General Approach for Introduction of Various Chemical Labels in Specific RNA Locations Based on Insertion of Amino Linkers
Abstract
:1. Introduction
2. Introduction of Amino Linker into RNA in the Course of Its Chemical Synthesis
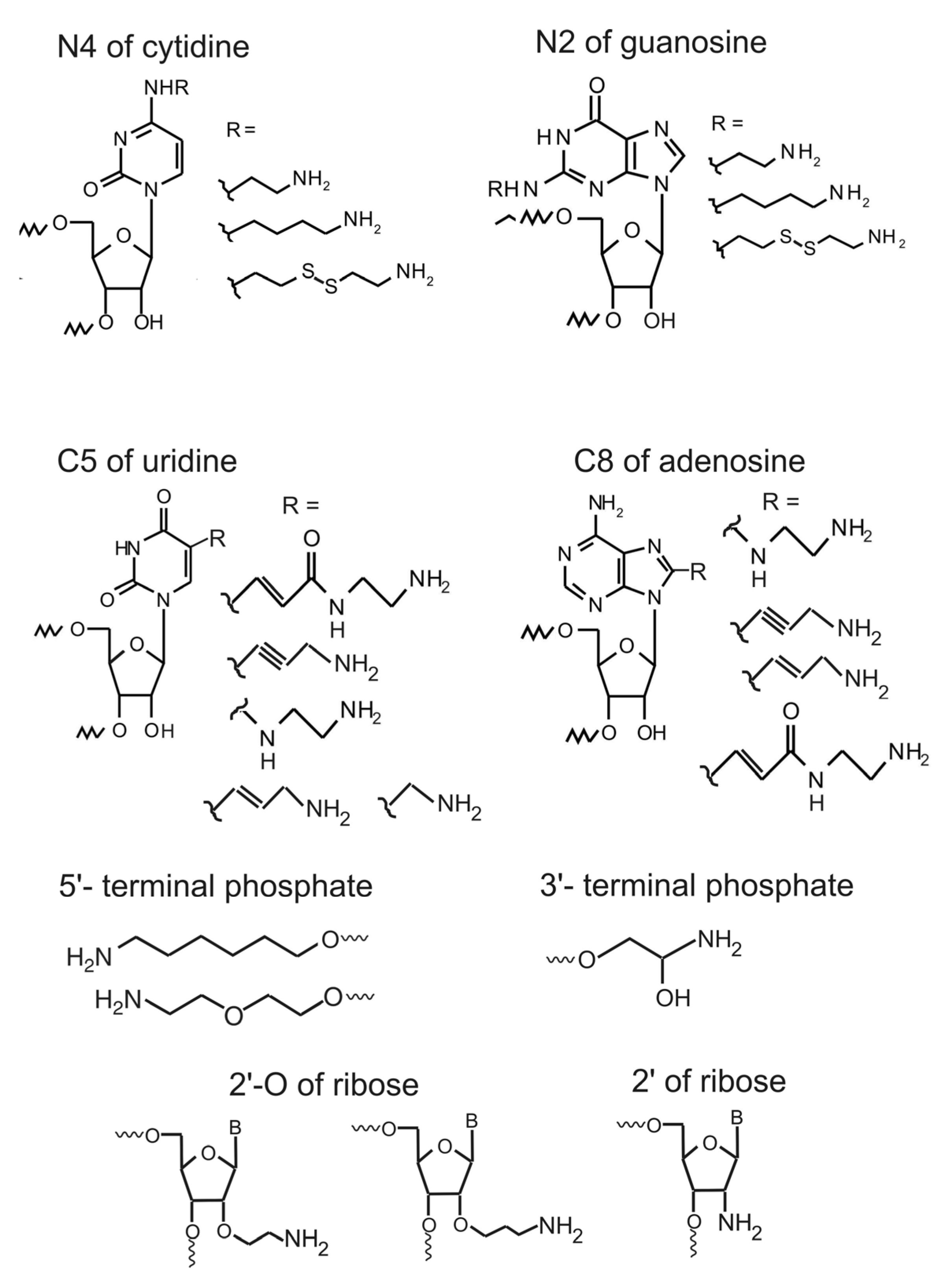
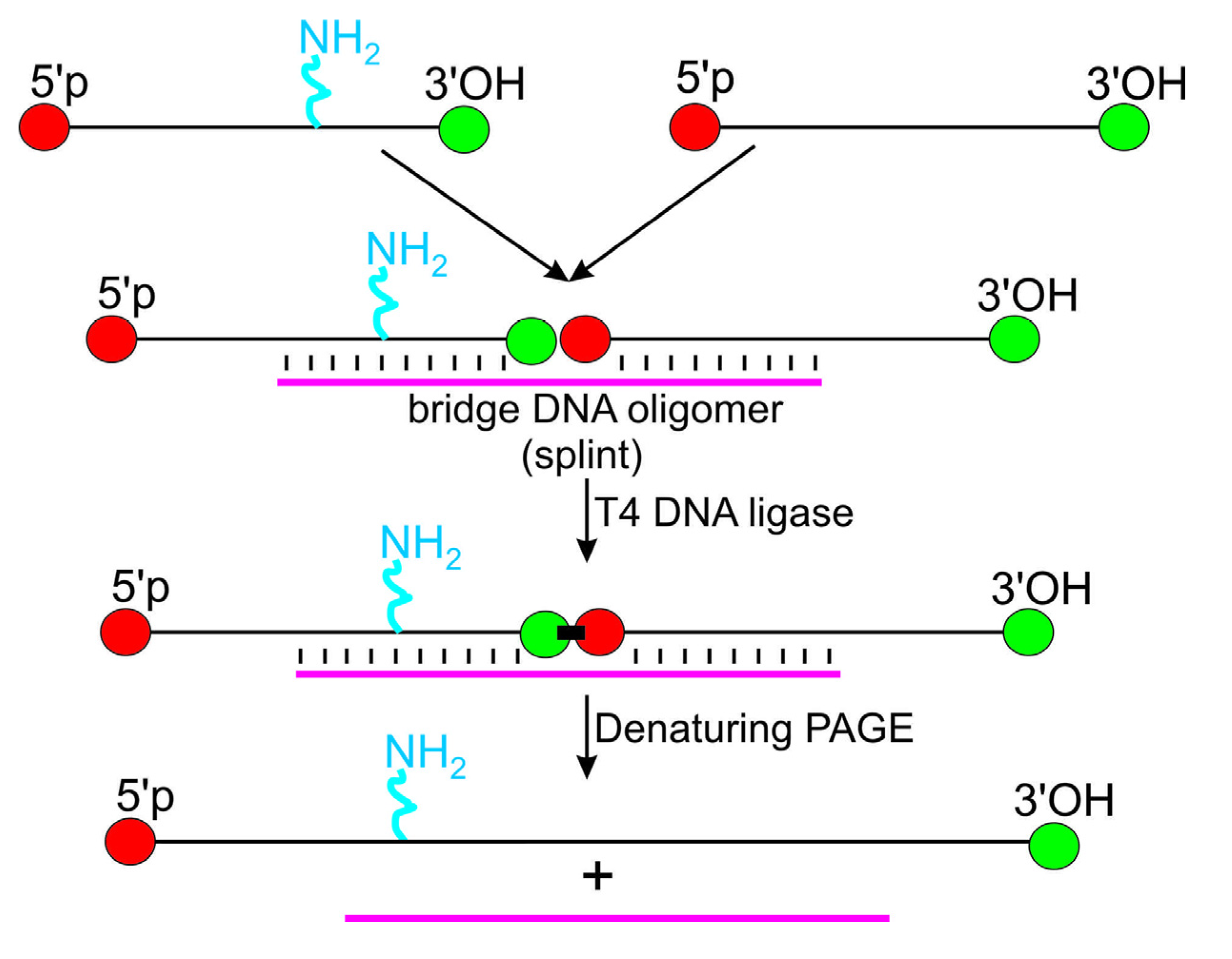
3. Introduction of Amino Linker into Internal RNA Positions by Assembly of the Modified RNA from Shorter RNA Segments
4. Synthesis of Amino Linker-Containing RNAs with the Use of T7 RNA Polymerase and Modified Ribonucleoside Triphosphates
5. Method for Introduction of Amino Linkers at the 5'-Terminus of Unmodified RNAs

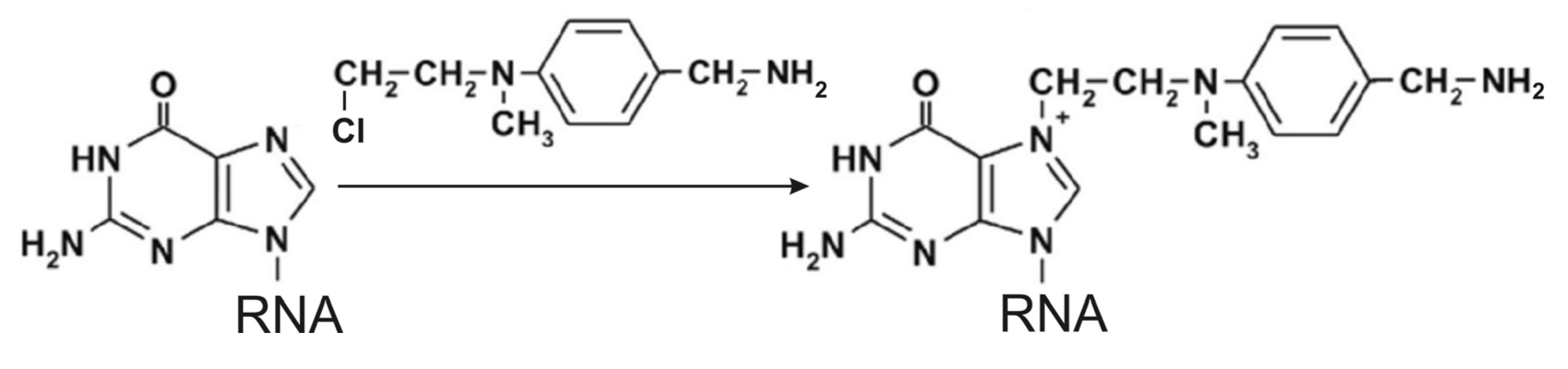
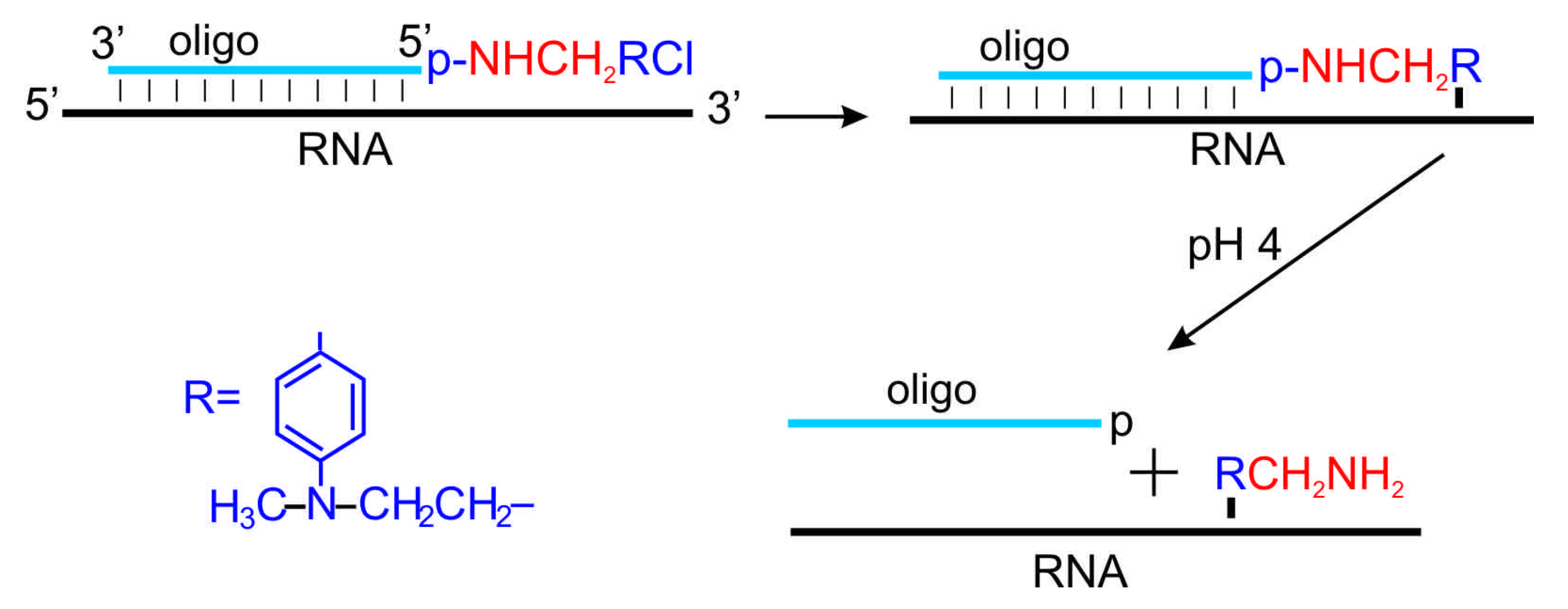
6. Introduction of Amino Linkers at Guanosine Residues of Unmodified RNAs
7. Approach for Site-Specific Introduction of Amino Linkers into Intact Relatively Long Structured RNAs
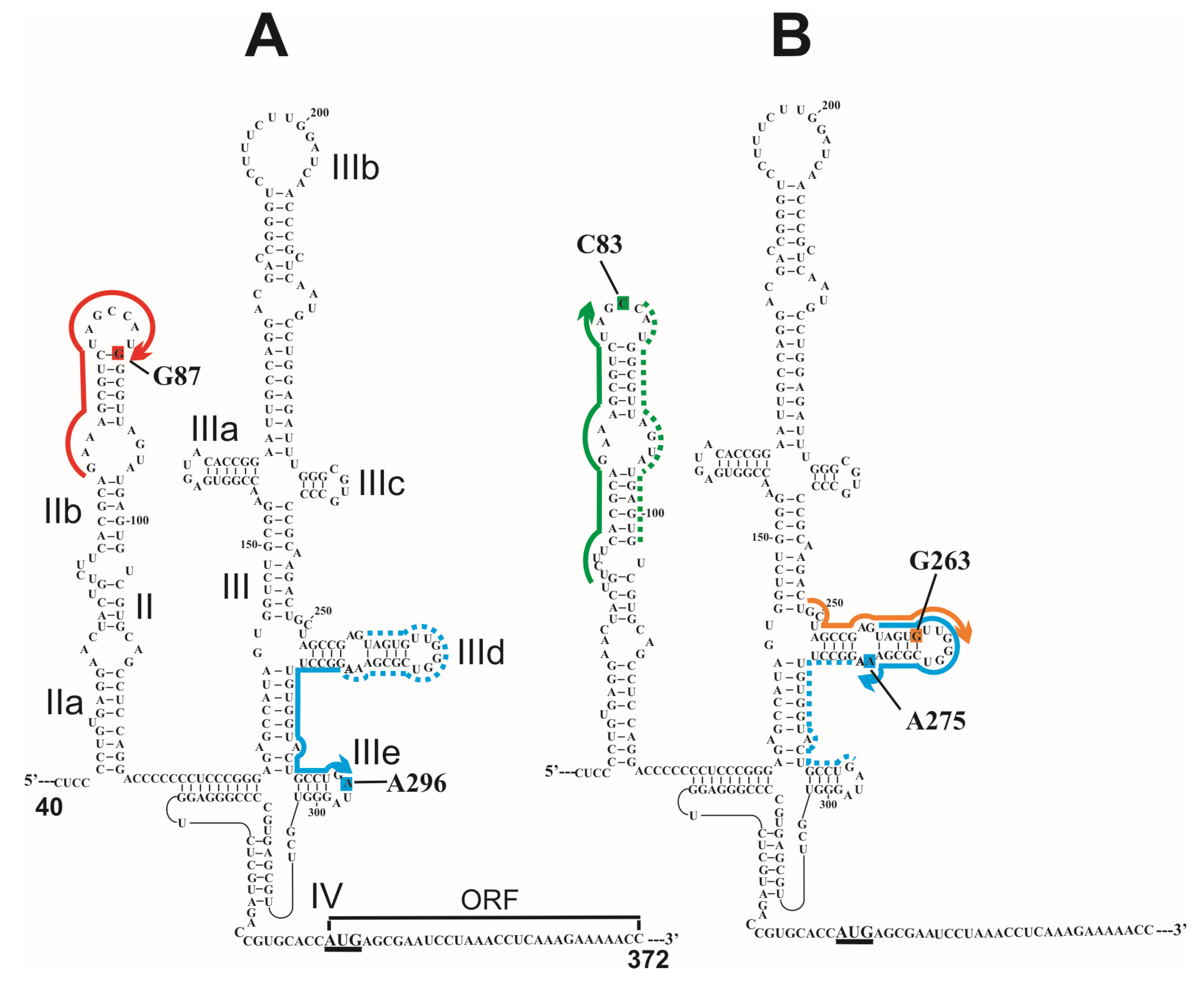
8. Coupling of Amino Linker with Reporter Groups and Application of Derivatized RNAs
9. Conclusions
Acknowledgments
Conflicts of Interest
References
- Muller, S. Synthesis of modified RNA for structure-function studies. Chim. Oggi 2012, 30, 42–46. [Google Scholar]
- Moore, M.; Query, C.C. Joining of RNAs by splinted ligation. Meth. Enzymol. 2000, 317, 109–123. [Google Scholar]
- Solomatin, S.; Herschlag, D. Methods of site-specific labeling of RNA with fluorescent dyes. Meth. Enzymol. 2009, 469, 47–68. [Google Scholar]
- Jayaprakash, K.N.; Peng, C.G.; Butler, D.; Varghese, J.P.; Maier, M.A.; Rajeev, K.G.; Manoharan, M. Non-nucleoside building blocks for copper-assisted and copper-free click chemistry for the efficient synthesis of RNA conjugates. Org. Lett. 2010, 12, 5410–5413. [Google Scholar] [CrossRef]
- Fauster, K.; Hartl, M.; Santner, T.; Aigner, M.; Kreutz, C.; Bister, K.; Ennifar, E.; Micura, R. 2-Azido RNA, a versatile tool for chemical biology: Synthesis, X-ray structure, siRNA applications, click labeling. ACS Chem. Biol. 2012, 7, 581–589. [Google Scholar] [CrossRef]
- El-Sagheer, A.H.; Brown, T. Click nucleic acid ligation: Applications in biology and nanotechnology. Acc. Chem. Res. 2012, 45, 1258–1267. [Google Scholar] [CrossRef]
- Sergiev, P.V.; Lavrik, I.N.; Wlasoff, V.A.; Dokudovskaya, S.S.; Dontsova, O.A.; Bogdanov, A.A.; Brimacombe, R. The path of mRNA through the bacterial ribosome: A site-directed crosslinking study using new photoreactive derivatives of guanosine and uridine. RNA 1997, 3, 464–475. [Google Scholar]
- Bulygin, K.N.; Graifer, D.M.; Repkova, M.N.; Smolenskaja, I.A.; Veniyaminova, A.G.; Karpova, G.G. Nucleotide G1207 of 18S rRNA is an essential component of the human 80S ribosomal decoding center. RNA 1997, 3, 1480–1485. [Google Scholar]
- Graifer, D.M.; Babkina, G.T.; Matasova, N.B.; Vladimirov, S.N.; Karpova, G.G.; Vlassov, V.V. Structural arrangement of tRNA binding site on Escherichia coli ribosomes, as revealed from data on affinity labelling with photoactivatable tRNA derivatives. Biochim. Biophys. Acta 1989, 1008, 146–156. [Google Scholar]
- demeshkina, N.; Laletina, E.; Meschaninova, M.; Ven’yaminova, A.; Graifer, D.; Karpova, G. Positioning of mRNA codons with respect to 18S rRNA at the P and E sites of human ribosome. Biochim. Biophys. Acta 2003, 1627, 39–46. [Google Scholar]
- Belikova, A.M.; Zarytova, V.F.; Grineva, N.I. Synthesis of ribonucleosides and diribonucleoside phosphates containing 2-chloroethylamine and nitrogen mustard residues. Tetrahedron Lett. 1967, 37, 3557–3562. [Google Scholar] [CrossRef]
- Knorre, D.G.; Vlassov, V.V. Complementary-addressed (sequence specific) modification of nucleic acids. Prog. Nucleic Acids Res. Mol. Biol. 1985, 32, 291–320. [Google Scholar] [CrossRef]
- Gimautdinova, O.I.; Gorshkova, I.I.; Karpova, G.G.; Kutiavin, I.V.; Graifer, D.M. Selective alkylation of G24 residue in tRNAPhe. Mol. Biol. 1984, 18, 1419–1423. [Google Scholar]
- Gimautdinova, O.I.; Gorn, V.V.; Gorshkova, I.I.; Graifer, D.M.; Karpova, G.G. Alkylation of tRNAPhe with 4-(N-2-chloroethyl-N-methylamino)benzyl-5'-phosphamide of d(ATTTTCA). Bioorg. Khim. 1986, 12, 490–498. [Google Scholar]
- Venkstern, T.V.; Graifer, D.M.; Karpova, G.G.; Morozov, I.A. Studies in the interaction of the tRNAPhe derivative bearing an arylazide group on its G24 residue with Escherichia coli ribosomes and tRNA-(adenine-l-)methyl transferase from Thermus thermophilus. Biopolym. Cell 1990, 6, 59–65. [Google Scholar]
- Zenkova, M.; Ehresmann, C.; Caillet, J.; Springer, M.; Karpova, G.G.; Ehresmann, B.; Romby, P. A novel approach to introduce site-directed specific cross-links within RNA-protein complexes. Eur. J. Biochem. 1995, 231, 726–735. [Google Scholar] [CrossRef]
- Laletina, E.; Graifer, D.; Malygin, A.; Ivanov, A.; Shatsky, I.; Karpova, G. Proteins surrounding hairpin IIIe of the hepatitis C virus internal ribosome entry site on the human 40S ribosomal subunit. Nucleic Acids Res. 2006, 34, 2027–2036. [Google Scholar] [CrossRef]
- Babaylova, E.; Graifer, D.; Malygin, A.; Stahl, J.; Shatsky, I.; Karpova, G. Positioning of subdomain IIId and apical loop of domain II of the hepatitis C IRES on the human 40S ribosome. Nucleic Acids Res. 2009, 37, 1141–1151. [Google Scholar]
- Venyaminova, A.G.; Repkova, M.N.; Ivanova, T.M.; Dobrikov, M.I.; Bulygin, K.N.; Graifer, D.M.; Karpova, G.G.; Zarytova, V.F. New photoreactive mRNA analogues for the affinity labeling of ribosomes. Nucleos. Nucleot. 1995, 14, 1069–1072. [Google Scholar]
- Allerson, C.R.; Chen, S.L.; Verdine, G.L. A chemical method for site-specific modification of RNA: The convertible nucleoside approach. J. Am. Chem. Soc. 1997, 119, 7423–7433. [Google Scholar] [CrossRef]
- Repkova, M.N.; Ivanova, T.M.; Komarova, N.I.; Meschaninova, M.I.; Kuznetsova, M.A.; Ven’yaminova, A.G. H-Phosphonate synthesis of oligoribonucleotides containing modified bases. I. Photoactivatable derivatives of oligoribonucleotides with perfluoroarylazide groups in heterocyclic bases. Russ. J. Bioorg. Chem. 1999, 25, 612–622. [Google Scholar]
- Repkova, M.N.; Meshchaninova, M.I.; Ivanova, T.M.; Komarova, N.I.; Pyshnyi, D.V.; Venyaminova, A.G. Oligoribonucleotides containing an aminoalkyl group at the N(4) atom of cytosine as precursors of new reagents for site-specific modifications of biopolymers. Russ. Chem. Bull. Int. Ed. 2002, 51, 1194–1197. [Google Scholar] [CrossRef]
- Repkova, M.; Meshchaninova, M.; Pyshnyi, D.; Venyaminova, A. Oligoribonucleotides with functionalized nucleobases as new modifiers of biopolymers. Nucleos. Nucleot. Nucleic Acids 2003, 22, 1509–1512. [Google Scholar] [CrossRef]
- Bulygin, K.N.; Repkova, M.N.; Ven’yaminova, A.G.; Graifer, D.M.; Karpova, G.G.; Frolova, L.Y.; Kisselev, L.L. Positioning of the mRNA stop signal with respect to polypeptide chain release factors and ribosomal proteins in 80S ribosomes. FEBS Lett. 2002, 514, 96–101. [Google Scholar] [CrossRef]
- Wu, X.; Pitsch, S. Functionalization of the sugar moiety of oligoribonucleotides on solid support. Bioconjug. Chem. 1999, 10, 921–924. [Google Scholar] [CrossRef]
- Smalley, M.K.; Silverman, S.K. Fluorescence of covalently attached pyrene as a general RNA folding probe. Nucleic Acids Res. 2006, 34, 152–166. [Google Scholar] [CrossRef]
- Bramsen, J.B.; Laursen, M.B.; Nielsen, A.F.; Hansen, T.B.; Bus, C.; Langkjaer, N.; Babu, B.R.; Højland, T.; Abramov, M.; van Aerschot, A. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009, 37, 2867–2881. [Google Scholar] [CrossRef]
- Herdewijn, P. Heterocyclic modifications of oligonucleotides and antisense technology. Antisense Nucleic Acid Drug. Dev. 2000, 10, 297–310. [Google Scholar] [CrossRef]
- Kojima, N.; Komatsu, Y. Synthesis and application of highly reactive amino linkers for functional oligonucleotides. Curr. Protoc. Nucleic Acid Chem. 2012. [Google Scholar] [CrossRef]
- Wachowius, F.; Höbartner, C. Chemical RNA modifications for studies of RNA structure and dynamics. ChemBioChem 2010, 11, 469–480. [Google Scholar] [CrossRef]
- Schoch, J.; Ameta, S.; Jäschke, A. Inverse electron-demand Diels–Alder reactions for the selective and efficient labeling of RNA. Chem. Commun. 2011, 47, 12536–12537. [Google Scholar] [CrossRef]
- Moore, M.J.; Sharp, P.A. Site-specific modification of pre-mRNA: The 2'-hydroxyl groups at the splice sites. Science 1992, 256, 992–997. [Google Scholar]
- Silverman, S.K. Deoxyribozymes: Selection design and serendipity in the development of DNA catalysts. Acc. Chem. Res. 2009, 42, 1521–1531. [Google Scholar] [CrossRef]
- Sergiev, P.; Dokudovskaya, S.; Romanova, E.; Topin, A.; Bogdanov, A.; Brimacombe, R.; Dontsova, O. The environment of 5S rRNA in the ribosome: Cross-links to the GTPase-associated area of 23S rRNA. Nucleic Acids Res. 1998, 26, 2519–2525. [Google Scholar]
- Kossinova, O.; Malygin, A.; Krol, A.; Karpova, G. A novel insight into the mechanism of mammalian selenoprotein synthesis. RNA 2013, 19, 1147–1158. [Google Scholar] [CrossRef]
- Bulygin, K.N.; Matasova, N.B.; Graifer, D.M.; Veniyaminova, A.G.; Yamkovoy, V.I.; Stahl, J.; Karpova, G.G. Protein environment of mRNA at the decoding site of 80S ribosomes from human placenta as revealed from affinity labeling with mRNA analogs—derivatives of oligoribonucleotides. Biochim. Biophys. Acta 1997, 1351, 325–332. [Google Scholar]
- Gimautdinova, O.I.; Karpova, G.G.; Knorre, D.G.; Kobetz, N.D. The proteins of the messenger RNA binding site of Escherichia coli ribosomes. Nucleic Acids Res. 1981, 9, 3465–3481. [Google Scholar] [CrossRef]
- Vladimirov, S.N.; Babkina, G.T.; Venijaminova, A.G.; Gimautdinova, O.I.; Zenkova, M.A.; Karpova, G.G. Structural arrangement of the decoding site of Escherichia coli ribosomes as revealed from the data on affinity labelling of ribosomes by analogs of mRNA—derivatives of oligoribonucleotides. Biochim. Biophys. Acta 1990, 1048, 245–256. [Google Scholar]
- Graifer, D.M.; Zenkova, M.A.; Malygin, A.A.; Mamaev, S.V.; Mundus, D.A.; Karpova, G.G. Identification of a site on 18S rRNA of human placenta ribosomes in the region of the mRNA binding centre. J. Mol. Biol. 1990, 214, 121–128. [Google Scholar] [CrossRef]
- Malygin, A.A.; Graifer, D.M.; Bulygin, K.N.; Zenkova, M.A.; Yamkovoy, V.I.; Stahl, J.; Karpova, G.G. Arrangement of mRNA at the decoding site of human ribosomes. 18S rRNA nucleotides and ribosomal proteins cross-linked to oligouridylate derivatives with alkylating groups at either the 3'- or the 5'-termini. Eur. J. Biochem. 1994, 226, 715–723. [Google Scholar] [CrossRef]
- Vlasov, V.V.; Lavrik, O.I.; Mamaev, S.V.; Khodyreva, S.N.; Chizhikov, V.E. Chemical modification of phenylalanyl-tRNA synthetase and ribosomes of Escherichia coli with derivatives of tRNA-Phe carrying photoreactive groups on guanosine residues. Mol. Biol. 1980, 14, 531–538. [Google Scholar]
- Knorre, D.G.; Vlassov, V.V.; Zarytova, V.F.; Karpova, G.G. Nucleotide and Oligonucleotide Derivatives as Enzymes and Nucleic Acids Targeted Irreversible Inhibitors. In Advances in Enzyme Regulation; Weber, G., Ed.; Pergamon Press: Oxford, UK, 1986; pp. 277–300. [Google Scholar]
- Bulygin, K.; Malygin, A.; Karpova, G.; Westermann, P. Site-specific modification of 4.5S RNA apical domain by complementary oligodeoxynucleotides carrying an alkylating group. Eur. J. Biochem. 1998, 251, 175–180. [Google Scholar]
- Malygin, A.A.; Karpova, G.G.; Westermann, P. Hybridization of two oligodeoxynucleotides to both strands of an RNA hairpin structure increases the efficiency of an RNA-DNA duplex formation. FEBS Lett. 1996, 392, 114–116. [Google Scholar] [CrossRef]
- Hellen, C.U.T.; Sarnow, P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001, 15, 1593–1612. [Google Scholar] [CrossRef]
- Graifer, D.; Karpova, G. Structural and functional topography of the human ribosome. Acta Biochim. Biophys. Sin. 2012, 44, 281–299. [Google Scholar] [CrossRef]
- Graifer, D.; Karpova, G. Photoactivatable RNA derivatives as tools for studying the structural and functional organization of complex cellular ribonucleoprotein machineries. RSC Adv. 2013, 3, 2858–2872. [Google Scholar] [CrossRef]
- Dittmar, K.A.; Goodenbour, J.M.; Pan, T. Tissue-specific differences in human transfer RNA expression. PloS Genet. 2006, 2, 2107–2115. [Google Scholar]
- Lee, T.-H.; Lapidus, L.J.; Zhao, W.; Travers, K.J.; Herschlag, D.; Chu, S. Measuring the folding transition time of single RNA molecules. Biophys. J. 2007, 92, 3275–3283. [Google Scholar] [CrossRef]
- Sattin, B.D.; Zhao, W.; Travers, K.; Chu, S.; Herschlag, D. Direct measurement of tertiary contact cooperativity in RNA folding. J. Am. Chem. Soc. 2008, 130, 6085–6087. [Google Scholar] [CrossRef]
- Jiao, Y.; Riechmann, J.L.; Meyerowitz, E.M. Transcriptome-wide analysis of uncapped mRNAs in arabidopsis reveals regulation of mRNA degradation. Plant Cell 2008, 20, 2571–2585. [Google Scholar] [CrossRef]
- Lemay, J.-F.; Penedo, J.C.; Tremblay, R.; Lilley, D.M.J.; Lafontaine, D.A. Folding of the adenine riboswitch. Chem. Biol. 2006, 13, 857–868. [Google Scholar] [CrossRef]
- Soulière, M.F.; Altman, R.B.; Schwarz, V.; Haller, A.; Blanchard, S.C.; Micura, R. Tuning a riboswitch response through structural extension of a pseudoknot. Proc. Natl. Acad. Sci. USA 2013, 110, E3256–E3264. [Google Scholar]
- Crawford, D.J.; Hoskins, A.A.; Friedmanc, L.J.; Gelles, J.; Moore, M.J. Single-molecule colocalization FRET evidence that spliceosome activation precedes stable approach of 5'-splice site and branch site. Proc. Natl. Acad. Sci. USA 2013, 110, 6783–6788. [Google Scholar]
- Graifer, D.; Karpova, G. Structural Aspects of Ribosomal Ligands Interactions Providing the Work of the Human Translational Machinery. In Ribosomes; Zhou, L., Wang, L., Eds.; Nova Science Publishers: Hauppauge, NY, USA, 2013; pp. 1–29. [Google Scholar]
- Sergiev, P.; Leonov, A.; Dokudovskaya, S.; Shpanchenko, O.; Dontsova, O.; Bogdanov, A.; Rinke-Appel, J.; Mueller, F.; Osswald, M.; von Knoblauch, K.; et al. Correlating the X-ray structures for halo- and thermophilic ribosomal subunits with biochemical data for the Escherichia coli ribosome. Cold Spring Harb. Symp. Quant. Biol. 2001, 66, 87–100. [Google Scholar] [CrossRef]
- Malygin, А.А.; Graifer, D.М.; Laletina, Е.S.; Shatsky, I.N.; Karpova, G.G. An approach to identify the functionally important RNA sites by complementary addressed modification. Mol. Biol. 2003, 37, 873–879. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Graifer, D.; Karpova, G. General Approach for Introduction of Various Chemical Labels in Specific RNA Locations Based on Insertion of Amino Linkers. Molecules 2013, 18, 14455-14469. https://doi.org/10.3390/molecules181214455
Graifer D, Karpova G. General Approach for Introduction of Various Chemical Labels in Specific RNA Locations Based on Insertion of Amino Linkers. Molecules. 2013; 18(12):14455-14469. https://doi.org/10.3390/molecules181214455
Chicago/Turabian StyleGraifer, Dmitri, and Galina Karpova. 2013. "General Approach for Introduction of Various Chemical Labels in Specific RNA Locations Based on Insertion of Amino Linkers" Molecules 18, no. 12: 14455-14469. https://doi.org/10.3390/molecules181214455



