Selective G-Quadruplex DNA Recognition by a New Class of Designed Cyanines
Abstract
:1. Introduction

2. Results
2.1. UV-Thermal Melting Screening (Tm)
| Compound/Drug:DNA Ratio | Tel22 | AATT duplex | ||||||
|---|---|---|---|---|---|---|---|---|
| 1:1 | 2:1 | 4:1 | 6:1 | 1:1 | 2:1 | 4:1 | 6:1 | |
| 24 | 2 | 4 | 9 | 14 | 0 | 1 | 1 | 2 |
| a | - | 2.1 | 4.6 | 8.7 | 0 | 0 | 0 | 0 |
| 25 | 1 | 1 | 1 | 0 | ND | |||
| 26 | 1 | 2 | 4 | 11 | 0 | 2 | 2 | 2 |
| 27 | 2 | 1 | 1 | 1 | ND | |||
| 28 | 1 | 1 | 3 | 7 | 1 | 1 | 2 | 3 |
| 29 | 3 | 8 | 12 | 16 | 0 | 1 | 1 | 2 |
| 30 | 3 | 7 | 15 | 19 | 0 | 0 | 1 | 1 |
| b | - | 3.1 | 8.9 | 13.2 | 0 | 0 | 0 | 0.5 |
| 31 | 1 | 5 | 14 | 24 | 0 | 0 | 0 | 0 |
| c | - | 2.5 | 15.6 | - | 0 | 0 | 0 | 1.1 |
| 32 | 3 | 7 | 17 | 21 | 1 | 1 | 3 | 3 |
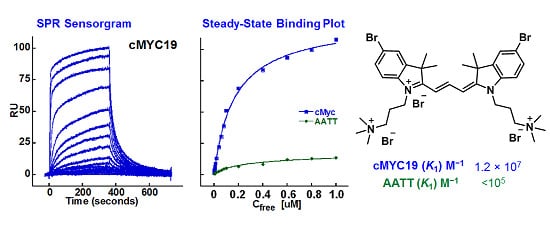
2.2. Biosensor-Surface Plasmon Resonance Studies (SPR)

| Compound | Tel22 (K1; K2) M−1 | cMYC19 (K1; K2) M−1 | AATT (K1) M−1 |
|---|---|---|---|
| 24 | 3.9 × 105; 3.9 × 104 | 3.8 × 106; 1.4 × 105 | <105 |
| 25 | No Binding | No Binding | No Binding |
| 26 | 1.3 × 105; 1.7 × 104 | 1.2 × 106; 6.6 × 105 | <105 |
| 27 | 8.2 × 105; 5.8 × 104 | 5.7 × 105; 4.5 × 104 | <105 |
| 28 | 8.3 × 105; 6.1 × 104 | 5.0 × 106; 5.6 × 105 | <105 |
| 29 | 5.9 × 106; 3.9 × 105 | 6.3 × 106; 4.3 × 105 | <105 |
| 30 | 2.7 × 106; 2.9 × 105 | 8.4 × 106; 3.3 × 105 | <105 |
| 31 | 5.5 × 106; 4.5 × 105 | 1.2 × 107; 2.7 × 106 | <105 |
| 32 | 2.1 × 106; 2.2 × 105 | 7.8 × 106; 2.5 × 106 | <105 |
2.3. Circular Dichroism Studies (CD)
2.4. Nuclear Magnetic Resonance Studies (NMR)



2.5. Fluorescence Studies

3. Discussion
4. Experimental
4.1. Synthesis of Cyanines
4.1.1. General Synthesis for the Alkylated Indolenine Salts 15–23 (Scheme 1)
4.1.2. General Synthesis for the Trimethine Cyanine Binding Agents 24–32 (Scheme 1)

4.2. Nucleic Acids
4.3. UV-Thermal Melting
4.4. Biosensor-Surface Plasmon Resonance
4.5. Circular Dichroism
4.6. Fluorescence Titrations
4.7. Nuclear Magnetic Resonance
5. Conclusions
Supplementary Materials
Acknowledgments
Conflicts of Interest
References
- Neidle, S.; Balasubramanian, S. Quadruplex Nucleic Acids; RSC Publishers: Cambridge, UK, 2006; pp. 301–315. [Google Scholar]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef]
- Paeschke, K.; Simonsson, T.; Postberg, J.; Rhodes, D.; Lipps, H.J. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005, 12, 847–854. [Google Scholar] [CrossRef]
- Schaffitzel, C.; Postberg, J.; Paeschke, K.; Lipps, H. Probing Telomeric G-Quadruplex DNA Structures in Cells with in Vitro Generated Single-Chain Antibody Fragments. In G-Quadruplex DNA; Baumann, P., Ed.; Humana Press: New York, NY, USA, 2010; Volume 608, pp. 159–181. [Google Scholar]
- Neidle, S.; Parkinson, G.N. Quadruplex DNA crystal structures and drug design. Biochimie 2008, 90, 1184–1196. [Google Scholar] [CrossRef]
- Hurley, L.; Wheelhouse, R.; Sun, D.; Kerwin, S.; Salazar, M.; Fedoroff, O.; Han, F.; Han, H.; Izbicka, E.; von Hoff, D. G-quadruplexes as targets for drug design. Pharmacol. Ther. 2000, 85, 141–158. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Hurley, L.H.; Neidle, S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef]
- Monchaud, D. A hitchhiker’s guide to G-quadruplex ligands. Org. Biomol. Chem. 2008, 6, 627–636. [Google Scholar] [CrossRef]
- Shin-ya, K.; Wierzba, K.; Matsuo, K.-I.; Ohtani, T.; Yamada, Y.; Furihata, K.; Hayakawa, Y.; Seto, H. Telomestatin, a novel telomerase inhibitor from Streptomyces anulatus. J. Am. Chem. Soc. 2001, 123, 1262–1263. [Google Scholar] [CrossRef]
- Drygin, D.; Siddiqui-Jain, A.; O’Brien, S.; Schwaebe, M.; Lin, A.; Bliesath, J.; Ho, C.B.; Proffitt, C.; Trent, K.; Whitten, J.P.; et al. Anticancer activity of CX-3543: A direct inhibitor of rRNA biogenesis. Cancer Res. 2009, 69, 7653–7661. [Google Scholar] [CrossRef]
- Henary, M.; Mojzych, M. Stability and Reactivity of Polymethine Dyes in Solution. In Heterocyclic Polymethine Dyes; Strekowski, L., Ed.; Springer: Berlin, Germany, 2008; Volume 14, pp. 221–238. [Google Scholar]
- Mojzych, M.; Henary, M. Synthesis of Cyanine Dyes. In Heterocyclic Polymethine Dyes; Strekowski, L., Ed.; Springer: Berlin, Germany, 2008; Volume 14, pp. 1–9. [Google Scholar]
- Datta, S.G.; Reynolds, C.; Goud, Y.K.; Datta, B. Interaction of YOYO-1 with guanine-rich DNA. J. Biomol. Struct. Dyn. 2013, 31, 1–9. [Google Scholar]
- Largy, E.; Granzhan, A.; Hamon, F.; Verga, D.; Teulade-Fichou, M.-P. Visualizing the Quadruplex: From Fluorescent Ligands to Light-Up Probes. In Quadruplex Nucleic Acids; Chaires, J.B., Graves, D., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 330, pp. 111–177. [Google Scholar]
- Mohanty, J.; Barooah, N.; Dhamodharan, V.; Harikrishna, S.; Pradeepkumar, P.I.; Bhasikuttan, A.C. Thioflavin T as an efficient inducer and selective fluorescent sensor for the human telomeric G-quadruplex DNA. J. Am. Chem. Soc. 2012, 135, 367–376. [Google Scholar]
- Vummidi, B.R.; Alzeer, J.; Luedtke, N.W. Fluorescent probes for G-quadruplex structures. ChemBioChem 2013, 14, 540–558. [Google Scholar] [CrossRef]
- Nanjunda, R.; Owens, E.A.; Mickelson, L.; Alyabyev, S.; Kilpatrick, N.; Wang, S.; Henary, M.; Wilson, W.D. Halogenated pentamethine cyanine dyes exhibiting high fidelity for G-quadruplex DNA. Bioorg. Med. Chem. 2012, 20, 7002–7011. [Google Scholar] [CrossRef]
- Fox, K.R. Drug-DNA Interaction Protocols, 2nd ed.; Humana Press: New York, NY, USA, 2010; pp. 318–329. [Google Scholar]
- Nanjunda, R.; Musetti, C.; Kumar, A.; Ismail, M.A.; Farahat, A.A.; Wang, S.; Sissi, C.; Palumbo, M.; Boykin, D.W.; Wilson, W.D. Heterocyclic dications as a new class of telomeric G-quadruplex targeting agents. Curr. Pharm. Des. 2012, 18, 1934–1947. [Google Scholar] [CrossRef]
- Nanjunda, R.; Munde, M.; Liu, Y.; Wilson, W.D. Real-Time Monitoring of Nucleic Acid Interactions with Biosensor-Surface Plasmon Resonance. In Methods for Studying Nucleic Acid/Drug Interactions; Tor, Y., Wanunu, M., Eds.; CRC Press: Boca Raton, FL, USA, 2011; pp. 91–134. [Google Scholar]
- Munde, M.; Kumar, A.; Nhili, R.; Depauw, S.; David-Cordonnier, M.-H.; Ismail, M.A.; Stephens, C.E.; Farahat, A.A.; Batista-Parra, A.; Boykin, D.W.; et al. DNA minor groove induced dimerization of heterocyclic cations: Compound structure, binding affinity, and specificity for a TTAA site. J. Mol. Biol. 2010, 402, 847–864. [Google Scholar] [CrossRef]
- Dai, J.; Carver, M.; Hurley, L.H.; Yang, D. Solution structure of a 2:1 quindoline–c-MYC G-quadruplex: Insights into G-quadruplex-interactive small molecule drug design. J. Am. Chem. Soc. 2011, 133, 17673–17680. [Google Scholar] [CrossRef]
- Barbieri, C.M.; Srinivasan, A.R.; Rzuczek, S.G.; Rice, J.E.; LaVoie, E.J.; Pilch, D.S. Defining the mode, energetics and specificity with which a macrocyclic hexaoxazole binds to human telomeric G-quadruplex DNA. Nucleic Acids Res. 2007, 35, 3272–3286. [Google Scholar] [CrossRef]
- Chung, W.J.; Heddi, B.; Tera, M.; Iida, K.; Nagasawa, K.; Phan, A.T. Solution structure of an intramolecular (3+1) human telomeric G-quadruplex bound to a telomestatin derivative. J. Am. Chem. Soc. 2013, 135, 13495–13501. [Google Scholar] [CrossRef]
- Hounsou, C.; Guittat, L.; Monchaud, D.; Jourdan, M.; Saettel, N.; Mergny, J.-L.; Teulade-Fichou, M.-P. G-quadruplex recognition by quinacridines: A SAR, NMR, and biological study. ChemMedChem 2007, 2, 655–666. [Google Scholar] [CrossRef]
- Campbell, N.H.; Parkinson, G.N.; Reszka, A.P.; Neidle, S. Structural basis of DNA quadruplex recognition by an acridine drug. J. Am. Chem. Soc. 2008, 130, 6722–6724. [Google Scholar] [CrossRef]
- Blankson, G.A.; Pilch, D.S.; Liu, A.A.; Liu, L.F.; Rice, J.E.; LaVoie, E.J. Macrocyclic biphenyl tetraoxazoles: Synthesis, evaluation as G-quadruplex stabilizers and cytotoxic activity. Bioorg. Med. Chem. 2013, 21, 4511–4520. [Google Scholar] [CrossRef]
- Rodger, A.; Norden, B. Circular Dichroism and Linear Dichroism; Oxford University Press: New York, NY, USA, 1997. [Google Scholar]
- Ambrus, A.; Chen, D.; Dai, J.; Jones, R.A.; Yang, D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry 2005, 44, 2048–2058. [Google Scholar] [CrossRef]
- Moyzis, R.K.; Buckingham, J.M.; Cram, L.S.; Dani, M.; Deaven, L.L.; Jones, M.D.; Meyne, J.; Ratliff, R.L.; Wu, J.R. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. USA 1988, 85, 6622–6626. [Google Scholar] [CrossRef]
- Makarov, V.L.; Hirose, Y.; Langmore, J.P. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 1997, 88, 657–666. [Google Scholar] [CrossRef]
- Yang, Q.; Xiang, J.; Yang, S.; Zhou, Q.; Li, Q.; Tang, Y.; Xu, G. Verification of specific G-quadruplex structure by using a novel cyanine dye supramolecular assembly: I. Recognizing mixed G-quadruplex in human telomeres. Chem. Commun. 2009, 9, 1103–1105. [Google Scholar]
- David, W.M.; Brodbelt, J.; Kerwin, S.M.; Thomas, P.W. Investigation of quadruplex oligonucleotide-drug interactions by electrospray ionization mass spectrometry. Anal. Chem. 2002, 74, 2029–2033. [Google Scholar] [CrossRef]
- Kerwin, S.M.; Sun, D.; Kern, J.T.; Rangan, A.; Thomas, P.W. G-quadruplex DNA binding by a series of carbocyanine dyes. Bioorg. Med. Chem. Lett. 2001, 11, 2411–2414. [Google Scholar] [CrossRef]
- Kovalska, V.; Losytskyy, M.; Yarmoluk, S.; Lubitz, I.; Kotlyar, A. Mono and trimethine cyanines cyan 40 and cyan 2 as probes for highly selective fluorescent detection of non-canonical DNA structures. J. Fluoresc. 2011, 21, 223–230. [Google Scholar] [CrossRef]
- Kanony, C.; Åkerman, B.; Tuite, E. Photobleaching of asymmetric cyanines used for fluorescence imaging of single DNA molecules. J. Am. Chem. Soc. 2001, 123, 7985–7995. [Google Scholar] [CrossRef]
- Mohammed, H.S.; Delos Santos, J.O.; Armitage, B. Noncovalent binding and fluorogenic response of cyanine dyes to DNA homoquadruplex and PNA-DNA heteroquadruplex structures. Artif. DNA PNA XNA 2011, 2, 43–49. [Google Scholar] [CrossRef]
- Dai, J.; Carver, M.; Yang, D. Polymorphism of human telomeric quadruplex structures. Biochimie 2008, 90, 1172–1183. [Google Scholar] [CrossRef]
- Phan, A.T.; Kuryavyi, V.; Patel, D.J. DNA architecture: From G to Z. Curr. Opin. Struct. Biol. 2006, 16, 288–298. [Google Scholar]
- Hamon, F.; Largy, E.; Guédin-Beaurepaire, A.; Rouchon-Dagois, M.; Sidibe, A.; Monchaud, D.; Mergny, J.-L.; Riou, J.-F.; Nguyen, C.-H.; Teulade-Fichou, M.-P. An acyclic oligoheteroaryle that discriminates strongly between diverse G-quadruplex topologies. Angew. Chem. 2011, 50, 8745–8749. [Google Scholar]
- Fasman, G.D. Handbook of Biochemistry and Molecular Biology, Nucleic Acids; CRC Press: Cleveland, OH, USA, 1975; Volume 1, p. 589. [Google Scholar]
- Sample Availability: Not available.
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Nanjunda, R.; Owens, E.A.; Mickelson, L.; Dost, T.L.; Stroeva, E.M.; Huynh, H.T.; Germann, M.W.; Henary, M.M.; Wilson, W.D. Selective G-Quadruplex DNA Recognition by a New Class of Designed Cyanines. Molecules 2013, 18, 13588-13607. https://doi.org/10.3390/molecules181113588
Nanjunda R, Owens EA, Mickelson L, Dost TL, Stroeva EM, Huynh HT, Germann MW, Henary MM, Wilson WD. Selective G-Quadruplex DNA Recognition by a New Class of Designed Cyanines. Molecules. 2013; 18(11):13588-13607. https://doi.org/10.3390/molecules181113588
Chicago/Turabian StyleNanjunda, Rupesh, Eric A. Owens, Leah Mickelson, Tyler L. Dost, Ekaterina M. Stroeva, Hang T. Huynh, Markus W. Germann, Maged M. Henary, and W. David Wilson. 2013. "Selective G-Quadruplex DNA Recognition by a New Class of Designed Cyanines" Molecules 18, no. 11: 13588-13607. https://doi.org/10.3390/molecules181113588