Characterisation of Phenolic Compounds in South African Plum Fruits (Prunus salicina Lindl.) using HPLC Coupled with Diode-Array, Fluorescence, Mass Spectrometry and On-Line Antioxidant Detection
Abstract
:1. Introduction
| Cultivars/selections | Skin colour (ripe) | Flesh Colour (ripe) | Harvest date |
|---|---|---|---|
| Sun Breeze | Yellow | Yellow | 14 February 2012 |
| Laetitia | Red | Yellow | 8 February 2012 |
| African Delight | Red | Yellow | 12 February 2012 |
| Sapphire | Red | Yellow | 13 December 2011 |
| Ruby Red | Red | Red | 3 January 2012 |
| Ruby Crunch (PR02-62) a | Red | Red | 31 January 2012 |
| PR02-55 | Red | Red | 21 December 2010 |
| PR03-34 | Red | Red | 20 December 2011 |
| PR04-19 | Red | Red | 13 December 2011 |
| PR04-32 | Red | Red | 17 January 2012 |
| PR04-35 | Red | Red | 20 December 2011 |
2. Results and Discussion
2.1. High Performance Liquid Chromatography with Diode-Array and Fluorescence Detection (HPLC-DAD-FLD) Method Optimisation
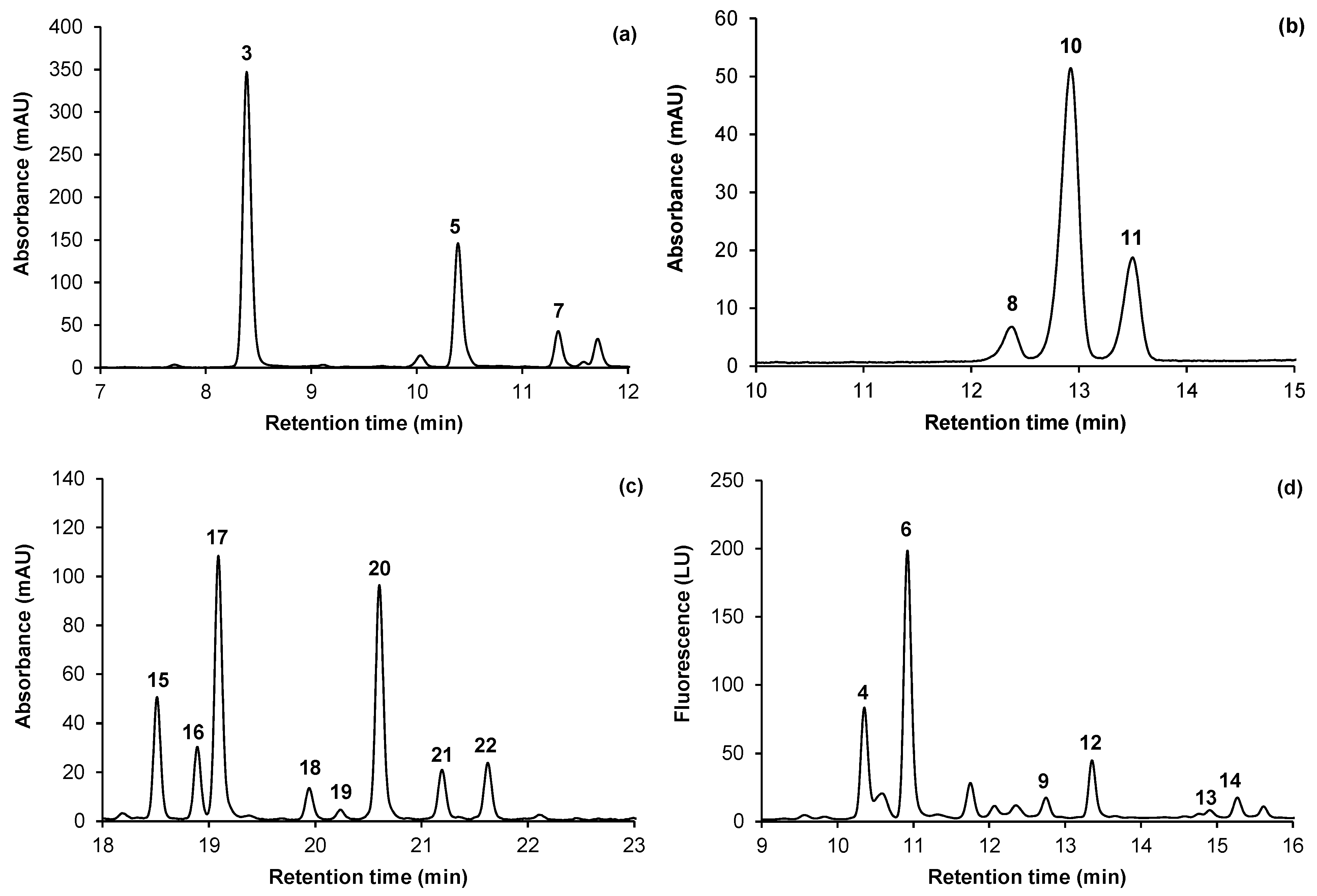
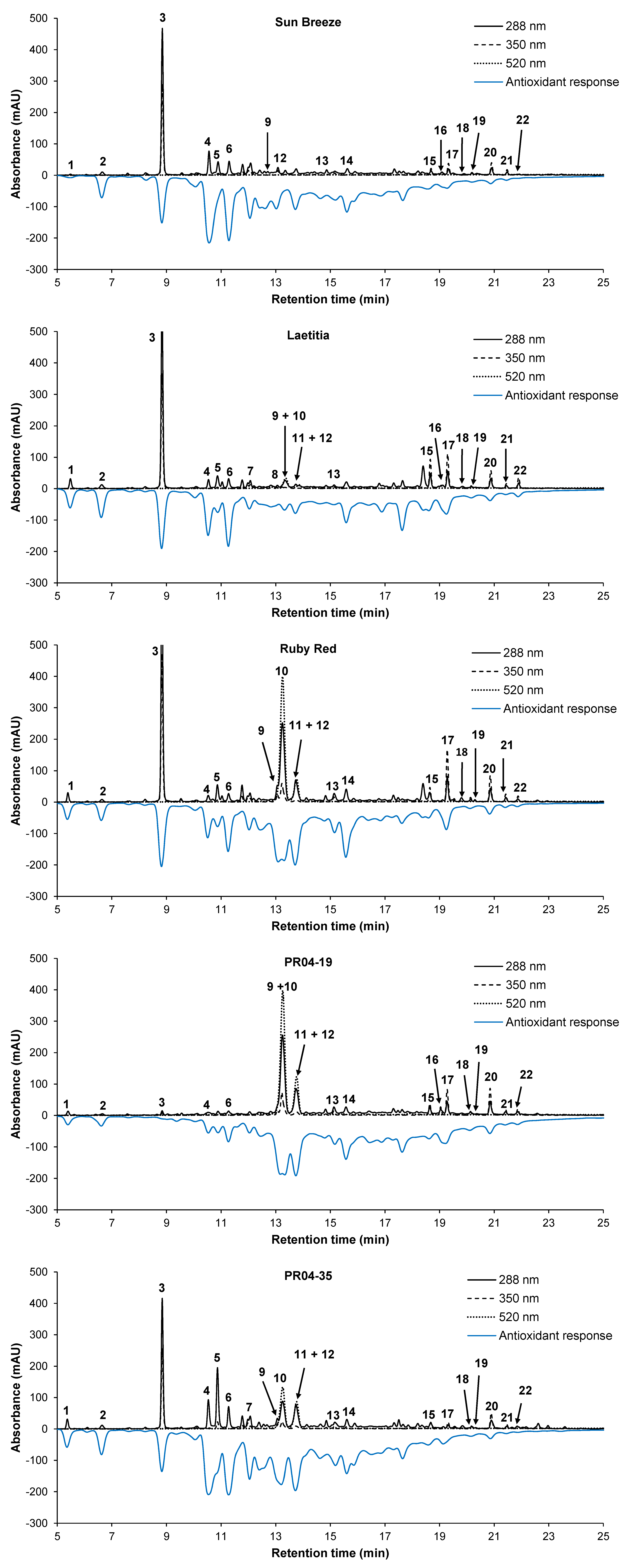
2.2. Identification and Quantification of Phenolic Compounds
| Peak no. | tR (min) a | λmax (nm) | [M+H]+/ M+ b | Na-adduct ions | Fragment ions | Identification |
|---|---|---|---|---|---|---|
| 1 | 5.5 | 275 | 353 | - | 177, 160 * | Unknown compound 1 |
| 2 | 6.6 | 279 | 867 | - | 579 *, 247 | B-type procyanidin trimer 1 |
| 3 | 8.4 | 324 | 355 | - | 163 | Neochlorogenic acid c |
| 4 | 10.3 | 278 | 579 | - | 427, 409 *, 291 | Procyanidin B1 c |
| 5 | 10.6 | 312 | 339 | - | 147 | 3 -O-p-Coumaroylquinic acid |
| 6 | 10.8 | 278 | 291 | - | 139 | (+)-Catechin c |
| 7 | 11.8 | 325 | 355 | - | 163 | Chlorogenic acid c |
| 8 | 12.4 | 276, 515 | 449 | - | 287 | Cyanidin-3 -O-galactoside c |
| 9 | 12.7 | 278 | 579 | - | - | Procyanidin B2 c |
| 10 | 12.9 | 276, 515 | 449 | - | 287 | Cyanidin-3 -O-glucoside c |
| 11 | 13.5 | 280, 515 | 595 | - | 287 | Cyanidin-3 -O-rutinoside c |
| 12 | 13.3 | 278 | 291 | - | 139 | (-)-Epicatechin c |
| 13 | 14.8 | 278 | 865 | - | 577, 425 *, 287 | A-type procyanidin trimer |
| 14 | 15.3 | 279 | 867 | - | 579 *, 409, 291, 247 | B-type procyanidin trimer 2 |
| a d | 16.8 | 278 | 577 | - | 425, 287 * | A-type procyanidin dimer 1 |
| 15 | 18.5 | 254, 352 | 611 | 633 | 465, 303 *, 229 | Quercetin-3 -O-rutinoside c |
| 16 | 18.9 | 254, 351 | 597 | 619 | 303 *, 229 | Quercetin pentosyl-hexoside |
| 17 | 19.1 | 254, 353 | 465 | - | 303 * | Quercetin-3 -O-glucoside c |
| b d | 19.1 | 278 | 577 | - | 425, 287 * | A-type procyanidin dimer 2 |
| 18 | 19.9 | 254, 351 | 435 | 891, 457 | 303 *, 229 | Quercetin-3 -O-xyloside |
| 19 | 20.1 | 254, 351 | 567 | 589 | 303 *, 229 | Quercetin pentosyl-pentoside |
| 20 | 20.6 | 255, 352 | 435 | 891, 457 | 303 *, 229 | Quercetin-3 -O-arabinoside c |
| 21 | 21.1 | 255, 347 | 449 | 919, 471 | 303 *, 229 | Quercetin-3 -O-rhamnoside c |
| 22 | 21.6 | 256, 351 | 507 | 529 | 303 *, 229 | Quercetin-acetylhexoside |

| Com-pound | Sun Breeze | Laetitia | African Delight | Sapphire | Ruby Red | Ruby Crunch | PR02-55 | PR03-34 | PR04-19 | PR04-32 | PR04-35 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phenolic acids | |||||||||||
| 3 | 218.0 | 366.6 | 391.1 | nd | 400.6 | 59.5 | 214.7 | 243.4 | 9.6 | 83.4 | 260.9 |
| 5 | 11.1 | 14.5 | nd | nd | 22.7 | nd | 94.8 | 111.0 | nd | nd | 108.0 |
| 7 | nd | nd | 24.7 | nd | nd | nd | nd | 11.3 | nd | nd | 21.6 |
| Anthocyanins | |||||||||||
| 8 | nd | 4.3 | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| 10 | nd | 47.9 | 82.1 | 222.6 | 456.8 | 221.3 | 38.3 | 238.0 | 461.8 | 84.6 | 190.9 |
| 11 | nd | 14.8 | 15.1 | 53.2 | 78.8 | 97.8 | 20.2 | nq | 145.2 | 31.8 | 114.4 |
| Flavonols | |||||||||||
| 15 | nq | 65.5 | 73.9 | 23.1 | 28.1 | 79.5 | 5.1 | 14.7 | 22.1 | 9.9 | 14.3 |
| 16 | 5.6 | 5.3 | 9.7 | 8.7 | nd | 4.3 | nd | 6.0 | 10.6 | nd | nd |
| 17 | 19.2 | 69.3 | 143.3 | 62.8 | 88.9 | 85.5 | 5.6 | 27.7 | 49.5 | 14.8 | 18.0 |
| 18 | 4.8 | 5.2 | 2.7 | 6.5 | 8.5 | 4.2 | 3.0 | 3.7 | 6.2 | 3.0 | 5.8 |
| 19 | 2.5 | 1.9 | 2.1 | 4.0 | 3.7 | nq | 2.4 | 3.0 | 1.3 | 3.8 | 3.3 |
| 20 | 18.7 | 32.8 | 12.7 | 42.4 | 48.2 | 23.8 | 21.3 | 22.3 | 38.9 | 20.2 | 36.9 |
| 21 | 9.7 | 9.9 | 3.5 | 8.7 | 16.2 | 6.7 | 5.6 | 5.4 | 9.4 | 4.0 | 12.2 |
| 22 | 2.1 | 24.8 | 20.5 | 13.4 | 13.2 | 2.8 | 4.8 | 5.7 | 11.4 | 4.6 | 5.8 |
| Flavan-3-ols | |||||||||||
| 4 | 203.7 | 100.8 | 119.8 | 87.1 | 62.5 | 271.4 | 189.6 | 200.7 | 24.3 | 218.2 | 318.4 |
| 6 | 65.6 | 61.5 | 65.5 | 71.3 | 53.9 | 171.5 | 106.2 | 114.0 | 33.6 | 172.0 | 177.0 |
| 9 | 16.1 | 7.6 | 12.4 | 53.6 | 50.0 | 32.7 | 13.4 | 14.5 | 18.9 | 101.8 | 46.7 |
| 12 | 8.3 | 10.2 | 6.3 | 58.0 | 54.6 | 41.3 | 14.8 | 20.8 | 42.3 | 93.2 | 36.3 |
2.3. Method Validation
| Compound | Calibration range (µg injected on-column) | Slope | Y-Intercept | r2 |
|---|---|---|---|---|
| Neochlorogenic | 0.03–1.60 | 2482.8 | −9.1 | 1.000 |
| Chlorogenic acid | 0.03–1.52 | 2680.0 | −16.6 | 1.000 |
| Cyanidin-3-O-glucoside | 0.07–3.57 | 1889.1 | 6.9 | 1.000 |
| Cyanidin-3-O-rutinoside | 0.04–2.00 | 1918.4 | 8.1 | 1.000 |
| Quercetin-3-O-rutinoside | 0.01–0.51 | 1622.9 | −1.4 | 1.000 |
| Quercetin-3-O-glucoside | 0.02–1.02 | 1692.1 | −4.1 | 1.000 |
| Quercetin-3-O-arabinoside | 0.20–0.98 | 1854.6 | −11.9 | 1.000 |
| Quercetin-3-O-rhamnoside | 0.02–0.99 | 1911.1 | −7.5 | 1.000 |
| (+)-Catechin | 0.20–0.99 | 1444.6 | 3.4 | 1.000 |
| (-)-Epicatechin | 0.02–1.00 | 1121.7 | 1.6 | 1.000 |
| Procyanidin B1 | 0.02–0.99 | 415.6 | 0.4 | 1.000 |
| Procyanidin B2 | 0.02–1.00 | 750.3 | −0.1 | 1.000 |
| Compound | Calibration mixture (1 µL) | Calibration mixture (30 µL) | Ruby Red | African Delight | ||||
|---|---|---|---|---|---|---|---|---|
| % RSD | % change | % RSD | % change | % RSD | % change | % RSD | % change | |
| Neochlorogenic acid | 1.1 | 2.3 | 0.1 | 0.1 | 0.3 | 0.8 | 0.3 | 1.1 |
| Chlorogenic acid | 1.0 | 0.0 | 0.1 | 0.4 | nd | nd | 1.3 | 2.8 |
| 3-O-p-Coumaroylquinic acid | n/a | n/a | n/a | n/a | 1.4 | −1.6 | nd | nd |
| Cyanidin-3-O-glucoside | 1.5 | −1.4 | 1.4 | −4.4 | 0.3 | −0.7 | 0.8 | −0.8 |
| Cyanidin-3-O-rutinoside | 2.4 | −4.3 | 1.9 | −5.7 | 0.7 | −1.6 | 2.2 | −1.3 |
| Quercetin-3-O-rutinoside | 2.7 | 6.8 | 0.1 | 0.1 | 0.2 | 0.2 | 0.3 | 1.1 |
| Quercetin pentosyl-hexoside | n/a | n/a | n/a | n/a | nd | nd | 2.1 | 1.9 |
| Quercetin-3-O-glucoside | 1.0 | 0.0 | 0.2 | 0.0 | 0.4 | 1.1 | 0.4 | 1.0 |
| Quercetin-3-O-arabinoside | 0.8 | 2.3 | 0.6 | 0.0 | 0.4 | −1.1 | 0.5 | 1.1 |
| Quercetin-3-O-rhamnoside | 0.9 | 1.3 | 0.6 | −0.1 | 0.3 | 0.8 | 1.5 | 2.5 |
| Quercetin-3-O-xyloside | n/a | n/a | n/a | n/a | 0.4 | 1.2 | 1.4 | 2.3 |
| Quercetin pentosyl-pentoside | n/a | n/a | n/a | n/a | 0.5 | 1.5 | 2.0 | −2.0 |
| Quercetin-acetylhexoside | n/a | n/a | n/a | n/a | 0.3 | 0.7 | 0.3 | 0.6 |
| (+)-Catechin | 1.3 | 0.3 | 1.3 | −0.6 | 1.1 | 2.6 | 1.1 | 2.1 |
| (-)-Epicatechin | 1.5 | 0.4 | 1.3 | −0.7 | 1.5 | 2.0 | 2.0 | 3.0 |
| Procyanidin B1 | 1.5 | 1.1 | 1.2 | −0.9 | 1.4 | 2.8 | 1.2 | 3.4 |
| Procyanidin B2 | 1.7 | 0.9 | 1.4 | −0.9 | 2.3 | 0.9 | 2.0 | 3.1 |
2.4. On-line ABTS•+ Scavenging Antioxidant Assay
3. Experimental
3.1. Chemicals
3.2. Harvesting of Fruit and Sample Preparation
3.3. High-Performance Liquid Chromatography with Diode-Array and Fluorescence Detection (HPLC-DAD-FLD)
3.4. Liquid Chromatography-Mass Spectrometry (HPLC-DAD-MS)
3.5. HPLC Method Validation
3.6. On-line Antioxidant Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Conflicts of Interest
References
- Crozier, A.; Jaganath, I.B.; Clifford, M.N. Dietary phenolics: chemistry, bioavailability and effects on health. Nat. Prod. Rep. 2009, 26, 1001–1043. [Google Scholar] [CrossRef]
- Kim, D.-O.; Chun, O.K.; Kim, Y.J.; Moon, H.-Y.; Lee, C.Y. Quantification of polyphenolics and their antioxidant capacity in fresh plums. J. Agric. Food Chem. 2003, 51, 6509–6515. [Google Scholar]
- Noratto, G.; Porter, W.; Byrne, D.D.H.; Cisneros-Zevallos, L. Identifying peach and plum polyphenols with chemopreventive potential against estrogen-independent breast cancer cells. J. Agric. Food Chem. 2009, 57, 5219–5226. [Google Scholar] [CrossRef]
- Tomás-Barberán, F.A.; Gil, M.I.; Cremin, P.; Waterhouse, A.L.; Hess-Pierce, B.; Kader, A.A. HPLC−DAD−ESIMS analysis of phenolic compounds in nectarines, peaches, and plums. J. Agric. Food Chem. 2001, 49, 4748–4760. [Google Scholar] [CrossRef]
- Fang, N.; Yu, S.; Prior, R.L. LC/MS/MS characterization of phenolic constituents in dried plums. J. Agric. Food Chem. 2002, 50, 3579–3585. [Google Scholar] [CrossRef]
- Slimestad, R.; Vangdal, E.; Brede, C. Analysis of phenolic compounds in six Norwegian plum cultivars (Prunus domestica L.). J. Agric. Food Chem. 2009, 57, 11370–11375. [Google Scholar] [CrossRef]
- Treutter, D.; Wang, D.; Farag, M.A.; Baires, G.D.A.; Rühmann, S.; Neumüller, M. Diversity of phenolic profiles in the fruit skin of Prunus domestica plums and related species. J. Agric. Food Chem. 2012, 60, 12011–12019. [Google Scholar] [CrossRef]
- Usenik, V.; Kastelec, D.; Veberič, R.; Štampar, F. Quality changes during ripening of plums (Prunus domestica L.). Food Chem. 2008, 111, 830–836. [Google Scholar] [CrossRef]
- Usenik, V.; Stampar, F.; Kastelec, D. Phytochemicals in fruits of two Prunus domestica L. plum cultivars during ripening. J. Sci. Food Agric. 2012, 93, 681–692. [Google Scholar] [CrossRef]
- Lozano, M.; Vidal-Aragón, M.C.; Hernández, M.T.; Ayuso, M.C.; Bernalte, M.J.; García, J.; Velardo, B. Physicochemical and nutritional properties and volatile constituents of six Japanese plum (Prunus salicina Lindl.) cultivars. Eur. Food Res. Technol. 2008, 228, 403–410. [Google Scholar]
- Pellegrini, N.; Del Rio, D.; Colombi, B.; Bianchi, M.; Brighenti, F. Application of the 2,2‘-azinobis(3-ethylbenzothiazoline-6-sulfonic acid) radical cation assay to a flow injection system for the evaluation of antioxidant activity of some pure compounds and beverages. J. Agric. Food Chem. 2003, 51, 260–264. [Google Scholar] [CrossRef]
- de Beer, D.; Steyn, N.; Joubert, E.; Muller, N. Enhancing the polyphenol content of a red-fleshed Japanese plum (Prunus salicina Lindl.) nectar by incorporating a polyphenol-rich extract from the skins. J. Sci. Food Agric. 2012, 92, 2741–2750. [Google Scholar] [CrossRef]
- Möller, B.; Herrmann, K. Quinic acid esters of hydroxycinnamic acids in stone and pome fruit. Phytochemistry 1983, 22, 477–481. [Google Scholar] [CrossRef]
- Nakatani, N.; Kayano, S.-I.; Kikuzaki, H.; Sumino, K.; Katagiri, K.; Mitani, T. Identification, quantitative determination, and antioxidative activities of chlorogenic acid isomers in prune (Prunus domestica L.). J. Agric. Food Chem. 2000, 48, 5512–5516. [Google Scholar] [CrossRef]
- Nunes, C.; Guyot, S.; Marnet, N.; Barros, A.S.; Saraiva, J.A.; Renard, C.M.G.C.; Coimbra, M.A. Characterization of plum procyanidins by thiolytic depolymerization. J. Agric. Food Chem. 2008, 56, 5188–5196. [Google Scholar]
- Donovan, J.L.; Meyer, A.S.; Waterhouse, A.L. Phenolic composition and antioxidant activity of prunes and prune juice (Prunus domestica). J. Agric. Food Chem. 1998, 46, 1247–1252. [Google Scholar] [CrossRef]
- Williams, B.L.; Wender, S.H. Isolation and identification of quercetin and some quercetin glycosides from plums (Prunus salicina). J. Am. Chem. Soc. 1953, 75, 4363–4364. [Google Scholar] [CrossRef]
- Barros, L.; Dueñas, M.; Carvalho, A.M.; Ferreira, I.C.F.R.; Santos-Buelga, C. Characterization of phenolic compounds in flowers of wild medicinal plants from Northeastern Portugal. Food Chem. Toxicol. 2012, 50, 1576–1582. [Google Scholar] [CrossRef]
- Slimestad, R.; Hostettmann, K. Characterisation of phenolic constituents from juvenile and mature needles of Norway spruce by means of high performance liquid chromatography-mass spectrometry. Phytochem. Anal. 1996, 7, 42–48. [Google Scholar] [CrossRef]
- Gu, L.; Kelm, M.A.; Hammerstone, J.F.; Zhang, Z.; Beecher, G.; Holden, J.; Haytowitz, D.; Prior, R.L. Liquid chromatographic/electrospray ionization mass spectrometric studies of proanthocyanidins in foods. J. Mass Spectrom. 2003, 38, 1272–1280. [Google Scholar] [CrossRef]
- De Pascual-Teresa, S.; Santos-Buelga, C.; Rivas-Gonzalo, C. Quantitative analysis of flavan-3-ols in Spanish foodstuffs and beverages. J. Agric. Food Chem. 2000, 48, 5331–5337. [Google Scholar] [CrossRef]
- Raudonis, R.; Raudone, L.; Jakstas, V.; Janulis, V. Comparative evaluation of post-column free radical scavenging and ferric reducing antioxidant power assays for screening of antioxidants in strawberries. J. Chromatogr., A 2012, 1233, 8–15. [Google Scholar] [CrossRef]
- Rzeppa, S.; Von Bargen, C.; Bittner, K.; Humpf, H.-U. Analysis of flavan-3-ols and procyanidins in food samples by reversed phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry (RP-HPLC-ESI-MS/MS). J. Agric. Food Chem. 2011, 59, 10594–10603. [Google Scholar] [CrossRef]
- Chun, O.K.; Kim, D.-O.; Moon, H.Y.; Kang, H.G.; Lee, C.Y. Contribution of individual polyphenolics to total antioxidant capacity of plums. J. Agric. Food Chem. 2003, 51, 7240–7245. [Google Scholar] [CrossRef]
- Anonymous. Agricultural Product Standards Act, Act No. 119 of 1990, G.N.R. 1983/1991; Lex Patria Publishers: Johannesburg, South Africa, 1991. [Google Scholar]
- Anonymous. Carrying temperature regimes for stone fruit. In Handling Procedure and Optimum Temperature Requirements for Sea Export of Stone Fruit; Perishable Products Export Control Board: Cape Town, South Africa, 2006. [Google Scholar]
- Pellegrini, N.; Re, R.; Yang, M.; Rice-Evans, C.A. Screening of dietary carotenoids and carotenoid-rich fruit extracts for antioxidant activities applying 2,2'-azinobis (3-ethylenebenzothiazoline-6-sulfonic acid) radical cation decolorization. Methods Enzymol. 1999, 299, 379–389. [Google Scholar] [CrossRef]
- Sample availability: Samples of the compounds are not available from the authors.
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Venter, A.; Joubert, E.; De Beer, D. Characterisation of Phenolic Compounds in South African Plum Fruits (Prunus salicina Lindl.) using HPLC Coupled with Diode-Array, Fluorescence, Mass Spectrometry and On-Line Antioxidant Detection. Molecules 2013, 18, 5072-5090. https://doi.org/10.3390/molecules18055072
Venter A, Joubert E, De Beer D. Characterisation of Phenolic Compounds in South African Plum Fruits (Prunus salicina Lindl.) using HPLC Coupled with Diode-Array, Fluorescence, Mass Spectrometry and On-Line Antioxidant Detection. Molecules. 2013; 18(5):5072-5090. https://doi.org/10.3390/molecules18055072
Chicago/Turabian StyleVenter, Alet, Elizabeth Joubert, and Dalene De Beer. 2013. "Characterisation of Phenolic Compounds in South African Plum Fruits (Prunus salicina Lindl.) using HPLC Coupled with Diode-Array, Fluorescence, Mass Spectrometry and On-Line Antioxidant Detection" Molecules 18, no. 5: 5072-5090. https://doi.org/10.3390/molecules18055072





