Changes in SeMSC, Glucosinolates and Sulforaphane Levels, and in Proteome Profile in Broccoli (Brassica oleracea var. Italica) Fertilized with Sodium Selenate
Abstract
:1. Introduction
2. Results and Discussion
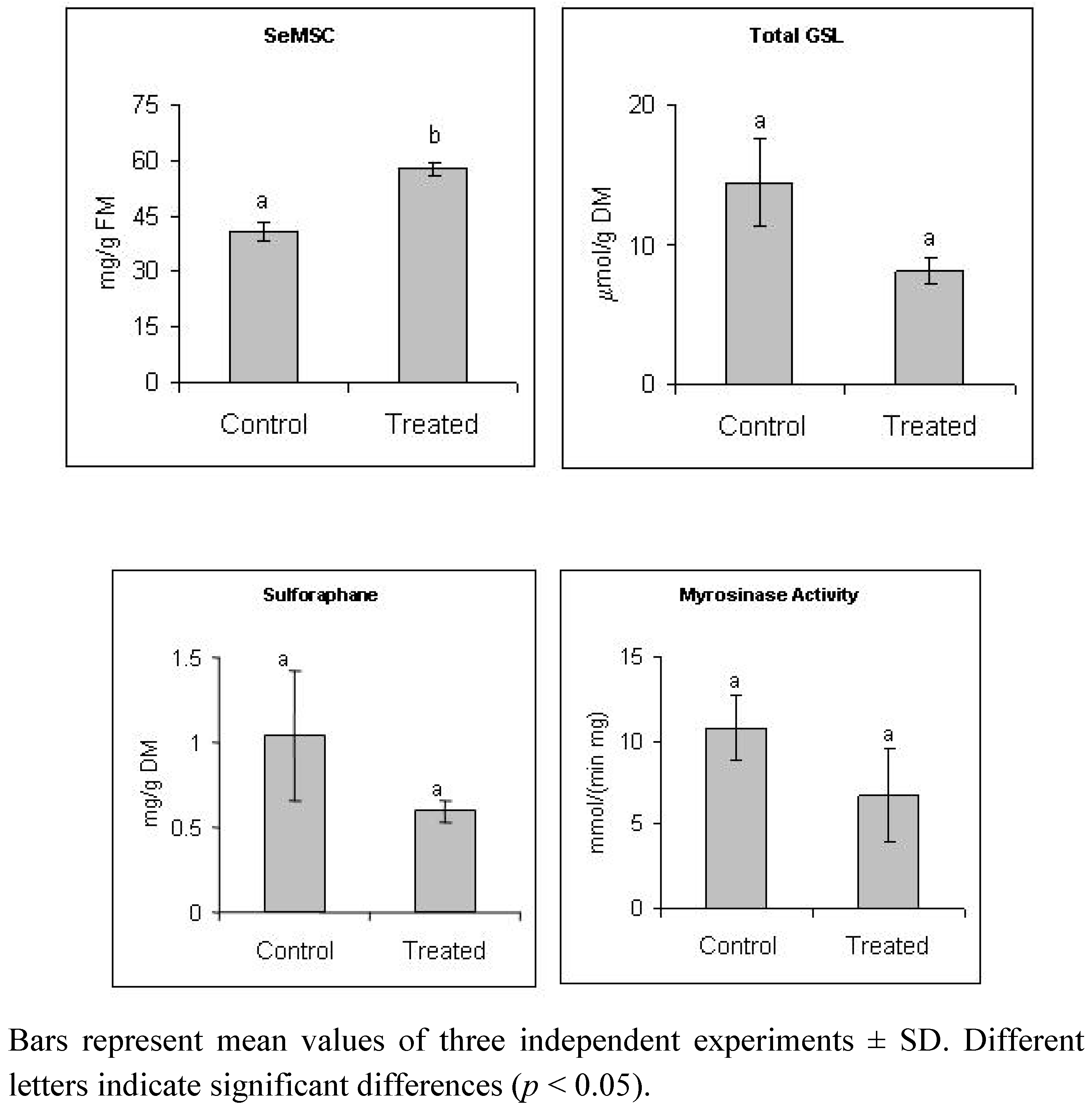
2.1. Selenate Fertilization Increases SeMSC Level But does not Affect Significantly Glucosinolates and Sulforaphane Levels in Broccoli
2.2. Selenate Fertilization Changes the Proteome Profile in Broccoli Florets
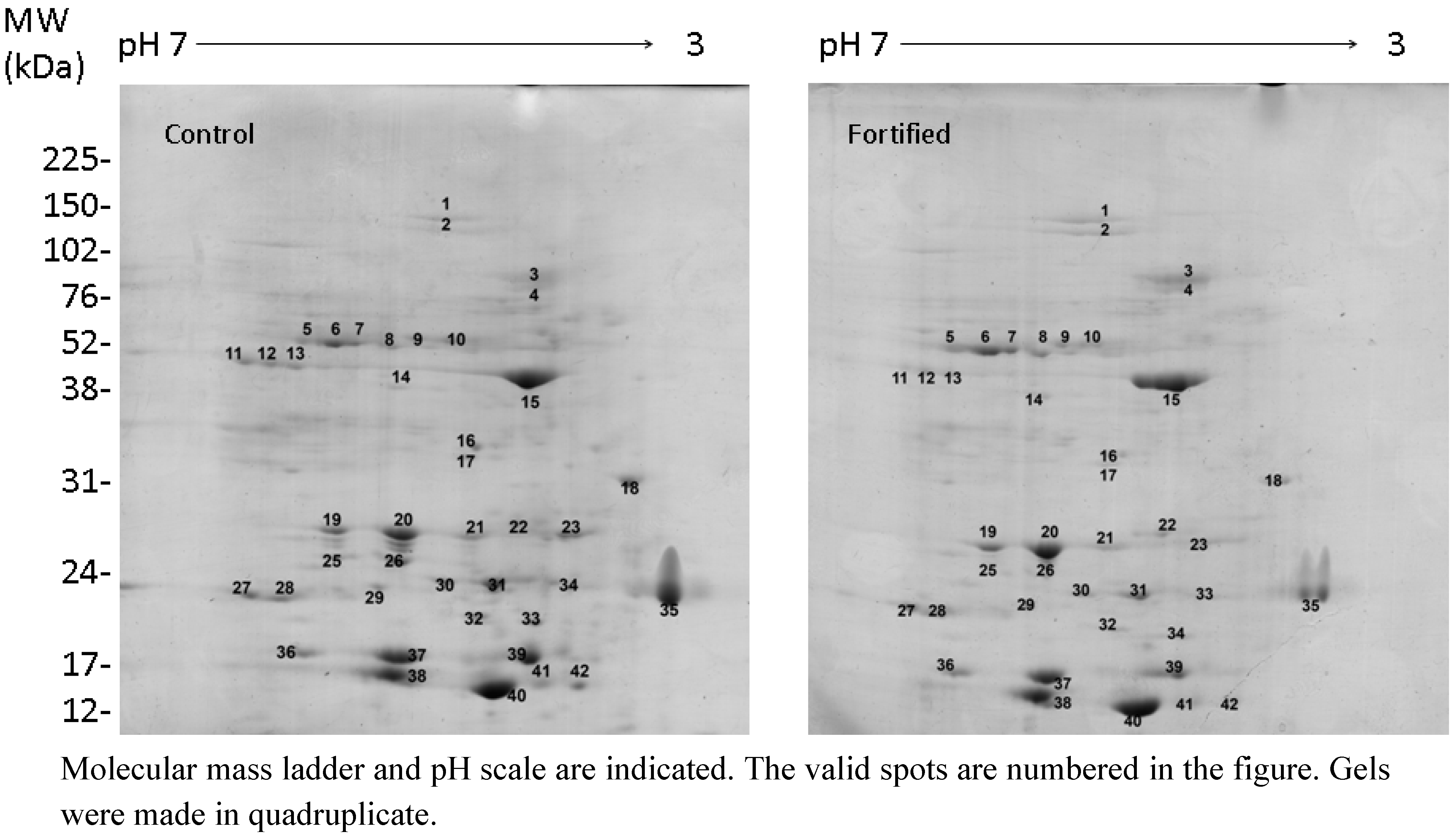
| Normalized Spot Volume | |||
|---|---|---|---|
| Spot Nr. | Control | Fortified | p-value |
| 1 | 6.3 ± 2.1 | 10.8 ± 5.4 | 0.172 |
| 2 | 5.5 ± 0.9 | 8.8 ± 2.9 | 0.073 |
| 3 * | 8.9 ± 3.1 | 32.8 ± 6.3 | 0.001 * |
| 4 | 6.1 ± 1.6 | 5.1 ± 1.6 | 0.449 |
| 5 | 9.0 ± 5.0 | 15.6 ± 2.3 | 0.055 |
| 6 | 34.3 ± 12.8 | 44.5 ± 9.8 | 0.252 |
| 7 | 13.9 ± 4.0 | 12.8 ± 4.8 | 0.727 |
| 8 | 8.2 ± 3.3 | 13.0 ± 2.5 | 0.059 |
| 9 | 10.5 ± 5.3 | 8.1 ± 2.1 | 0.437 |
| 10 * | 10.0 ± 1.1 | 14.5 ± 1.1 | 0.001 * |
| 11 | 12.0 ± 2.7 | 6.7 ± 3.6 | 0.077 |
| 12 | 10.4 ± 3.5 | 6.4 ± 2.2 | 0.105 |
| 13 | 9.7 ± 2.0 | 6.8 ± 1.8 | 0.079 |
| 14 | 3.6 ± 1.01 | 2.5 ± 1.2 | 0.296 |
| 15 * | 104.1 ± 11.0 | 132.3 ± 8.8 | 0.007 * |
| 16 | 5.8 ± 1.9 | 6.4 ± 1.7 | 0.633 |
| 17 | 4.9 ± 1.7 | 3.5 ± 0.8 | 0.206 |
| 18 * | 21.2 ± 2.1 | 12.3 ± 1.2 | 0.000 * |
| 19 | 21.8 ± 2.7 | 23.9 ± 2.3 | 0.332 |
| 20 * | 57.0 ± 8.1 | 76.1 ± 4.8 | 0.015 * |
| 21 | 18.5 ± 6.0 | 12.6 ± 2.7 | 0.123 |
| 22 | 9.8 ± 3.0 | 8.3 ± 1.7 | 0.403 |
| 23 * | 25.6 ± 0.9 | 11.4 ± 5.2 | 0.002 * |
| 24 | 5.9 ± 0.9 | 5.7 ± 0.6 | 0.397 |
| 25 | 7.9 ± 5.7 | - | - |
| 26 | 23.8 ± 11.2 | 1.8 ± 0.4 | 0.059 |
| 27 | 13.5 ± 3.5 | 16.1 ± 5.1 | 0.432 |
| 28 * | 9.5 ± 6.5 | 19.5 ± 2.3 | 0.028 * |
| 29 | 7.0 ± 1.7 | 5.6 ± 1.4 | 0.371 |
| 30 | 10.7 ± 2.4 | 9.1 ± 0.3 | 0.332 |
| 31 * | 34.9 ± 2.7 | 26.9 ± 3.9 | 0.014 * |
| 32 | 8.4 ± 3.7 | 5.2 ± 1.8 | 0.227 |
| 33 | 7.5 ± 4.6 | 5.1 ± 2.6 | 0.390 |
| 34 | 7.7 ± 4.4 | 3.9 ± 0.5 | 0.205 |
| 35 | 115.5 ± 17.1 | 93.8 ± 43.1 | 0.387 |
| 36 | 18.0 ± 1.1 | 19.2 ± 4.4 | 0.608 |
| 37 | 70.7 ± 7.6 | 60.8 ± 4.5 | 0.104 |
| 38 | 73.8 ± 8.3 | 84.4 ± 15.6 | 0.276 |
| 39 | 44.7 ± 8.6 | 38.3 ± 10.8 | 0.388 |
| 40 | 104.6 ± 22.7 | 125.8 ± 21.8 | 0.227 |
| 41 | 18.1 ± 7.6 | 15.0 ± 5.6 | 0.531 |
| 42 * | 11.2 ± 2.0 | 6.6 ± 0.1 | 0.012 * |
| SN | RC | Protein identification | Peptide sequences | Accession number | Theoretical | PS | |
|---|---|---|---|---|---|---|---|
| MW (KDa) | pI | ||||||
| 3 | + | Heat shock protein 70KDa (A. thaliana) | FEELNMDLFR | gi|9294373 | 71.3 | 5 | 640 |
| NALENYAYNMR | |||||||
| TTPSYVAFTDSER | |||||||
| ARFEELNMDLFR | |||||||
| NAVVTVPAYFNDSQR | |||||||
| GVWEGQPHADIGRIDLGTTYSCVG | |||||||
| EYQEGIFESRTYSDNQPGVLIQV | |||||||
| 10 | + | Mitochondrial F1 ATP synthase beta subunit (A. thaliana) | VVDLLAPYQR | gi|6686269 | 54.2 | 5.4 | 1060 |
| AHGGFSVFAGVGER | |||||||
| VGLTGLTVAEYFR | |||||||
| FTQANSEVSALLGR | |||||||
| LVLEVSHHLGQNVVR | |||||||
| DAEGQDVLLFIDNIFR | |||||||
| QISELGIYPAVDPLDSTSR | |||||||
| IQPESRAVGYQPTLASDLGAL | |||||||
| FHEPDTQREGLPPIMTSLEVQD | |||||||
| FHEPDTQREGLPPIMTSLEVQD | |||||||
| 15 | + | Myrosinase- binding protein (A. thaliana) | VYVGQGDSGVVYVK | gi|9279646 | 32.3 | 4.8 | 97 |
| 18 | − | β-1,3-glucanase (O. europaea) | AIETYIFAMFDENQK | gi|51507325 | 23.4 | 8.8 | 68 |
| 20 | + | Chlorophyll protein (B. oleracea) | KEPFYGGIAYK | gi|27530934 | 22.2 | 6 | 322 |
| TTAQYLILPLSPR | |||||||
| LQPLCPLGISQSSVK | |||||||
| YGWGKEVDESSSASEEPAILV | |||||||
| 23 | − | Photosystem II protein (B. oleracea) | FLVPSYR | gi|49359169 | 36.8 | 6.8 | 469 |
| EREDGIDYAAVTVQLPGGE | |||||||
| SDKTPDELGTGAEKVIGVFQSLQPS | |||||||
| 28 | + | Glutathione S-transferase (A. thaliana) | VYGPHFASPK | gi|3201613 | 24.9 | 5.6 | 121 |
| YDLYALGVGDFPVIGSKLADLAHLPFT | |||||||
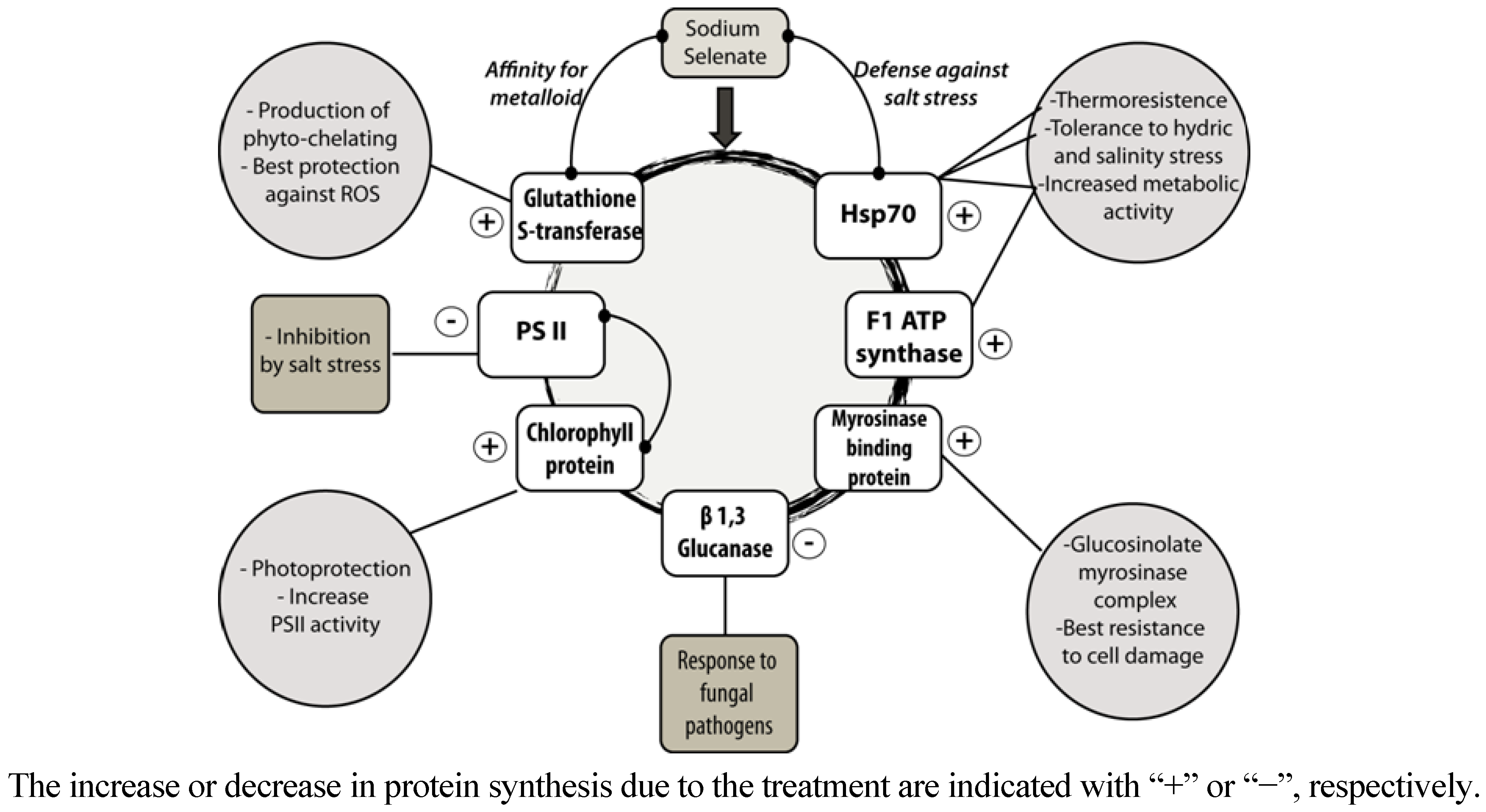
3. Experimental
3.1. Broccoli Culture
3.2. Quantification of SeMSC
3.3. Quantification of Total Glucosinolates
3.4. Quantification of Sulforaphane
3.5. Protein Extraction and Quantification of Myrosinase Activity
3.6. Protein Extraction for 2-D Gel Electrophoresis
3.7. Two-dimensional Gel Electrophoresis
3.8. Image Acquisition and Analysis
3.9. Protein Identification
3.10. Statistical Analyses
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Birringer, M.; Pilawa, S.; Flohé, L. Trends in selenium biochemistry. Nat. Prod. Rep. 2002, 6, 693–718. [Google Scholar] [CrossRef]
- Rayman, M.P. Selenium and human health. Lancet 2012, 379, 1256–1268. [Google Scholar] [CrossRef]
- Pedrero, Z.; Madrid, Y. Novel approaches for selenium speciation in foodstuffs and biological specimens: a review. Anal. Chim. Acta 2009, 634, 135–152. [Google Scholar] [CrossRef]
- Pilon-Smits, A.H.; LeDuc, D. Phytoremediation of selenium using transgenic plants. Curr. Opin. Biotechnol. 2009, 20, 207–212. [Google Scholar] [CrossRef]
- Lyi, S.M.; Heller, L.I.; Rutzke, M.; Welch, R.M.; Kochian, L.V.; Li, L. Molecular and biochemical characterization of the selenocysteine Se-methyl transferase gene and Se-methylselenocysteine synthesis in broccoli. Plant Physiol. 2005, 138, 409–420. [Google Scholar] [CrossRef]
- Davis, C.D.; Zheng, H.; Finley, J.W. Selenium-enriched broccoli decreases intestinal tumorogenesis in multiple intestinal neoplasia mice. J. Nutr. 2002, 132, 307–309. [Google Scholar]
- Finley, J.W. Reduction of cancer risk by consumption of selenium-enriched plants: Enrichment of broccoli with selenium increases the anticarcinogenic properties of broccoli. J. Med. Food 2003, 6, 19–26. [Google Scholar] [CrossRef]
- Halkier, B.A.; Gershenzon, J. Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 2006, 57, 303–333. [Google Scholar] [CrossRef]
- Mumm, M.; Burrow, M.; Bukovinszkine´kiss, G.; Kazanzidou, E.; Wittstock, U.; Dicke, M.; Gershenzon, J. Formation of simple nitriles upon glucosinolates hydrolysis affect direct and indirect defense against the specialist herbivore Pieris rapae. J. Chem. Ecol. 2008, 34, 1311–1321. [Google Scholar] [CrossRef]
- Clarke, J.D.; Dashow, R.H.; Ho, E. Multi-targeted prevention of cancer by sulforaphane. Cancer Lett. 2008, 269, 291–304. [Google Scholar] [CrossRef]
- Matich, A.J.; McKenzie, M.J.; Lill, R.E.; Brummel, D.A.; McGhie, T.K.; Chen, R.K.Y. Selenoglucosinolates and their metabolites produced in Brassica sp. fertilised with sodium selenate. Phytochemistry 2012, 75, 140–152. [Google Scholar] [CrossRef]
- Charron, C.S.; Kopsell, D.A.; Randle, W.M.; Sams, C.E. Sodium selenite fertilization increases selenium accumulation and decreases glucosinolates concentration in rapid-cycling Brassica oleraceae. J. Sci. Food Agric. 2001, 81, 962–966. [Google Scholar] [CrossRef]
- Robbins, R.J.; Keck, A.S.; Banuelos, G.; Finley, J.W. Cultivation condition and selenium fertilization alter phenolic profile, glucosinolates and sulforaphane content in broccoli. J. Med. Food 2005, 8, 204–214. [Google Scholar] [CrossRef]
- Hsu, F.C.; Wirtz, M.; Heppel, S.C.; Bogs, J.; Krämer, U.; Khan, M.S.; Bub, A.; Hell, R.; Raush, T. Generation of Se-fortified broccoli as functional food: impact of Se fertilization on S metabolism. Plant Cell Environ. 2011, 34, 192–207. [Google Scholar] [CrossRef]
- Barickman, T.C.; Kopsell, D.A.; Sams, C.E. Selenium influences glucosinolates and isothiocyanates and increases sulphur uptake in Arabidopsis thaliana and rapid-cycling Brassica oleracea. J. Agric. Food Chem. 2013, 61, 202–209. [Google Scholar] [CrossRef]
- Yao, Y.A.; Sun, H.Y.; Xu, F.S.; Zhang, X.J.; Liu, S.Y. Comparative proteomic analysis changes by low phosphorus stress in two Brassica napus genotypes. Planta 2011, 233, 523–537. [Google Scholar] [CrossRef]
- Kong, F.; Mao, S.J.; Du, K.; Wu, M.A.; Zhou, X.Y.; Chu, C.C.; Wang, Y.P. Comparative proteomic analysis of OsNAS1 transgenic Brassica napus under salt stress. Chin. Sci. Bull. 2011, 56, 2343–2350. [Google Scholar] [CrossRef]
- Wang, Z.H.; Wang, Z.F.; Chen, S.S.; Shi, L.; Xu, F.S. Proteomic reveals the adaptability mechanism of Brassica napus to short-term boron deprivation. Plant Soil 2011, 347, 195–210. [Google Scholar] [CrossRef]
- Liu, M.S.; Li, H.C.; Chang, Y.M.; Wu, M.T.; Chen, L.F.O. Proteomic analysis of stress-related proteins in transgenic broccoli harboring a gene for cytokinin production during postharvest senescence. Plant Sci. 2011, 181, 288–299. [Google Scholar] [CrossRef]
- Chan, S.; Cheng, S. The Prp19-associated complex is required for specifying interactions of U5 and U6 with Pre-mRNA during spliceosome activation. J. Biol. Chem. 2005, 280, 31190–31199. [Google Scholar] [CrossRef]
- Hubbard, M.J.; McHugh, N.J. Mitochondrial ATP synthase Fl-13-subunit is a calcium-binding protein. FEBS Lett. 1996, 391, 323–329. [Google Scholar] [CrossRef]
- Sanchez-Ballesta, M.T.; Gosalbes, M.J.; Rodrigo, M.J.; Granell, A.; Zacarias, L.; Lafuente, M.T. Characterization of a β-1,3-glucanase from citrus fruit as related to chilling-induced injury and ethylene production. Postharvets Biol. Technol. 2006, 40, 133–140. [Google Scholar]
- Morant, A.V.; Jørgensen, K.; Jørgensen, C.; Paquette, S.M.; Sánchez-Pérez, R.; Møller, B.L.; Bak, S. β-Glucosidases as detonators of plant chemical defense. Phytochemistry 2008, 69, 1795–1813. [Google Scholar]
- Martin, N.; Muller, C. Induction of plant responses by a sequestering insect: Relationship of glucosinolate concentration and myrosinase activity. Basic App. Ecol. 2007, 8, 13–25. [Google Scholar] [CrossRef]
- Damaraju, S.; Schlede, S.; Eckhardt, U.; Lokstein, H.; Grima, B. Functions of the water soluble chlorophyll-binding protein in plants. J. Plant Physiol. 2011, 168, 1444–1451. [Google Scholar] [CrossRef]
- Dixon, D.P.; Skipsey, M.; Edwards, R. Roles for glutathione transferases in plant secondary metabolism. Phytochemistry 2010, 71, 338–350. [Google Scholar]
- Liang, H.; Yuan, Q.P.; Dong, H.R.; Liu, Y.M. Determination of sulforaphane in broccoli and cabbage by high-performance liquid chromatography. J. Food Comp. Anal. 2006, 19, 473–476. [Google Scholar] [CrossRef]
- Rakariyathan, N.; Butr-Indr, B.; Niansup, H.; Shank, L. Improvement of myrosinase activity of Aspergillus sp. NR4617 by chemical mutagenesis. Open J. Biotechnol. 2006, 9, 379–385. [Google Scholar]
- Wang, W.; Scali, M.; Vignani, R.; Spadafora, A.; Sensi, E.; Mazzuca, S.; Cresti, M. Protein extraction for two-dimensional electrophoresis from olive leaf, a plant tissue containing high levels of interfering compounds. Electrophoresis 2003, 24, 2369–2375. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Mahn, A.; Ismail, M. Depletion of highly abundant proteins in blood plasma by ammonium sulfate precipitation for 2-D PAGE analysis. J. Chromathogr. B Analyt. Technol. Biomed. Life Sci. 2011, 879, 3645–3648. [Google Scholar] [CrossRef]
- Grove, H.; Hollung, K.; Uhlen, A.K.; Martens, H.; Faergestad, E.M. Challenges related to analysis of protein spot volumes from two-dimensional gel electrophoresis as revealed by replicate gels. J. Proteome Res. 2006, 5, 3399–3410. [Google Scholar] [CrossRef]
- Mahn, A.; Toledo, H.; Ruz, M. Dietary supplementation with selenomethylselenocysteine produces a differential proteomic response. J. Nutr. Biochem. 2009, 20, 791–799. [Google Scholar] [CrossRef]
- Sample Availability: Not available.
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Sepúlveda, I.; Barrientos, H.; Mahn, A.; Moenne, A. Changes in SeMSC, Glucosinolates and Sulforaphane Levels, and in Proteome Profile in Broccoli (Brassica oleracea var. Italica) Fertilized with Sodium Selenate. Molecules 2013, 18, 5221-5234. https://doi.org/10.3390/molecules18055221
Sepúlveda I, Barrientos H, Mahn A, Moenne A. Changes in SeMSC, Glucosinolates and Sulforaphane Levels, and in Proteome Profile in Broccoli (Brassica oleracea var. Italica) Fertilized with Sodium Selenate. Molecules. 2013; 18(5):5221-5234. https://doi.org/10.3390/molecules18055221
Chicago/Turabian StyleSepúlveda, Ignacio, Herna Barrientos, Andrea Mahn, and Alejandra Moenne. 2013. "Changes in SeMSC, Glucosinolates and Sulforaphane Levels, and in Proteome Profile in Broccoli (Brassica oleracea var. Italica) Fertilized with Sodium Selenate" Molecules 18, no. 5: 5221-5234. https://doi.org/10.3390/molecules18055221




