Phosphorylase Reverse Reaction: Variable Ribosylation Sites" Molecules 18, no. 10: 12587-12598. https://doi.org/10.3390/molecules181012587





