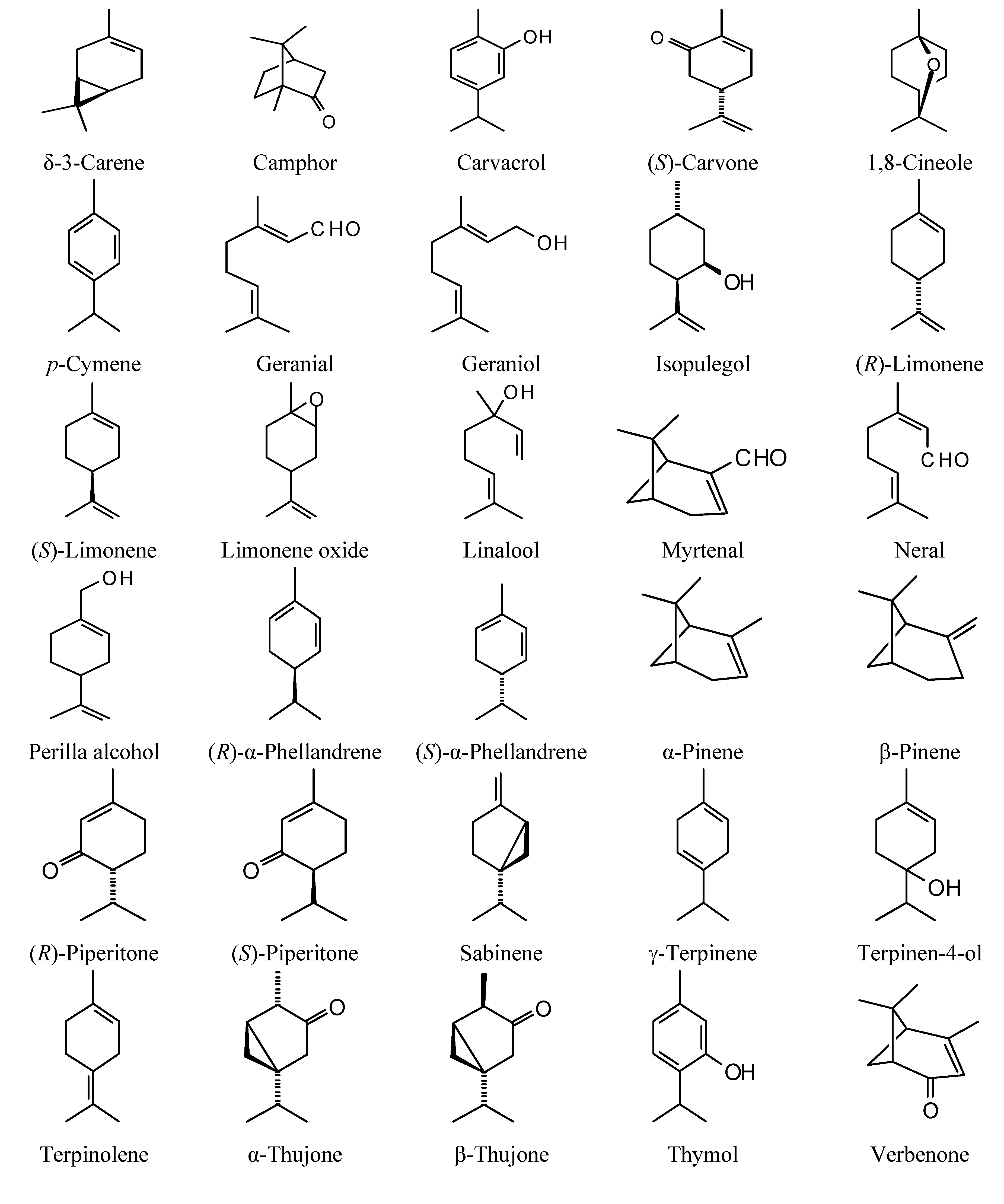
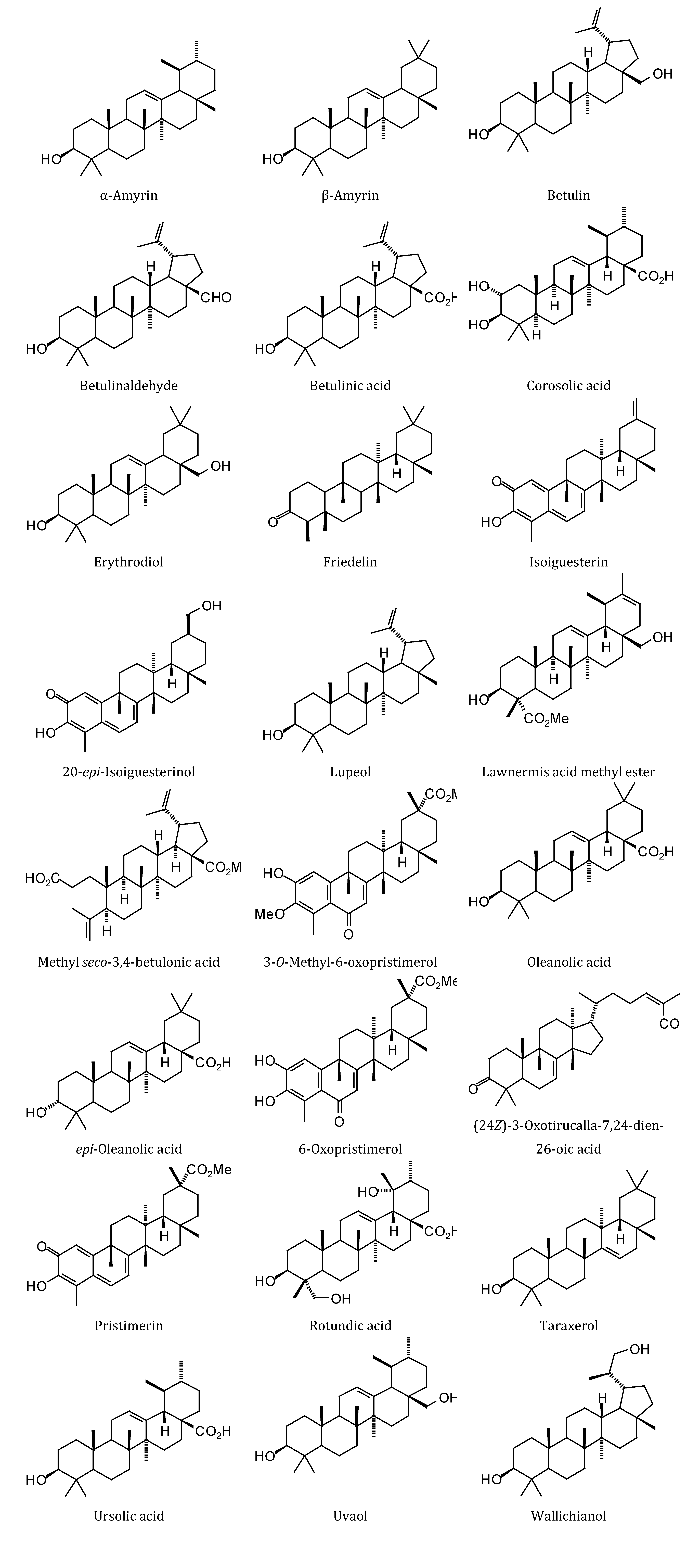
2.1. Monoterpenoid Docking
The structures of the monoterpenoids examined in this study are shown in
Figure 1, while the corresponding docking energies are summarized in
Table 1,
Table 2 and
Table 3. The overall strongest docking monoterpenoid ligands were the acyclic geranial, geraniol, and neral, probably owing to their flexibility. These ligands, however, did not show docking selectivity to any of the
Leishmania protein targets, but rather docked strongly to most of the proteins investigated. The protein targets that showed predominantly strong docking by monoterpenoids were
L. major uridine diphosphate-glucose pyrophosphorylase (LmajUGPase),
L. major methionyl t-RNA synthetase (LmajMetRS), and
L. infantum nicotinamidase (LinfPnC1). Geranial had a docking energy of −76.9 kJ/mol with LmajUGPase, comparable in docking energy with several other proteins. Both enantiomers of piperitone showed significantly stronger docking to Lmaj UGPase (−68.0 kJ/mol) than the other targets, suggesting selectivity for that protein. Geranial was also the strongest docking ligand with LmajMetRS (−76.8 kJ/mol), but perilla alcohol (−73.6 kJ/mol) was selective for that protein target. Carvone, piperitone, and α-thujone showed significantly selective docking to LinfPnC1 (docking energies less than −73 kJ/mol). Interestingly, although the monoterpenoids showed a docking propensity for LinfPnC1, higher terpenoids (sesquiterpenoids, diterpenoids, and triterpenoids) showed very little inclination to dock to this protein, generally with positive docking energies (see below).
Monoterpenoids represents a very small percentage of terpene-derived compounds that have been reported to have antileishmanial activity, and the docking energies of monoterpenoids were generally weaker than those obtained for limonoids, withanolides, triterpenoids, steroids and diterpenoids with these same targets (see below). Their docking energies were much higher than the energies obtained for the co-crystallized ligands of those protein targets. The higher docking energies of these compounds correlate with their small size (and molecular weight), and the minimal intermolecular interactions they are able to have with the protein targets. So, comparatively, it appears that monoterpenoids will not be prime leads for structure-based antileishmanial drug discovery. However, they may be useful in fragment-based drug discovery [
62,
63]. Additionally, several terpene-derived compounds are used in topical formulations. Therefore, those monoterpenoids that have antileishmanial activity and no reported toxicity at physiologically relevant concentration/dosage should be evaluated as possible components of topical polytherapy for leishmaniasis.
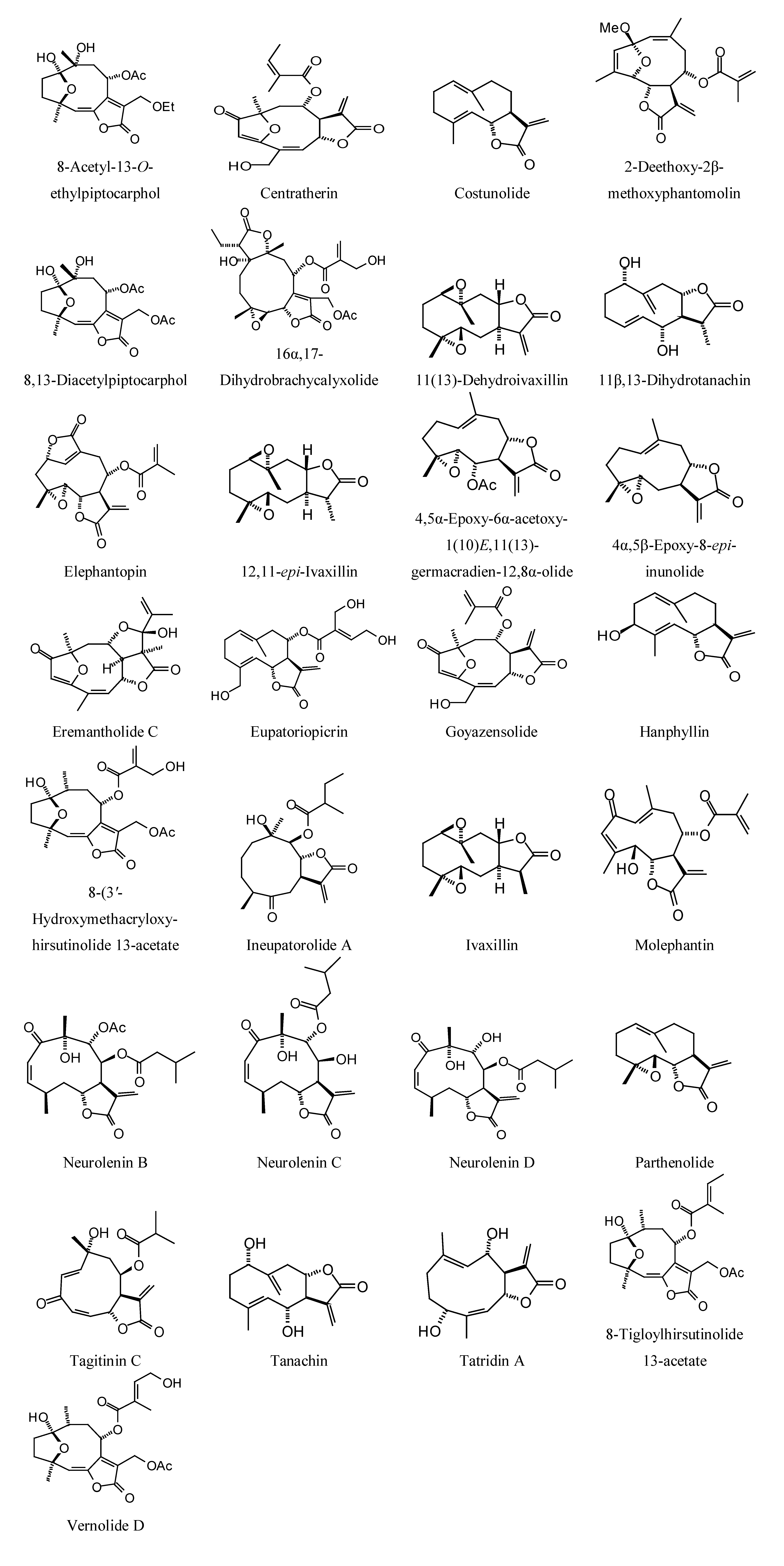
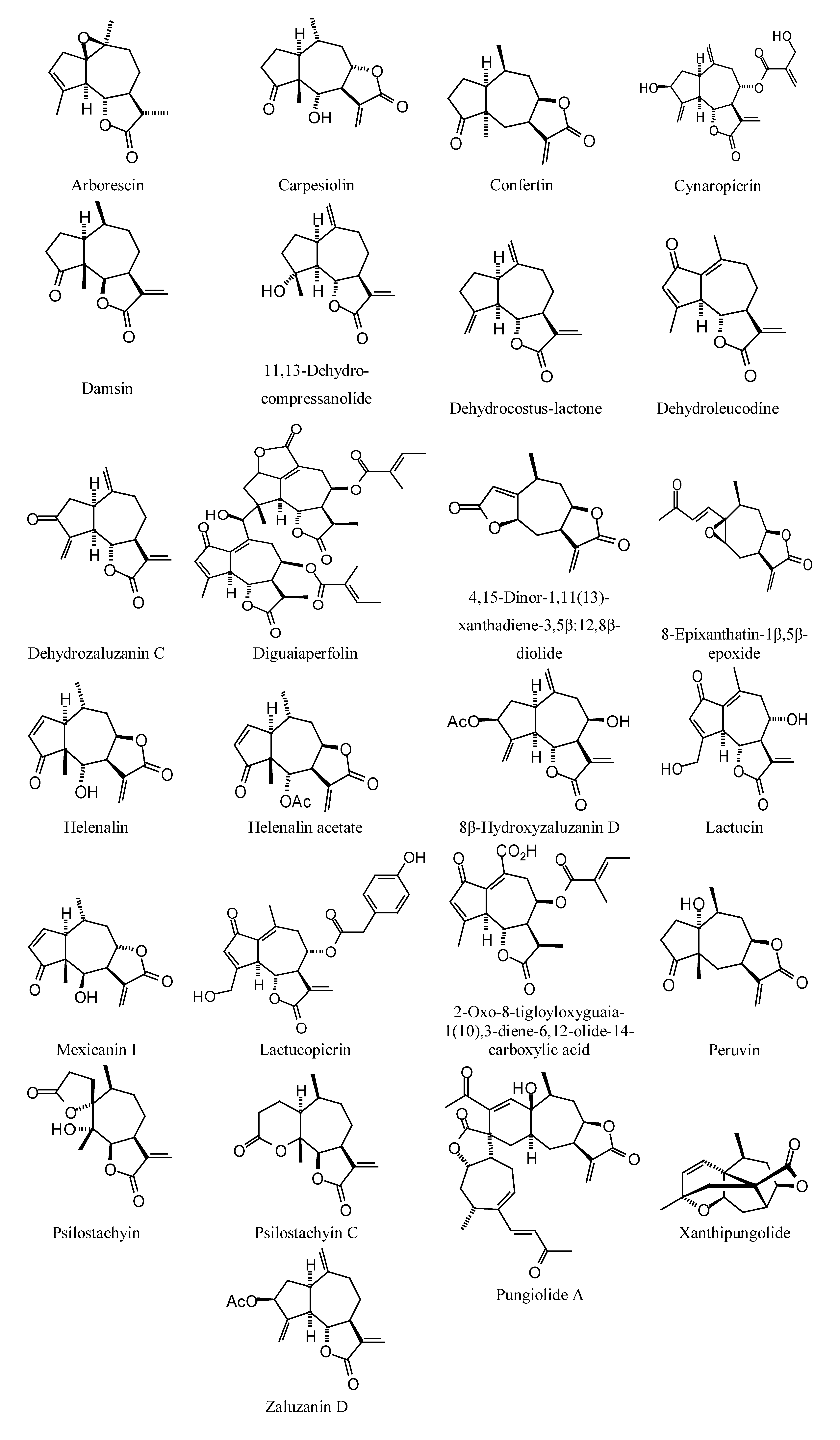
2.2. Sesquiterpenoid Docking
Sesquiterpenoids examined in this work are shown in
Figure 2,
Figure 3,
Figure 4 and
Figure 5; docking energies of sesquiterpenoids are summarized in
Table 4,
Table 5,
Table 6,
Table 7,
Table 8,
Table 9,
Table 10,
Table 11,
Table 12,
Table 13,
Table 14 and
Table 15. The germacranolide sesquiterpenoids exhibited the overall strongest docking energies toward the
Leishmania protein targets, with 16,17-dihydrobrachycalyxolide the strongest-docking germacranolide. This ligand showed docking selectivity toward LmajMetRS (docking energy = −152.9 kJ/mol) and
L. mexicana phosphor-mannomutase (LmexPMM) (docking energy = −136.3 kJ/mol). The two proteins most selectively targeted by the germacranolides in terms of docking energies were LmajMetRS and
L. major dihydroorotate dehydrogenase (LmajDHODH). Although most germacranolides did not dock with LinfPnC1, tatridin A did show docking selectivity for this protein target. Based on molecular weight, the strongest-docking germacranolide was 4α,5β-epoxy-8-
epi-inunolide, and this ligand showed docking selectivity toward both LmajMet RS and LmajDHODH.
Figure 2.
Germacranolide sesquiterpenoids examined in this work.
Figure 2.
Germacranolide sesquiterpenoids examined in this work.
Figure 3.
Guaianolide sesquiterpenoids examined in this work.
Figure 3.
Guaianolide sesquiterpenoids examined in this work.
Figure 4.
Eudesmanolide sesquiterpenoids examined in this work.
Figure 4.
Eudesmanolide sesquiterpenoids examined in this work.
Figure 5.
Miscellaneous sesquiterpenoids examined in this work.
Figure 5.
Miscellaneous sesquiterpenoids examined in this work.
Table 4.
MolDock docking energies (kJ/mol) of germacranolide sesquiterpenoids with Leishmania major protein targets.
Table 4.
MolDock docking energies (kJ/mol) of germacranolide sesquiterpenoids with Leishmania major protein targets.
| Germacranolides | LmajCatB | LmajDHODH | LmajdUTPase | LmajNDKb | LmajNH | LmajNMT | LmajOPB | LmajPDE1 | LmajPTR1 | LmajMetRS | LmajTyrRS | LmajUGPase |
|---|
| 8-Acetyl-13-O-ethylpiptocarphol | −90.8 | −113.1 | −99.0 | −87.6 | −84.0 | −108.3 | −114.1 | −98.2 | −105.3 | −127.1 | −99.1 | −105.1 |
| Centratherin | −85.4 | −115.1 | −92.2 | −95.7 | −96.6 | −106.8 | −101.2 | −102.8 | −101.5 | −125.8 | −112.9 | −108.0 |
| Costunolide | −62.3 | −86.5 | −73.8 | −71.8 | −68.1 | −79.8 | −78.4 | −75.0 | −79.9 | −75.6 | −76.3 | −81.8 |
| 2-Deethoxy-2β-methoxyphantomolin | −94.9 | −121.1 | −96.1 | −85.7 | −88.2 | −95.4 | −96.6 | −101.1 | −102.4 | −114.6 | −105.2 | −95.3 |
| 8,13-Diacetylpiptocarphol | −92.2 | −115.1 | −105.5 | −93.4 | −88.9 | −106.7 | −100.2 | −102.6 | −107.3 | −131.5 | −99.6 | −110.6 |
| 16α,17-Dihydrobrachycalyxolide | −100.1 | −129.1 | −112.5 | −117.3 | −111.0 | −121.5 | −100.2 | −122.8 | −105.0 | −152.9 | −117.7 | −115.7 |
| 11(13)-Dehydroivaxillin | −78.0 | −108.3 | −78.6 | −80.6 | −81.5 | −84.4 | −91.4 | −81.9 | −89.1 | −101.9 | −88.3 | −95.7 |
| 11β,13-Dihydrotanachin | −66.9 | −96.3 | −73.5 | −67.8 | −79.6 | −78.7 | −77.2 | −75.3 | −66.8 | −77.2 | −78.9 | −89.6 |
| Elephantopin | −86.7 | −114.6 | −98.8 | −90.3 | −91.5 | −105.0 | −97.8 | −106.8 | −103.9 | −119.3 | −105.0 | −105.7 |
| 11-epi-Ivaxillin | −76.9 | −109.9 | −80.5 | −81.7 | −83.5 | −85.1 | −94.9 | −86.9 | −90.1 | −102.4 | −80.7 | −85.9 |
| 4,5α-Epoxy-6α-acetoxy-1(10)E,11(13)-germacradien-12,8α-olide | −77.1 | −92.4 | −94.2 | −80.4 | −82.5 | −99.6 | −95.4 | −89.1 | −86.5 | −94.8 | −87.9 | −92.5 |
| 4α,5β-Epoxy-8-epi-inunolide | −74.1 | −100.5 | −76.5 | −76.8 | −80.3 | −82.5 | −86.2 | −81.0 | −88.5 | −99.1 | −84.8 | −88.2 |
| Eremantholide C | −89.2 | −103.4 | −86.5 | −100.2 | −86.2 | −96.6 | −95.3 | −98.1 | −83.1 | −100.9 | −87.2 | −101.8 |
| Eupatoriopicrin | −83.7 | −107.8 | −106.2 | −99.2 | −104.9 | −114.5 | −111.1 | −105.7 | −100.3 | −120.7 | −97.3 | −102.0 |
| Goyazensolide | −73.5 | −107.0 | −90.0 | −97.6 | −93.1 | −108.4 | −101.1 | −107.7 | −98.2 | −116.4 | −110.9 | −107.4 |
| Hanphyllin | −71.2 | −91.5 | −73.8 | −76.9 | −77.7 | −80.8 | −79.5 | −76.3 | −80.7 | −85.7 | −84.5 | −84.4 |
| 8-(3'-Hydroxymethacryloxy)-hirsutinolide 13-acetate | −94.0 | −107.3 | −106.8 | −105.6 | −104.4 | −107.8 | −105.0 | −110.7 | −109.6 | −134.9 | −105.1 | −108.6 |
| Ineupatorolide A | −83.1 | −117.3 | −90.0 | −92.4 | −91.0 | −98.7 | −100.4 | −97.2 | −112.8 | −119.4 | −112.1 | −105.9 |
| Ivaxillin | −76.6 | −104.2 | −79.3 | −78.3 | −75.2 | −83.2 | −90.6 | −82.6 | −83.4 | −98.1 | −82.8 | −88.3 |
| Molephantin | −80.8 | −105.0 | −87.4 | −80.0 | −87.4 | −101.6 | −87.9 | −99.0 | −94.7 | −106.0 | −99.3 | −91.9 |
| Neurolenin B | −86.0 | −116.4 | −88.0 | −100.0 | −88.7 | −95.6 | −77.3 | −103.0 | −86.7 | −99.8 | −90.3 | −99.5 |
| Neurolenin C | −82.7 | −116.3 | −93.0 | −94.4 | −86.2 | −102.1 | −89.6 | −95.8 | −102.6 | −112.6 | −99.0 | −117.6 |
| Neurolenin D | −90.5 | −111.1 | −95.9 | −99.2 | −92.4 | −97.5 | −87.9 | −101.4 | −94.6 | −104.7 | −90.7 | −100.6 |
| Parthenolide | −65.8 | −98.7 | −80.6 | −75.2 | −81.4 | −78.8 | −83.1 | −80.1 | −84.4 | −89.3 | −79.3 | −89.6 |
| Tagitinin C | −87.4 | −102.0 | −90.4 | −89.0 | −96.1 | −96.8 | −95.1 | −105.2 | −102.8 | −111.3 | −94.5 | −102.2 |
| Tanachin | −71.3 | −94.1 | −71.1 | −78.3 | −76.4 | −80.6 | −82.5 | −78.9 | −75.1 | −91.4 | −80.0 | −86.8 |
| Tatridin A | −74.1 | −96.6 | −69.7 | −75.7 | −76.3 | −80.7 | −88.6 | −78.4 | −89.8 | −91.2 | −80.8 | −90.7 |
| 8-Tigloylhirsutinolide 13-acetate | −93.8 | −116.4 | −105.4 | −98.0 | −99.9 | −114.6 | −110.3 | −113.6 | −113.6 | −124.8 | −105.5 | −119.9 |
| Vernolide D | −97.8 | −124.5 | −110.9 | −106.3 | −121.8 | −117.9 | −112.4 | −118.3 | −114.4 | −128.1 | −111.8 | −121.5 |
Table 5.
MolDock docking energies (kJ/mol) of germacranolide sesquiterpenoids with Leishmania donovani and L. mexicana protein targets.
Table 5.
MolDock docking energies (kJ/mol) of germacranolide sesquiterpenoids with Leishmania donovani and L. mexicana protein targets.
| Germacranolides | LdonCatB | LdonCyp | LdonDHODH | LdonNMT | LmexGAPDH | LmexGPDH | LmexPGI | LmexPMM | LmexPYK | LmexPYK | LmexPYK | LmexTIM |
|---|
| | | | | | | | | | Site 1 | Site 2 | Site 2 | |
|---|
| 8-Acetyl-13-O-ethylpiptocarphol | −92.9 | −97.6 | −79.4 | −84.2 | −93.2 | −99.2 | −88.6 | −113.3 | −102.5 | −94.9 | −99.2 | −95.1 |
| Centratherin | −89.0 | −−88.7 | −86.0 | −90.9 | −82.2 | −126.6 | −87.5 | −108.9 | −100.3 | −96.0 | −107.5 | −100.6 |
| Costunolide | −78.9 | −80.5 | −61.1 | −65.8 | −63.5 | −80.6 | −64.5 | −76.0 | −84.8 | −73.8 | −87.1 | −77.6 |
| 2-Deethoxy-2β-methoxyphantomolin | −101.6 | −88.0 | −80.7 | −87.6 | −88.7 | −110.2 | −87.4 | −110.3 | −103.1 | −85.8 | −105.8 | −79.2 |
| 8,13-Diacetylpiptocarphol | −86.4 | −99.7 | −88.0 | −92.8 | −96.2 | −102.6 | −97.4 | −118.1 | −103.0 | −100.4 | −94.7 | −100.0 |
| 16α,17-Dihydrobrachycalyxolide | −104.2 | −104.4 | −100.0 | −96.1 | −108.9 | −130.0 | −107.9 | −136.3 | −125.1 | −108.8 | −110.3 | −114.6 |
| 11(13)-Dehydroivaxillin | −78.1 | −85.0 | −64.3 | −83.7 | −72.9 | −99.4 | −68.0 | −90.6 | −91.0 | −79.8 | −106.6 | −81.8 |
| 11β,13-Dihydrotanachin | −73.4 | −77.2 | −52.3 | −75.2 | −69.6 | −81.9 | −67.2 | −79.1 | −82.3 | −74.9 | −78.1 | −66.1 |
| Elephantopin | −83.4 | −90.2 | −86.6 | −88.1 | −82.6 | −116.1 | −89.7 | −115.8 | −110.5 | −92.9 | −103.0 | −92.4 |
| 11-epi-Ivaxillin | −77.0 | −84.8 | −66.5 | −88.4 | −74.1 | −96.3 | −66.4 | −88.9 | −93.2 | −79.4 | −106.9 | −78.0 |
| 4,5α-Epoxy-6α-acetoxy-1(10)E,11(13)-germacradien-12,8α-olide | −77.0 | −90.7 | −66.4 | −82.3 | −73.0 | −98.9 | −79.5 | −91.7 | −97.8 | −88.0 | −88.3 | −81.5 |
| 4α,5β-Epoxy-8-epi-inunolide | −73.3 | −85.8 | −76.9 | −78.8 | −67.6 | −90.7 | −62.3 | −86.8 | −87.8 | −75.2 | −95.4 | −78.6 |
| Eremantholide C | −87.6 | −98.8 | −86.4 | −76.2 | −77.4 | −114.2 | −79.9 | −103.2 | −102.0 | −104.2 | −85.1 | −94.2 |
| Eupatoriopicrin | −87.0 | −94.3 | −98.3 | −96.1 | −99.5 | −116.5 | −92.5 | −102.7 | −119.4 | −103.5 | −96.0 | −97.7 |
| Goyazensolide | −86.2 | −85.7 | −83.9 | -82.3 | −87.8 | −124.1 | −88.4 | −107.9 | −99.0 | −95.9 | −103.4 | −98.7 |
| Hanphyllin | −85.4 | −85.0 | −32.6 | −67.3 | −69.8 | −81.3 | −66.5 | −80.5 | −86.6 | −75.1 | −87.5 | −83.5 |
| 8-(3'-Hydroxymethacryloxy-hirsutinolide 13-acetate | −98.6 | −98.4 | −80.0 | −98.5 | −88.4 | −113.9 | −96.1 | −117.6 | −112.4 | −101.4 | −110.7 | −103.4 |
| Ineupatorolide A | −97.4 | −95.2 | −94.2 | −86.5 | −72.5 | −102.4 | −82.2 | −99.7 | −100.1 | −91.2 | −100.4 | −104.6 |
| Ivaxillin | −73.9 | −84.9 | −74.6 | −78.3 | −77.9 | −90.4 | −68.4 | −91.3 | −93.5 | −77.9 | −105.0 | −80.7 |
| Molephantin | −87.9 | −94.5 | −36.3 | −89.4 | −79.4 | −92.9 | −78.8 | −96.6 | −106.8 | −88.8 | −99.8 | −99.9 |
| Neurolenin B | −88.1 | −82.6 | −66.6 | −76.5 | −77.7 | −98.4 | −80.1 | −110.7 | −103.3 | −92.0 | −100.4 | −85.9 |
| Neurolenin C | −93.6 | −89.8 | −49.0 | −94.7 | −86.5 | −94.4 | −79.3 | −106.0 | −95.0 | −89.9 | −91.6 | −95.2 |
| Neurolenin D | −94.8 | −78.1 | −77.6 | −95.3 | −87.1 | −94.1 | −80.8 | −111.4 | −97.7 | −89.1 | −92.4 | −92.0 |
| Parthenolide | −73.2 | −81.2 | −75.6 | −75.4 | −63.6 | −89.6 | −64.8 | −84.4 | −88.5 | −79.3 | −88.9 | −78.1 |
| Tagitinin C | −88.0 | −84.6 | −52.0 | −87.5 | −85.2 | −92.8 | -78.0 | −112.3 | −101.4 | −91.5 | −98.0 | −91.4 |
| Tanachin | −77.7 | −82.1 | −58.7 | −71.0 | −70.6 | −89.8 | −69.9 | −81.7 | −86.0 | −83.4 | −77.9 | −83.4 |
| Tatridin A | −84.7 | −84.5 | −67.6 | −72.8 | −59.3 | −85.7 | −61.6 | −83.8 | −83.5 | −81.2 | −89.7 | −76.9 |
| 8-Tigloylhirsutinolide 13-acetate | −104.5 | −83.9 | −94.8 | −97.3 | −91.0 | −110.3 | −99.2 | −117.5 | −111.3 | −98.3 | −108.0 | −102.8 |
| Vernolide D | −107.5 | −94.3 | −89.6 | −102.3 | −94.8 | −120.3 | −96.8 | −125.4 | −117.4 | −113.7 | −99.8 | −109.1 |
Table 6.
MolDock docking energies (kJ/mol) of germacranolide sesquiterpenoids with Leishmania infantum protein targets.
Table 6.
MolDock docking energies (kJ/mol) of germacranolide sesquiterpenoids with Leishmania infantum protein targets.
| Germacranolides | LinfCYP51 | LinfGLO2 | LinfPnC1 | LinfTDR1 | LinfTR |
|---|
| 8-Acetyl-13-O-ethylpiptocarphol | −100.4 | −89.8 | no dock | −82.3 | −99.7 |
| Centratherin | −109.0 | −87.1 | no dock | −88.5 | −104.9 |
| Costunolide | −74.4 | −63.6 | no dock | −69.9 | −71.2 |
| 2-Deethoxy-2β-methoxyphantomolin | −101.4 | −83.7 | no dock | −83.2 | −95.3 |
| 8,13-Diacetylpiptocarphol | −97.8 | −88.8 | no dock | −91.7 | −101.6 |
| 16α,17-Dihydrobrachycalyxolide | −126.4 | −108.8 | no dock | −104.2 | −112.9 |
| 11(13)-Dehydroivaxillin | −81.4 | −69.4 | −86.5 | −79.4 | −81.2 |
| 11β,13-Dihydrotanachin | −73.7 | −63.2 | −86.6 | −72.2 | −76.3 |
| Elephantopin | −110.7 | −90.5 | no dock | −92.2 | −104.4 |
| 11-epi-Ivaxillin | −83.4 | −73.9 | −79.8 | −77.7 | −81.2 |
| 4,5α-Epoxy-6α-acetoxy-1(10)E,11(13)-germacradien-12,8α-olide | −92.9 | −79.5 | no dock | −80.9 | −89.6 |
| 4α,5β-Epoxy-8-epi-inunolide | −77.1 | −67.0 | −80.3 | −70.2 | −77.2 |
| Eremantholide C | −91.3 | −70.7 | no dock | −81.6 | −95.3 |
| Eupatoriopicrin | −100.1 | −91.3 | no dock | −96.5 | −105.1 |
| Goyazensolide | −104.7 | −83.9 | no dock | −84.3 | −98.8 |
| Hanphyllin | −79.3 | −64.9 | −64.6 | −75.7 | −76.4 |
| 8-(3'-Hydroxymethacryloxy)-hirsutinolide 13-acetate | −120.2 | −90.6 | no dock | −100.1 | −114.7 |
| Ineupatorolide A | −102.5 | −84.2 | no dock | −89.1 | −95.0 |
| Ivaxillin | −82.2 | −70.1 | no dock | −75.0 | −84.9 |
| Molephantin | −101.0 | −82.4 | no dock | −82.3 | −101.2 |
| Neurolenin B | −95.7 | −76.0 | no dock | −81.0 | −92.5 |
| Neurolenin C | −95.9 | −76.9 | no dock | −82.0 | −93.3 |
| Neurolenin D | −99.7 | −76.0 | no dock | −80.7 | −86.9 |
| Parthenolide | −82.1 | −63.5 | −84.6 | −75.2 | −76.9 |
| Tagitinin C | −97.0 | −79.4 | no dock | −77.6 | −90.7 |
| Tanachin | −82.0 | −61.0 | −62.5 | −73.7 | −74.5 |
| Tatridin A | −87.0 | −65.2 | −97.2 | −70.9 | −77.8 |
| 8-Tigloylhirsutinolide 13-acetate | −124.6 | −88.1 | no dock | −91.5 | −109.1 |
| Vernolide D | −131.9 | −94.9 | no dock | −100.0 | −110.4 |
Table 7.
MolDock docking energies (kJ/mol) of guaianolide sesquiterpenoids with Leishmania major protein targets.
Table 7.
MolDock docking energies (kJ/mol) of guaianolide sesquiterpenoids with Leishmania major protein targets.
| Guaianolides | LmajCatB | LmajDHODH | LmajdUTPase | LmajNDKb | LmajNH | LmajNMT | LmajOPB | LmajPDE1 | LmajPTR1 | LmajMetRS | LmajTyrRS | LmajUGPase |
|---|
| Arborescin | −66.2 | −93.6 | −78.3 | −83.0 | −72.7 | −78.2 | −85.3 | −77.7 | −95.1 | −90.4 | −91.8 | −79.5 |
| Carpesiolin | −73.3 | −97.3 | −73.5 | −79.0 | −70.7 | −79.4 | −84.1 | −75.6 | −87.3 | −86.7 | −86.0 | −88.0 |
| Confertin | −62.4 | −90.0 | −72.4 | −73.5 | −75.8 | −72.7 | −81.5 | −70.1 | −74.8 | −94.4 | −78.3 | −83.7 |
| Cynaropicrin | −78.6 | −117.2 | −104.1 | −101.0 | −106.4 | −109.8 | −97.6 | −109.4 | −103.0 | −115.0 | −107.2 | −108.6 |
| Damsin | −64.9 | −102.1 | −74.1 | −77.0 | −77.4 | −83.2 | −79.4 | −78.2 | −74.8 | −89.1 | −77.5 | −85.1 |
| 11,13-Dehydrocompressanolide | −72.6 | −94.1 | −77.6 | −78.6 | −74.9 | −76.8 | −78.7 | −80.1 | −85.0 | −93.3 | −81.7 | −92.6 |
| Dehydrocostuslactone | −63.2 | −86.9 | −76.2 | −73.1 | −72.7 | −69.5 | −75.1 | −73.5 | −82.1 | −86.5 | −83.8 | −79.8 |
| Dehydroleucodine | −66.9 | −89.5 | −85.2 | −84.2 | −80.6 | −79.9 | −83.7 | −84.2 | −94.2 | −90.9 | −93.3 | −83.2 |
| Dehydrozaluzanin C | −73.3 | −90.4 | −74.9 | −78.7 | −69.8 | −79.0 | −86.3 | −77.7 | −85.4 | −92.2 | −88.4 | −85.4 |
| Diguaiaperfolin | −127.8 | −154.5 | −130.6 | −116.8 | −135.7 | −129.4 | −114.3 | −129.7 | −124.3 | −129.9 | −144.6 | −151.8 |
| 4,15-Dinor-1,11(13)-xanthadiene-3,5β:12,8β-diolide | −76.3 | −92.7 | −88.4 | −83.9 | −70.7 | −80.2 | −87.5 | −84.9 | −95.0 | −95.8 | −83.4 | −92.6 |
| 8-Epixanthatin-1β,5β-epoxide | −78.0 | −108.0 | −77.8 | −89.0 | −84.5 | −88.3 | −87.1 | −88.3 | −92.0 | −106.8 | −89.5 | −95.4 |
| Helenalin | −67.2 | −92.8 | −74.6 | −74.6 | −80.7 | −77.5 | −80.1 | −79.0 | −84.0 | −86.4 | −87.5 | −87.2 |
| Helenalin acetate | −72.4 | −88.8 | −84.1 | −81.6 | −86.2 | −88.9 | −92.6 | −91.0 | −76.8 | −98.7 | −90.3 | −89.8 |
| 8β-Hydroxyzaluzanin D | −81.2 | −102.8 | −95.6 | −87.0 | −89.7 | −91.1 | −98.7 | −90.2 | −88.3 | −94.1 | −93.9 | −98.9 |
| Lactucin | −68.8 | −100.7 | −84.2 | −91.9 | −93.4 | −87.9 | −89.7 | −91.7 | −104.1 | −92.2 | −103.6 | −89.3 |
| Lactucopicrin | −97.7 | −123.3 | −113.8 | −107.4 | −110.9 | −112.6 | −116.8 | −122.0 | −129.9 | −137.7 | −116.5 | −120.1 |
| Mexicanin I | −75.9 | −92.6 | −77.9 | −80.4 | −79.7 | −80.1 | −83.8 | −76.5 | −83.8 | −88.1 | −89.1 | −87.0 |
| 2-Oxo-8-tigloyloxyguaia-1(10),3-diene-6,12-olide-14-carboxylic acid | −82.1 | −117.0 | −99.2 | −96.9 | −95.9 | −96.7 | −90.1 | −105.0 | −108.3 | −114.1 | −103.1 | −110.8 |
| Peruvin | −62.0 | −91.6 | −71.8 | −72.0 | −78.4 | −81.3 | −79.5 | −76.2 | −79.1 | −85.4 | −78.2 | −101.9 |
| Psilostachyin | −71.8 | −89.5 | −75.0 | −83.3 | −75.7 | −78.7 | −78.1 | −79.8 | −76.7 | −90.3 | −81.7 | −87.2 |
| Psilostachyin C | −72.4 | −91.5 | −74.3 | −75.8 | −79.5 | −72.8 | −82.2 | −78.3 | −79.9 | −92.5 | −82.3 | −89.7 |
| Pungiolide A | −91.6 | −96.7 | −96.8 | −116.1 | −106.0 | −109.6 | −111.8 | −122.8 | −108.3 | −122.6 | −107.3 | −117.5 |
| Xanthipungolide | −52.4 | −78.4 | −50.0 | −55.4 | −62.7 | −64.5 | −62.6 | −66.8 | −48.3 | −69.8 | −60.4 | −74.8 |
| Zaluzanin D | −68.6 | −103.0 | −93.1 | −86.7 | −80.0 | −93.1 | −90.8 | −93.7 | −89.1 | −96.5 | −97.1 | −95.3 |
Table 8.
MolDock docking energies (kJ/mol) of guaianolide sesquiterpenoids with Leishmania donovani and L. mexicana protein targets.
Table 8.
MolDock docking energies (kJ/mol) of guaianolide sesquiterpenoids with Leishmania donovani and L. mexicana protein targets.
| Guaianolides | LdonCatB | LdonCyp | LdonDHODH | LdonNMT | LmexGAPDH | LmexGPDH | LmexPGI | LmexPMM | LmexPYK | LmexPYK | LmexPYK | LmexTIM |
|---|
| | | | | | | | | | Site 1 | Site 2 | Site 3 | |
|---|
| Arborescin | −74.7 | −82.7 | −71.6 | −73.6 | −68.3 | −−91.9 | −65.5 | −82.7 | −88.1 | −75.1 | −86.5 | −73.8 |
| Carpesiolin | −76.7 | −78.5 | −61.5 | −69.7 | −66.5 | −91.8 | −61.9 | −83.4 | −85.7 | −88.6 | −94.1 | −82.1 |
| Confertin | −67.1 | −85.2 | −12.0 | −68.1 | −62.1 | -85.1 | −61.5 | −79.0 | −85.9 | −77.1 | −82.1 | −70.8 |
| Cynaropicrin | −80.5 | −99.7 | −57.3 | −92.8 | −88.0 | −121.8 | −97.2 | −114.7 | −127.0 | −91.8 | −94.1 | −90.1 |
| Damsin | −73.3 | −88.7 | −0.9 | −73.6 | −69.4 | −85.7 | −64.6 | −79.5 | −88.1 | −79.9 | -84.8 | −68.3 |
| 11,13-Dehydrocompressanolide | −74.2 | −87.8 | −73.4 | −72.8 | −70.3 | −93.1 | −64.9 | −86.0 | −84.0 | −82.1 | −92.3 | −79.6 |
| Dehydrocostuslactone | −71.2 | −83.5 | -64.8 | −66.8 | −60.4 | −84.8 | −57.7 | −76.1 | −82.5 | −74.6 | −88.7 | −69.5 |
| Dehydroleucodine | −78.0 | −84.2 | −63.9 | −72.1 | −73.5 | −88.0 | −66.2 | −83.0 | −91.4 | −74.7 | −89.7 | −72.3 |
| Dehydrozaluzanin C | −74.4 | −86.1 | −79.0 | −76.0 | −68.3 | −91.0 | −70.8 | −82.9 | −85.4 | −80.5 | −99.5 | −77.9 |
| Diguaiaperfolin | −126.1 | −124.8 | −121.6 | −135.5 | −122.1 | −138.1 | −129.7 | −145.6 | −146.9 | −141.4 | −138.3 | −116.5 |
| 4,15-Dinor-1,11(13) -xanthadiene−3,5β:12,8β-diolide | −80.7 | −89.5 | −78.7 | −75.7 | −69.9 | −84.6 | −67.7 | −82.2 | −86.4 | −81.3 | −90.6 | −76.7 |
| 8-Epixanthatin-1β,5β-epoxide | −87.2 | −90.1 | −64.8 | −78.4 | −71.9 | −103.4 | −72.8 | −85.4 | −97.2 | −90.3 | −90.9 | −80.6 |
| Helenalin | −70.7 | −90.8 | −47.7 | −76.1 | −69.5 | −79.3 | −64.9 | −80.8 | −83.1 | −85.2 | −83.6 | −79.0 |
| Helenalin acetate | −74.2 | −77.2 | −76.9 | −74.9 | −78.5 | −105.5 | −74.7 | −87.6 | −99.3 | −86.9 | −78.5 | −79.9 |
| 8β-Hydroxyzaluzanin D | −82.3 | −85.7 | −84.0 | −84.3 | −76.7 | −101.7 | −77.6 | −101.1 | −110.6 | −82.4 | −90.4 | −79.3 |
| Lactucin | −87.0 | −87.4 | −70.5 | −72.2 | −84.2 | −94.7 | −74.0 | −92.4 | −100.7 | −87.8 | −94.2 | −79.1 |
| Lactucopicrin | −114.3 | −111.2 | −65.0 | −114.4 | −99.5 | −126.7 | −108.5 | −119.9 | −120.4 | −115.0 | −116.1 | −107.3 |
| Mexicanin I | −76.3 | −80.5 | −74.7 | −71.3 | −70.4 | −88.9 | −63.0 | −81.9 | −86.6 | −88.8 | −92.3 | −76.5 |
| 2-Oxo-8-tigloyloxyguaia-1(10),3-diene-6,12-olide-14-carboxylic acid | −97.4 | −97.0 | −91.1 | −101.5 | −81.8 | −117.3 | −82.4 | −111.8 | −118.9 | −96.8 | −115.4 | −86.6 |
| Peruvin | −66.7 | −83.6 | −64.1 | −74.6 | −62.7 | −83.0 | −61.5 | −83.0 | −88.5 | −79.1 | −83.6 | −74.1 |
| Psilostachyin | −69.6 | −85.7 | −6.0 | −73.5 | −65.8 | −91.5 | −63.4 | −85.3 | −92.7 | −78.1 | −88.8 | −70.5 |
| Psilostachyin C | −71.9 | −81.4 | −14.5 | −67.6 | −64.8 | −85.2 | −62.0 | −81.6 | −85.1 | −77.9 | −85.6 | -70.9 |
| Pungiolide A | −97.3 | −102.9 | −83.7 | −93.9 | −89.1 | −110.0 | −98.8 | −105.0 | −123.5 | −111.6 | −81.4 | −98.4 |
| Xanthipungolide | -50.4 | −63.3 | −3.8 | −55.4 | −57.3 | −75.0 | −43.1 | −67.5 | −66.9 | −57.2 | -83.7 | −54.3 |
| Zaluzanin D | −83.6 | −87.2 | −59.0 | −79.4 | −77.1 | −102.6 | −75.1 | −96.2 | −106.8 | −83.6 | −97.7 | −86.4 |
Table 9.
MolDock docking energies (kJ/mol) of guaianolide sesquiterpenoids with Leishmania infantum protein targets.
Table 9.
MolDock docking energies (kJ/mol) of guaianolide sesquiterpenoids with Leishmania infantum protein targets.
| Guaianolides | LinfCYP51 | LinfGLO2 | LinfPnC1 | LinfTDR1 | LinfTR |
|---|
| Arborescin | −79.3 | −64.1 | −59.5 | −70.7 | −95.1 |
| Carpesiolin | −81.0 | −64.2 | −65.0 | −72.2 | −82.4 |
| Confertin | −79.8 | −61.0 | −41.9 | −67.9 | −79.8 |
| Cynaropicrin | −106.2 | −85.8 | no dock | −91.6 | −105.4 |
| Damsin | −78.9 | −68.5 | −70.3 | −68.3 | −79.2 |
| 11,13-Dehydrocompressanolide | −77.9 | −66.7 | −68.8 | −74.1 | −78.2 |
| Dehydrocostuslactone | −73.5 | −61.4 | −68.8 | −69.1 | −82.4 |
| Dehydroleucodine | −79.6 | −65.7 | −72.1 | −71.5 | −92.8 |
| Dehydrozaluzanin C | −81.1 | −67.1 | −81.8 | −73.2 | −85.3 |
| Diguaiaperfolin | −141.0 | −118.8 | no dock | −118.6 | −147.5 |
| 4,15-Dinor-1,11(13)-xanthadiene-3,5β:12,8β-diolide | −79.7 | −67.8 | −70.8 | −76.8 | −84.4 |
| 8-Epixanthatin-1β,5β-epoxide | −84.2 | −77.5 | −52.1 | −80.4 | −88.8 |
| Helenalin | −89.0 | −62.4 | −24.6 | −70.1 | −79.7 |
| Helenalin acetate | −85.9 | −71.7 | no dock | −76.1 | −81.6 |
| 8β-Hydroxyzaluzanin D | −97.3 | −75.2 | no dock | −79.1 | −93.3 |
| Lactucin | −88.9 | −72.7 | no dock | −85.0 | −99.8 |
| Lactucopicrin | −114.8 | −103.6 | no dock | −106.7 | −101.6 |
| Mexicanin I | −79.1 | −64.4 | −83.3 | −73.2 | −79.3 |
| 2-Oxo-8-tigloyloxyguaia-1(10),3-diene-6,12-olide-14-carboxylic acid | −100.1 | −94.1 | no dock | −92.0 | −100.8 |
| Peruvin | −85.4 | −59.4 | no dock | −68.9 | −75.4 |
| Psilostachyin | −85.7 | −69.7 | no dock | −67.2 | −84.5 |
| Psilostachyin C | −74.1 | −59.9 | −55.5 | −67.0 | −82.1 |
| Pungiolide A | −109.4 | −97.5 | −69.9 | −105.6 | −114.8 |
| Xanthipungolide | −64.8 | −52.0 | −32.1 | −47.9 | −59.0 |
| Zaluzanin D | −92.4 | −75.7 | no dock | −77.4 | −86.5 |
Table 10.
MolDock docking energies (kJ/mol) of eudesmanolide sesquiterpenoids with Leishmania major protein targets.
Table 10.
MolDock docking energies (kJ/mol) of eudesmanolide sesquiterpenoids with Leishmania major protein targets.
| Eudesmanolides | LmajCatB | LmajDHODH | LmajdUTPase | LmajNDKb | LmajNH | LmajNMT | LmajOPB | LmajPDE1 | LmajPTR1 | LmajMetRS | LmajTyrRS | LmajUGPase |
|---|
| Alantolactone | −58.1 | −90.7 | −61.4 | −79.0 | −73.8 | −72.4 | −70.1 | −67.2 | −65.8 | −83.2 | −74.2 | −87.9 |
| Anthecularin | −56.1 | −77.7 | −62.7 | −68.7 | −67.2 | −65.7 | −73.7 | −72.3 | −59.6 | −70.9 | −72.6 | −85.9 |
| Arbusculin B | −54.8 | −79.8 | −67.6 | −64.4 | −66.2 | −71.8 | −75.8 | −64.1 | −66.6 | −75.9 | −73.3 | −79.8 |
| α−Cyclocostunolide | −63.7 | −85.3 | −75.1 | −72.6 | −69.7 | −70.6 | −74.7 | −69.1 | −78.5 | −86.1 | −77.1 | −77.2 |
| β−Cyclocostunolide | −64.6 | −85.9 | −67.0 | −73.5 | −67.9 | −69.7 | −76.1 | −66.7 | −81.9 | −83.2 | −73.0 | −77.9 |
| Deacetyl−β−cyclopryethrosin | −69.1 | −97.8 | −73.4 | −76.4 | −72.9 | −80.2 | −82.3 | −76.9 | −83.3 | −93.6 | −76.0 | −91.1 |
| 11,13−Dihydrovernodalin | −78.0 | −110.6 | −91.9 | −100.3 | −89.7 | −99.7 | −99.0 | −106.2 | −103.4 | −114.7 | −104.0 | −94.5 |
| Douglanin | −66.7 | −90.4 | −76.9 | −71.6 | −71.9 | −69.6 | −77.8 | −74.7 | −79.4 | −88.2 | −77.1 | −78.5 |
| Frullanolide | −60.6 | −81.1 | −71.3 | −73.2 | −69.2 | −73.1 | −76.9 | −77.3 | −73.5 | −80.4 | −75.1 | −82.6 |
| 4−Hydroxyanthecotulide | −76.1 | −104.1 | −90.1 | −103.0 | −88.3 | −101.7 | −95.0 | −100.5 | −100.8 | −103.2 | −98.1 | −112.3 |
| 8β−[4−Hydroxy−5−(5−hydroxytigloyloxy)-tigloyl]santamarin | −109.5 | −128.9 | −116.3 | −125.3 | −117.8 | −115.9 | −113.3 | −117.5 | −130.2 | −153.1 | −128.3 | −127.1 |
| Isoalantolactone | −60.2 | −89.6 | −65.0 | −82.0 | −74.4 | −74.4 | −70.3 | −72.2 | −66.4 | −87.3 | −74.2 | −88.6 |
| Ivalin | −66.1 | −94.6 | −68.6 | −78.4 | −74.3 | −80.7 | −74.8 | −74.6 | −73.3 | −88.8 | −73.7 | −92.0 |
| Ivalin acetate | −77.4 | −93.6 | −87.4 | −87.4 | −83.2 | −89.5 | −90.8 | −89.6 | −91.6 | −104.5 | −87.8 | −83.4 |
| Onoseriolide | −72.4 | −92.4 | −82.8 | −81.4 | −78.5 | −79.8 | −88.5 | −83.8 | −88.6 | −101.4 | −81.3 | −82.5 |
| 2−Oxoalantolactone | −59.1 | −94.0 | −64.8 | −79.6 | −76.3 | −78.0 | −75.0 | −69.7 | −68.4 | −85.5 | −75.0 | −91.1 |
| Oxyonoseriolide | −77.5 | −94.5 | −82.3 | −80.4 | −81.5 | −88.9 | −88.7 | −88.0 | −87.8 | −104.7 | −92.7 | −99.1 |
| 4−Peroxy−1,2,4,5−tetrahydro−α−santonin | −63.3 | −97.6 | −72.6 | −71.0 | −72.5 | −74.1 | −87.3 | −80.3 | −90.1 | −85.2 | −76.6 | −82.4 |
| Santamarin | −65.3 | −86.0 | −74.9 | −72.7 | −72.6 | −71.6 | −80.5 | −73.1 | −79.0 | −89.3 | −79.4 | −79.4 |
| α−Santonin | −63.8 | −92.0 | −73.1 | −73.4 | −68.6 | −71.3 | −84.1 | −78.0 | −82.7 | −85.6 | −85.1 | −84.1 |
| Sivasinolide | −69.8 | −90.0 | −68.9 | −84.5 | −72.9 | −78.7 | −84.5 | −72.6 | −86.8 | −98.1 | −80.6 | −82.9 |
| Trilobolide 6−isobutyrate | −86.0 | −108.8 | −90.3 | −93.8 | −95.6 | −103.7 | −102.2 | −97.6 | −82.6 | −87.4 | −95.1 | −94.7 |
| Trilobolide 6−methacrylate | −78.4 | −103.8 | −87.9 | −85.0 | −94.2 | −111.3 | −87.6 | −97.6 | −78.9 | −87.5 | −92.1 | −92.2 |
| Vernangulide B | −91.2 | −118.3 | −108.7 | −105.8 | −105.6 | −113.0 | −99.8 | −107.1 | −122.9 | −125.3 | −105.0 | −111.9 |
| Vernodalin | −80.8 | −107.5 | −94.4 | −101.0 | −92.4 | −105.7 | −89.4 | −92.4 | −104.5 | −116.3 | −95.8 | −102.2 |
| Vernodalol | −82.5 | −112.8 | −100.1 | −99.1 | −96.4 | −97.3 | −94.5 | −99.4 | −100.0 | −107.8 | −94.6 | −90.2 |
| Wedelolide A | −88.3 | −109.9 | −95.0 | −83.5 | −94.0 | −109.6 | −84.2 | −103.7 | −83.1 | −131.6 | −96.2 | −98.4 |
| Wedelolide B | −88.0 | −113.0 | −91.2 | −87.0 | −94.2 | −107.4 | −99.0 | −107.4 | −82.8 | −134.0 | −102.8 | −94.9 |
Table 11.
MolDock docking energies (kJ/mol) of eudesmanolide sesquiterpenoids with Leishmania donovani and L. mexicana protein targets.
Table 11.
MolDock docking energies (kJ/mol) of eudesmanolide sesquiterpenoids with Leishmania donovani and L. mexicana protein targets.
| Eudesmanolides | LdonCatB | LdonCyp | LdonDHODH | LdonNMT | LmexGAPDH | LmexGPDH | LmexPGI | LmexPMM | LmexPYK | LmexPYK | LmexPYK | LmexTIM |
|---|
| | | | | | | | | | Site 1 | Site 2 | Site 3 | |
|---|
| Alantolactone | −67.1 | −78.3 | −55.0 | −64.4 | −62.6 | −79.8 | −57.8 | −74.6 | −78.6 | −72.2 | −79.7 | −77.9 |
| Anthecularin | −62.4 | −65.2 | −57.0 | −63.9 | −60.1 | −80.3 | −59.8 | −70.9 | −79.7 | −64.4 | −81.3 | −69.7 |
| Arbusculin B | −60.0 | −74.6 | −58.2 | −63.5 | −64.3 | −84.1 | −55.6 | −75.3 | −73.9 | −69.6 | −74.8 | −70.3 |
| α−Cyclocostunolide | −67.2 | −78.6 | −62.6 | −62.9 | −60.5 | −83.6 | −57.3 | −73.7 | −78.8 | −73.8 | −80.1 | −68.0 |
| β−Cyclocostunolide | −69.9 | −78.0 | −69.9 | −67.0 | −60.2 | −85.0 | −58.9 | −78.3 | −76.3 | −75.2 | −87.9 | −73.4 |
| Deacetyl−β−cyclopryethrosin | −73.4 | −82.9 | −63.3 | −72.8 | −71.5 | −92.2 | −63.3 | −84.0 | −84.6 | −78.1 | −88.1 | −81.4 |
| 11,13−Dihydrovernodalin | −89.3 | −94.8 | −59.5 | −84.3 | −79.3 | −107.0 | −82.3 | −102.5 | −104.4 | −94.9 | −100.7 | −87.1 |
| Douglanin | −69.0 | −80.6 | −71.4 | −66.3 | −63.7 | −88.6 | −58.9 | −76.6 | −82.7 | −77.2 | −82.1 | −65.1 |
| Frullanolide | −61.5 | −77.8 | −65.2 | −63.1 | −67.0 | −79.4 | −58.7 | −75.6 | −82.1 | −72.5 | −86.1 | −72.8 |
| 4−Hydroxyanthecotulide | −87.9 | −94.0 | −93.9 | −86.1 | −86.4 | −103.7 | −85.0 | −100.3 | −98.8 | −91.5 | −98.0 | −97.0 |
| 8β−[4−Hydroxy−5−(5−hydroxytigloyloxy)-tigloyl]santamarin | −112.4 | −115.3 | −37.4 | −121.4 | −96.5 | −128.2 | −113.8 | −132.1 | −124.3 | −119.9 | −112.8 | −105.4 |
| Isoalantolactone | −69.9 | −83.1 | −70.6 | −64.4 | −59.3 | −82.4 | −56.8 | −77.4 | −80.0 | −69.7 | −85.1 | −74.0 |
| Ivalin | −77.3 | −84.5 | −74.6 | −73.2 | −62.5 | −84.1 | −62.0 | −80.9 | −83.6 | −73.4 | −88.9 | −70.0 |
| Ivalin acetate | −91.5 | −92.5 | −63.2 | −66.9 | −73.8 | −97.0 | −70.5 | −92.4 | −97.2 | −80.3 | −81.4 | −86.8 |
| Onoseriolide | −73.8 | −88.1 | −84.8 | −77.0 | −72.8 | −93.9 | −66.8 | −84.1 | −87.9 | −78.0 | −95.0 | −77.1 |
| 2−Oxoalantolactone | −64.0 | −80.8 | −58.1 | −64.1 | −65.3 | −85.5 | −61.4 | −77.8 | −81.7 | −74.7 | −83.5 | −77.9 |
| Oxyonoseriolide | −77.9 | −84.5 | −61.8 | −78.5 | −78.5 | −95.4 | −70.4 | −84.6 | −98.4 | −80.6 | −103.9 | −79.3 |
| 4−Peroxy−1,2,4,5−tetrahydro−α−santonin | −68.0 | −72.4 | −68.6 | −72.3 | −62.0 | −92.4 | −58.1 | −82.3 | −85.2 | −73.3 | −83.0 | −73.0 |
| Santamarin | −69.3 | −79.8 | −56.3 | −67.4 | −65.6 | −89.5 | −61.2 | −81.2 | −82.6 | −75.9 | −82.0 | −71.6 |
| α−Santonin | −68.9 | −78.3 | −71.0 | −68.6 | −65.5 | −85.6 | −57.8 | −82.5 | −80.7 | −69.1 | −85.4 | −72.0 |
| Sivasinolide | −71.6 | −85.1 | −61.8 | −73.5 | −68.1 | −94.9 | −62.3 | −84.7 | −88.4 | −77.9 | −91.5 | −76.4 |
| Trilobolide 6−isobutyrate | −82.4 | −83.4 | −71.3 | −80.7 | −79.8 | −99.2 | −84.5 | −103.2 | −96.5 | −89.4 | −92.1 | −72.4 |
| Trilobolide 6−methacrylate | −83.4 | −80.8 | −20.7 | −82.7 | −70.3 | −99.2 | −88.3 | −107.3 | −94.9 | −92.7 | −87.2 | −74.7 |
| Vernangulide B | −93.8 | −106.6 | −126.8 | −96.1 | −90.5 | −121.5 | −100.6 | −120.1 | −116.6 | −105.6 | −107.5 | −104.7 |
| Vernodalin | −84.6 | −94.2 | −79.2 | −87.4 | −80.6 | −110.8 | −81.5 | −106.2 | −104.5 | −106.1 | −99.5 | −94.0 |
| Vernodalol | −88.4 | −96.3 | −50.6 | −83.9 | −84.1 | −105.2 | −93.5 | −99.8 | −103.5 | −99.1 | −103.4 | −83.2 |
| Wedelolide A | −87.9 | −68.5 | −85.5 | −91.6 | −84.0 | −96.0 | −90.3 | −114.9 | −103.2 | −107.4 | −92.9 | −92.7 |
| Wedelolide B | −81.5 | −64.3 | −88.3 | −97.5 | −79.2 | −96.0 | −92.2 | −127.4 | −102.4 | −108.8 | −102.6 | −96.0 |
Table 12.
MolDock docking energies (kJ/mol) of eudesmanolide sesquiterpenoids with Leishmania infantum protein targets.
Table 12.
MolDock docking energies (kJ/mol) of eudesmanolide sesquiterpenoids with Leishmania infantum protein targets.
| Eudesmanolides | LinfCYP51 | LinfGLO2 | LinfPnC1 | LinfTDR1 | LinfTR |
|---|
| Alantolactone | −71.3 | −60.0 | no dock | −66.1 | −75.5 |
| Anthecularin | −70.8 | −47.5 | −58.4 | −62.3 | −71.0 |
| Arbusculin B | −71.0 | −58.8 | −12.7 | −62.1 | −73.6 |
| α−Cyclocostunolide | −70.4 | −60.3 | −82.7 | −69.1 | −79.0 |
| β−Cyclocostunolide | −75.9 | −62.0 | −79.0 | −71.6 | −73.1 |
| γ−Cyclocostunolide | −71.0 | −57.5 | −12.1 | −62.2 | −67.4 |
| Deacetyl−β−cyclopryethrosin | −74.8 | −64.8 | −75.5 | −71.5 | −82.2 |
| 11,13−Dihydrovernodalin | −105.4 | −83.0 | no dock | −85.8 | −95.4 |
| Douglanin | −76.4 | −60.7 | −10.6 | −67.9 | −80.1 |
| Frullanolide | −75.2 | −61.3 | −55.4 | −70.5 | −74.5 |
| 4−Hydroxyanthecotulide | −90.2 | −82.6 | −68.4 | −85.4 | −92.1 |
| 8β−[4−Hydroxy−5−(5−hydroxytigloyloxy)-tigloyl]santamarin | −117.8 | −98.9 | no dock | −103.4 | −114.9 |
| Isoalantolactone | −69.1 | −57.8 | −31.1 | −64.3 | −74.2 |
| Ivalin | −72.9 | −65.0 | −16.1 | −65.6 | −80.5 |
| Ivalin acetate | −88.7 | −72.0 | no dock | −76.6 | −91.2 |
| Onoseriolide | −82.0 | −70.0 | −24.6 | −71.8 | −82.4 |
| 2−Oxoalantolactone | −73.4 | −57.6 | no dock | −68.1 | −79.4 |
| Oxyonoseriolide | −86.7 | −71.3 | −11.8 | −78.4 | −87.9 |
| 4−Peroxy−1,2,4,5−tetrahydro−α−santonin | −80.6 | −73.2 | −66.0 | −66.5 | −78.2 |
| Santamarin | −67.8 | −59.5 | −85.2 | −71.3 | −81.3 |
| α−Santonin | −74.2 | −69.4 | −44.0 | −65.5 | −86.4 |
| Sivasinolide | −75.2 | −63.3 | −72.1 | −72.5 | −81.1 |
| Trilobide 6−isobutyrate | −97.9 | −82.0 | no dock | −80.3 | −90.2 |
| Trilobide 6−methacrylate | −96.0 | −86.3 | no dock | −77.2 | −90.2 |
| Vernangulide B | −105.2 | −98.6 | no dock | −99.3 | −99.0 |
| Vernodalin | −105.9 | −88.9 | no dock | −86.6 | −96.9 |
| Vernodalol | −103.1 | −79.4 | no dock | −88.9 | −96.8 |
| Wedelolide A | −97.6 | −80.8 | no dock | −90.4 | −99.4 |
| Wedelolide B | −105.8 | −77.6 | no dock | −92.8 | −96.6 |
Table 13.
MolDock docking energies (kJ/mol) of miscellaneous sesquiterpenoids with Leishmania major protein targets.
Table 13.
MolDock docking energies (kJ/mol) of miscellaneous sesquiterpenoids with Leishmania major protein targets.
| Miscellaneous Sesquiterpenoids | LmajCatB | LmajDHODH | LmajdUTPase | LmajNDKb | LmajNH | LmajNMT | LmajOPB | LmajPDE1 | LmajPTR1 | LmajMetRS | LmajTyrRS | LmajUGPase |
|---|
| Alloaromadendrene | −56.8 | −81.7 | −67.0 | −68.2 | −66.1 | −71.1 | −74.3 | −68.2 | −91.9 | −70.7 | −70.0 | −76.6 |
| Aromadendrene | −56.9 | −78.5 | −68.4 | −66.9 | −64.7 | −70.2 | −73.2 | −67.9 | −99.4 | −73.2 | −72.4 | −72.2 |
| 1,10−Bisaboladiene−3,4−diol | −67.4 | −92.7 | −81.0 | −82.8 | −79.8 | −87.5 | −89.2 | −82.9 | −102.6 | −91.1 | −80.7 | −89.5 |
| α−Bisabolol | −75.2 | −92.5 | −76.3 | −77.5 | −75.6 | −89.9 | −74.7 | −79.9 | −110.6 | −93.8 | −80.3 | −83.2 |
| Corymbolone | −56.5 | −82.2 | −59.1 | −67.5 | −70.2 | −66.5 | −69.5 | −65.5 | −59.5 | −77.2 | −68.3 | −85.9 |
| α−Eudesmol | −60.9 | −80.3 | −64.8 | −70.7 | −68.1 | −66.0 | −66.0 | −74.2 | −86.8 | −83.1 | −76.9 | −74.7 |
| β−Eudesmol | −58.8 | −83.4 | −69.6 | −69.6 | −66.5 | −68.2 | −72.7 | −67.3 | −89.6 | −76.9 | −68.1 | −79.1 |
| 1(10),5−Germacradien−4−ol | −62.0 | −88.1 | −68.8 | −69.1 | −73.3 | −70.1 | −76.0 | −72.2 | −100.0 | −87.1 | −75.0 | −88.6 |
| Germacrene D | −57.9 | −81.4 | −66.6 | −65.1 | −70.8 | −68.8 | −69.7 | −69.9 | −96.7 | −77.7 | −70.6 | −81.8 |
| Gossypol | −59.1 | −106.1 | −85.2 | −89.5 | −83.9 | −104.6 | −98.4 | −111.5 | −120.6 | −92.7 | −108.0 | −90.7 |
| Gossypol−6,6'−dimethylether | −90.8 | −108.3 | −84.1 | −95.0 | −85.6 | −95.5 | −84.6 | −114.3 | −117.5 | −88.0 | −108.6 | −100.3 |
| Gossypol−6−methylether | −93.1 | −109.1 | −85.9 | −93.7 | −103.2 | −116.8 | −102.1 | −113.3 | −122.2 | −94.1 | −111.6 | −95.6 |
| Homalomenol C | −67.8 | −88.7 | −65.1 | −69.4 | −72.6 | −72.4 | −76.6 | −74.7 | −92.6 | −83.9 | −75.5 | −88.6 |
| 1−Hydroperoxy−10(14),11−guaiadiene | −60.7 | −85.4 | −70.4 | −70.7 | −69.0 | −74.0 | −77.5 | −78.5 | −96.5 | −83.6 | −76.5 | −83.7 |
| 10−Hydroperoxy−1,11−guaiadiene | −64.6 | −82.2 | −76.5 | −73.3 | −74.6 | −77.5 | −77.5 | −82.7 | −109.1 | −86.4 | −80.1 | −79.9 |
| 14−Hydroperoxy−1(10),11−guaiadiene | −71.5 | −88.0 | −88.1 | −79.2 | −78.4 | −78.5 | −80.0 | −82.3 | −111.6 | −87.9 | −79.1 | −86.6 |
| Kudtriol | −60.6 | −86.7 | −65.3 | −68.9 | −72.5 | −71.5 | −66.2 | −69.2 | −81.5 | −82.3 | −68.0 | −80.5 |
| 5−epi−Kudtriol | −59.9 | −86.5 | −69.2 | −68.4 | −70.4 | −78.2 | −75.8 | −75.8 | −65.9 | −77.9 | −65.1 | −83.0 |
| Longifolene | −52.1 | −75.8 | −50.0 | −62.3 | −59.6 | −58.9 | −63.0 | −66.7 | −61.3 | −62.3 | −63.1 | −70.8 |
| Mukaadial | −67.0 | −89.5 | −71.1 | −72.7 | −67.3 | −75.0 | −76.6 | −72.0 | −80.7 | −79.2 | −82.3 | −81.8 |
| Mustakone | −47.5 | −78.0 | −61.9 | −62.0 | −64.5 | −64.2 | −67.6 | −67.5 | −66.1 | −71.7 | −65.4 | −81.7 |
| Muzigadial | −64.6 | −93.9 | −66.8 | −70.6 | −65.0 | −70.6 | −74.1 | −66.3 | −81.5 | −81.9 | −68.5 | −76.8 |
| Nerolidol | −69.5 | −91.4 | −86.4 | −86.5 | −81.2 | −93.0 | −84.3 | −89.8 | −117.0 | −100.5 | −87.9 | −94.2 |
| Oplopanone | −59.4 | −81.9 | −69.4 | −66.1 | −68.2 | −70.8 | −73.0 | −71.7 | −106.4 | −76.9 | −74.0 | −74.2 |
| 10,12−Peroxycalamenene | −41.0 | −71.0 | −57.9 | −57.4 | −60.8 | −65.9 | −66.3 | −74.3 | −70.6 | −69.3 | −68.4 | −70.7 |
| Plagiochilin A | −83.4 | −103.9 | −94.1 | −94.1 | −93.6 | −88.5 | −95.7 | −92.7 | −132.3 | −102.9 | −106.0 | −105.9 |
| Polygodial | −61.3 | −86.0 | −70.7 | −71.6 | −64.8 | −71.0 | −76.9 | −66.6 | −91.1 | −86.3 | −71.5 | −80.4 |
| Zingiberene−3,6−α−peroxide | −62.3 | −89.4 | −75.8 | −75.4 | −76.0 | −82.6 | −68.3 | −84.9 | −87.4 | −92.0 | −76.8 | −88.2 |
| Zingiberene−3,6−β−peroxide | −62.2 | −86.9 | −73.6 | −75.4 | −74.9 | −78.7 | −72.4 | −84.7 | −89.9 | −82.0 | −73.7 | −84.9 |
Table 14.
MolDock docking energies (kJ/mol) of miscellaneous sesquiterpenoids with Leishmania donovani and L. mexicana protein targets.
Table 14.
MolDock docking energies (kJ/mol) of miscellaneous sesquiterpenoids with Leishmania donovani and L. mexicana protein targets.
| Miscellaneous Sesquiterpenoids | LdonCatB | LdonCyp | LdonDHODH | LdonNMT | LmexGAPDH | LmexGPDH | LmexPGI | LmexPMM | LmexPYK | LmexPYK | LmexPYK | LmexTIM |
|---|
| | | | | | | | | | Site 1 | Site 2 | Site 3 | |
| Alloaromadendrene | −66.6 | −77.8 | −46.3 | −58.8 | −61.3 | −78.3 | −57.1 | −71.4 | −79.0 | −62.3 | −79.7 | −68.9 |
| Aromadendrene | −66.1 | −72.0 | −56.6 | −62.4 | −57.1 | −75.0 | −54.4 | −71.9 | −79.4 | −65.5 | −77.1 | −65.1 |
| 1,10−Bisaboladiene−3,4−diol | −73.0 | −82.3 | −74.6 | −85.5 | −70.9 | −94.2 | −71.9 | −78.6 | −88.2 | −76.9 | −84.5 | −80.0 |
| α−Bisabolol | −75.2 | −76.8 | −68.1 | −81.0 | −65.2 | −85.2 | −67.0 | −84.8 | −90.3 | −75.7 | −79.7 | −77.7 |
| Corymbolone | −60.9 | −72.2 | −62.0 | −63.5 | −60.6 | −74.4 | −56.1 | −70.5 | −74.6 | −72.4 | −80.2 | −69.4 |
| 6α,9α−Dihydroxypolygodial | −65.8 | −86.7 | −61.9 | −72.3 | −65.5 | −88.4 | −58.7 | −77.1 | −83.6 | −74.4 | −91.4 | −69.6 |
| α−Eudesmol | −62.3 | −74.4 | −49.2 | −57.4 | −57.2 | −78.4 | −53.9 | −76.3 | −73.5 | −64.3 | −82.9 | −67.2 |
| β−Eudesmol | −66.1 | −74.1 | −57.0 | −63.4 | −58.8 | −75.1 | −56.5 | −77.8 | −72.4 | −65.5 | −83.0 | −68.3 |
| 1(10),5−Germacradien−4−ol | −69.7 | −78.5 | −49.2 | −70.5 | −62.2 | −81.5 | −58.6 | −75.2 | −78.1 | −68.4 | −84.1 | −74.8 |
| Germacrene D | −65.0 | −74.6 | −64.4 | −68.3 | −60.1 | −77.0 | −58.3 | −70.1 | −73.5 | −64.9 | −82.0 | −71.3 |
| Gossypol | −79.2 | −86.4 | −88.4 | −64.5 | −81.7 | −117.4 | −76.4 | −116.4 | −110.5 | −96.9 | −93.4 | −86.4 |
| Gossypol−6,6'−dimethylether | −89.6 | −82.6 | −90.2 | −82.3 | −71.9 | −112.2 | −81.8 | −119.8 | −113.9 | −103.5 | −96.5 | −84.3 |
| Gossypol−6−methylether | −82.2 | −90.0 | −87.5 | −82.4 | −69.4 | −113.0 | −75.3 | −115.2 | −111.7 | −103.6 | −101.0 | −83.4 |
| Homalomenol C | −66.5 | −75.6 | −26.8 | −65.5 | −64.4 | −89.4 | −63.4 | −74.5 | −87.0 | −68.4 | −84.5 | −73.1 |
| 1−Hydroperoxy−10(14),11−guaiadiene | −59.3 | −78.1 | −59.8 | −61.2 | −63.7 | −85.3 | −61.7 | −73.4 | −81.5 | −74.5 | −79.2 | −68.5 |
| 10−Hydroperoxy−1,11−guaiadiene | −77.0 | −82.0 | −67.9 | −66.3 | −66.3 | −83.7 | −60.1 | −80.2 | −86.0 | −73.5 | −85.7 | −78.9 |
| 14−Hydroperoxy−1(10),11−guaiadiene | −78.4 | −92.8 | −85.2 | −69.2 | −70.4 | −93.9 | −67.8 | −79.5 | −87.7 | −75.2 | −91.0 | −79.6 |
| Kudtriol | −69.5 | −78.5 | −66.2 | −65.6 | −61.3 | −79.2 | −58.9 | −73.8 | −78.6 | −69.7 | −93.0 | −70.4 |
| 5−epi−Kudtriol | −59.0 | −78.2 | −65.4 | −66.2 | −69.3 | −79.6 | −55.7 | −76.8 | −77.6 | −70.7 | −83.7 | −68.5 |
| Longifolene | −52.4 | −69.7 | −35.7 | −56.3 | −57.6 | −74.0 | −52.4 | −63.5 | −68.1 | −63.1 | −66.8 | −60.8 |
| Mukaadial | −65.9 | −86.9 | −61.9 | −72.3 | −72.0 | −92.6 | −58.7 | −77.1 | −83.7 | −74.5 | −91.5 | −69.6 |
| Mustakone | −57.7 | −57.0 | −61.7 | −59.3 | −56.1 | −76.2 | −53.7 | −69.0 | −72.7 | −63.2 | −73.7 | −63.2 |
| Muzigadial | −70.3 | −85.5 | −15.2 | −62.1 | −66.3 | −78.6 | −57.9 | −76.5 | −80.4 | −73.1 | −81.2 | −71.5 |
| Nerolidol | −79.6 | −82.0 | −87.3 | −76.3 | −74.5 | −91.1 | −79.5 | −84.3 | −87.3 | −76.5 | −91.7 | −85.8 |
| Oplopanone | −68.5 | −78.9 | −64.1 | −65.2 | −59.8 | −78.2 | −58.4 | −71.5 | −77.0 | −67.2 | −78.5 | −68.5 |
| 10,12−Peroxycalamenene | −51.6 | −70.8 | −62.9 | −54.6 | −58.3 | −71.8 | −49.9 | −71.3 | −66.5 | −61.4 | −69.9 | −62.4 |
| Plagiochilin A | −96.2 | −89.1 | −95.1 | −81.9 | −78.8 | −105.9 | −75.5 | −99.4 | −99.8 | −97.9 | −87.5 | −89.2 |
| Polygodial | −60.0 | −80.5 | −57.6 | −68.6 | −62.1 | −81.9 | −55.2 | −73.3 | −78.4 | −69.2 | −84.7 | −76.3 |
| Zingiberene−3,6−α−peroxide | −75.1 | −71.7 | −80.1 | −74.3 | −67.0 | −83.7 | −67.1 | −80.7 | −79.9 | −74.0 | −81.1 | −76.1 |
| Zingiberene−3,6−β−peroxide | −71.6 | −75.1 | −77.1 | −76.2 | −60.7 | −84.3 | −66.0 | −74.9 | −83.0 | −67.0 | −87.3 | −74.6 |
Table 15.
MolDock docking energies (kJ/mol) of miscellaneous sesquiterpenoids with Leishmania infantum protein targets.
Table 15.
MolDock docking energies (kJ/mol) of miscellaneous sesquiterpenoids with Leishmania infantum protein targets.
| Miscellaneous Sesquiterpenoids | LinfCYP51 | LinfGLO2 | LinfPnC1 | LinfTDR1 | LinfTR |
|---|
| Alloaromadendrene | −66.8 | −57.0 | −52.9 | −61.7 | −67.2 |
| Aromadendrene | −60.8 | −59.7 | −60.8 | −61.6 | −76.8 |
| 1,10−Bisaboladiene−3,4−diol | −75.6 | −70.8 | −63.5 | −72.1 | −85.0 |
| α−Bisabolol | −76.5 | −74.1 | −73.4 | −75.0 | −74.7 |
| Corymbolone | −65.9 | −54.6 | −49.1 | −59.4 | −65.3 |
| 6α,9α−Dihydroxypolygodial | −75.7 | −62.3 | −23.0 | −63.6 | −78.0 |
| α−Eudesmol | −62.6 | −58.1 | −28.2 | −59.1 | −69.5 |
| β−Eudesmol | −68.0 | −59.6 | −23.9 | −64.3 | −69.7 |
| 1(10),5−Germacradien−4−ol | −70.4 | −58.4 | −52.6 | −63.6 | −70.2 |
| Germacrene D | −64.6 | −57.4 | −62.7 | −68.0 | −70.2 |
| Gossypol | −109.9 | −99.6 | no dock | −86.6 | −97.0 |
| Gossypol−6,6'−dimethylether | −109.5 | −95.1 | no dock | −88.7 | −101.1 |
| Gossypol−6−methylether | −113.1 | −99.6 | no dock | −94.5 | −100.9 |
| Homalomenol C | −72.3 | −57.3 | −41.9 | −64.4 | −68.5 |
| 1−Hydroperoxy−10(14),11−guaiadiene | −74.5 | −56.7 | −15.4 | −62.9 | −72.8 |
| 10−Hydroperoxy−1,11−guaiadiene | −79.3 | −66.5 | −38.7 | −67.3 | −75.4 |
| 14−Hydroperoxy−1(10),11−guaiadiene | −81.4 | −67.2 | −69.9 | −74.2 | −74.4 |
| Kudtriol | −68.1 | −51.5 | no dock | −65.3 | −72.4 |
| 5−epi−Kudtriol | −67.9 | −61.6 | no dock | −63.0 | −71.5 |
| Longifolene | −64.9 | −49.4 | −48.0 | −54.6 | −61.5 |
| Mukaadial | −74.9 | −61.5 | −21.0 | −63.7 | −78.0 |
| Mustakone | −63.5 | −57.5 | −18.1 | −55.3 | −63.3 |
| Muzigadial | −72.6 | −56.5 | −66.6 | −65.1 | −84.6 |
| Nerolidol | −79.3 | −72.3 | −72.0 | −79.1 | −82.5 |
| Oplopanone | −65.7 | −59.4 | −55.6 | −63.2 | −69.2 |
| 10,12−Peroxycalamenene | −65.2 | −46.6 | no dock | −58.2 | −70.5 |
| Plagiochilin A | −95.9 | −79.6 | no dock | −80.5 | −89.3 |
| Polygodial | −72.1 | −57.6 | −45.3 | −62.8 | −72.3 |
| Zingiberene−3,6−α−peroxide | −77.7 | −65.4 | −65.8 | −66.7 | −75.1 |
| Zingiberene−3,6−β−peroxide | −69.8 | −65.5 | −51.2 | −62.7 | −72.9 |
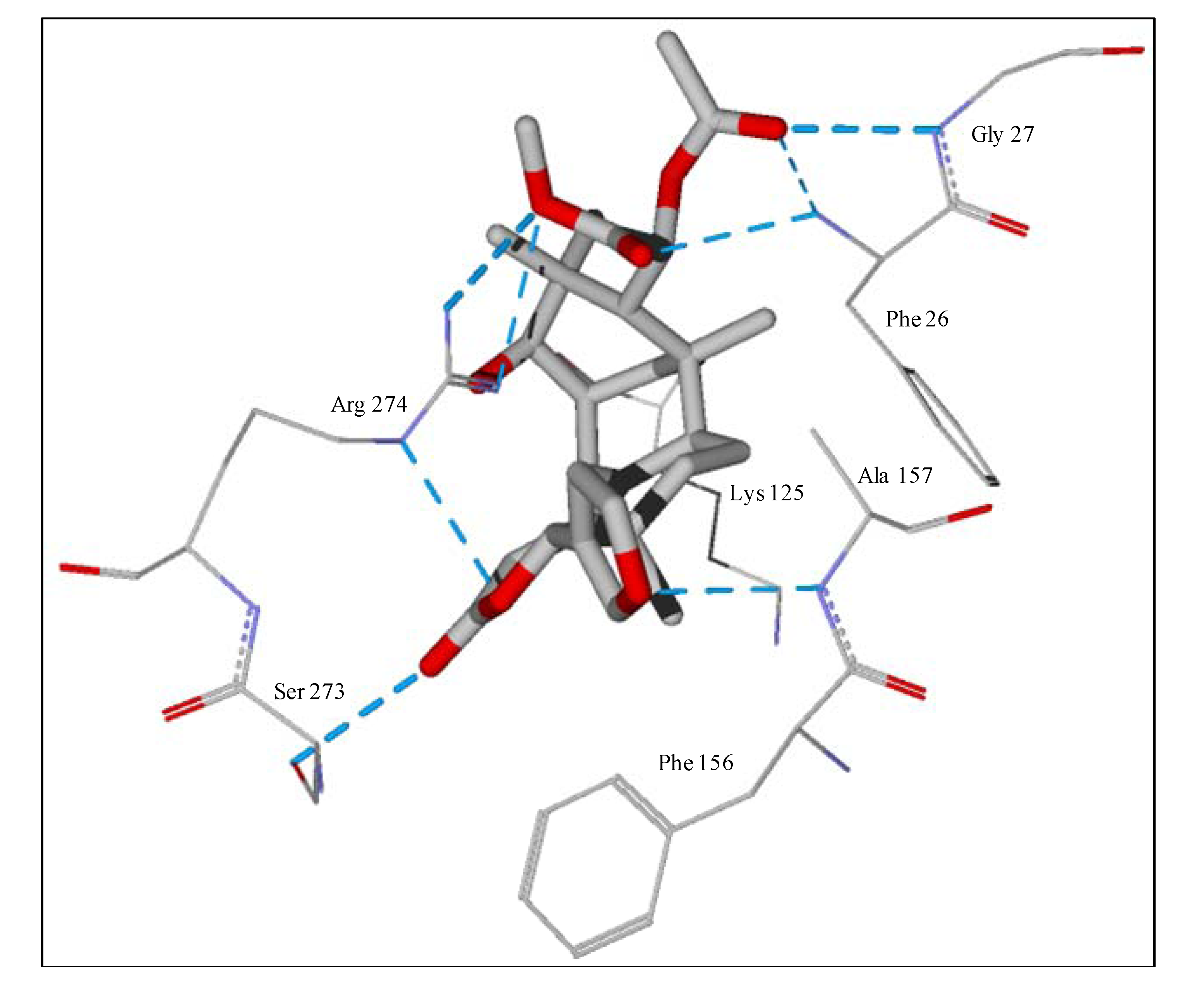
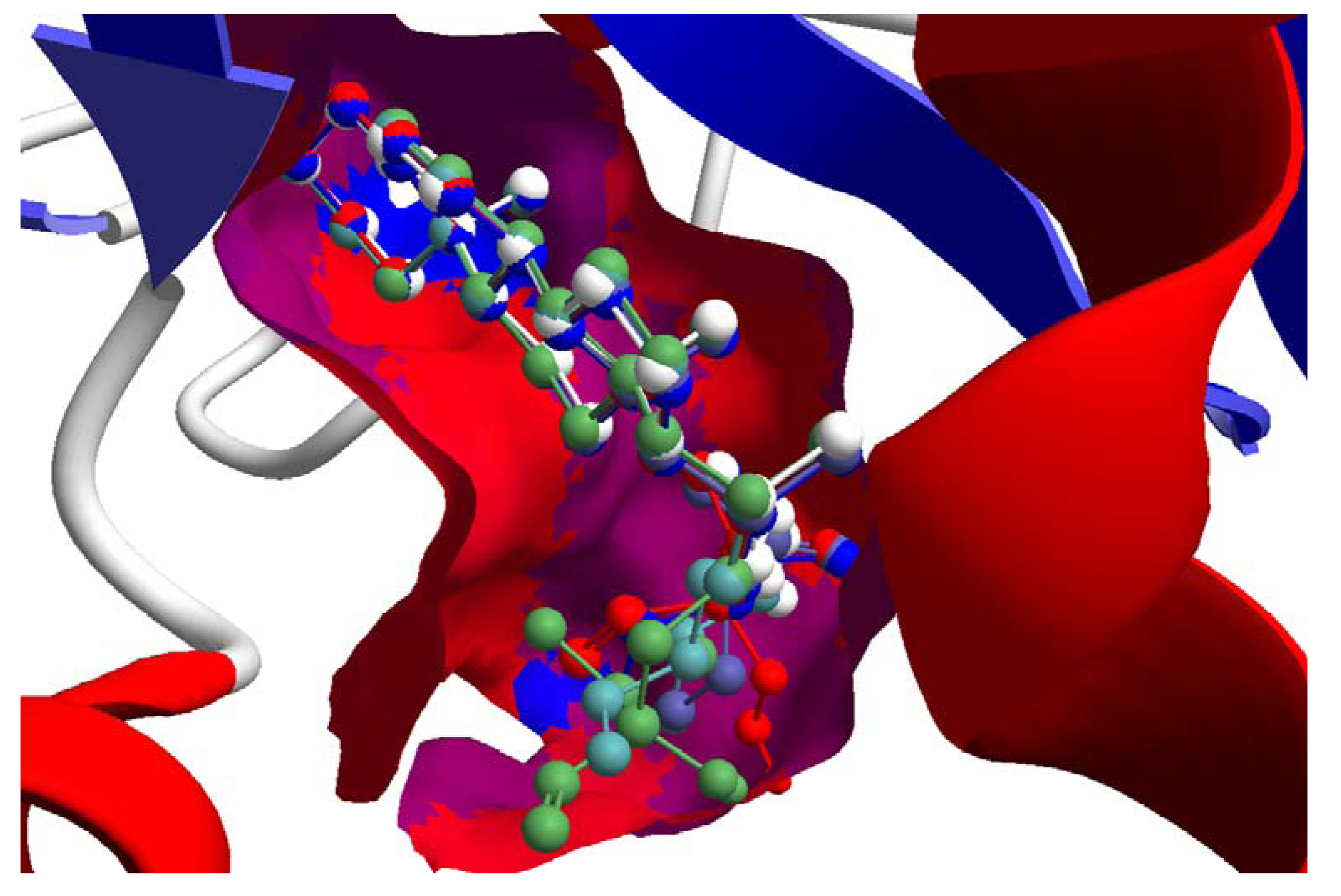
The guaianolide with the strongest docking energy was diguaiaperfolin, probably owing to its dimeric structure and larger molecular weight (716.77 amu). This ligand did show notable docking (−154.5 kJ/mol) with LmajDHODH as well as with LmajUGPase (docking energy = −151.8 kJ/mol). 8β-[4-Hydroxy-5-(5-hydroxytigloyloxy)tigloyl]santamarin was the strongest-docking eudesmanolide, and this ligand showed docking selectivity to LmajMetRS (docking energy = −153.1 kJ/mol). The proteins most strongly targeted by both the guaianolides and the eudesmaolides were also LmajMetRS and LmajDHODH. Interestingly, the miscellaneous sesquiterpenoids preferentially targeted L. major pteridine reductase 1 (LmajPTR1), and plagiochilin A showed notable selectivity (docking energy = −132.2 kJ/mol) for this protein.
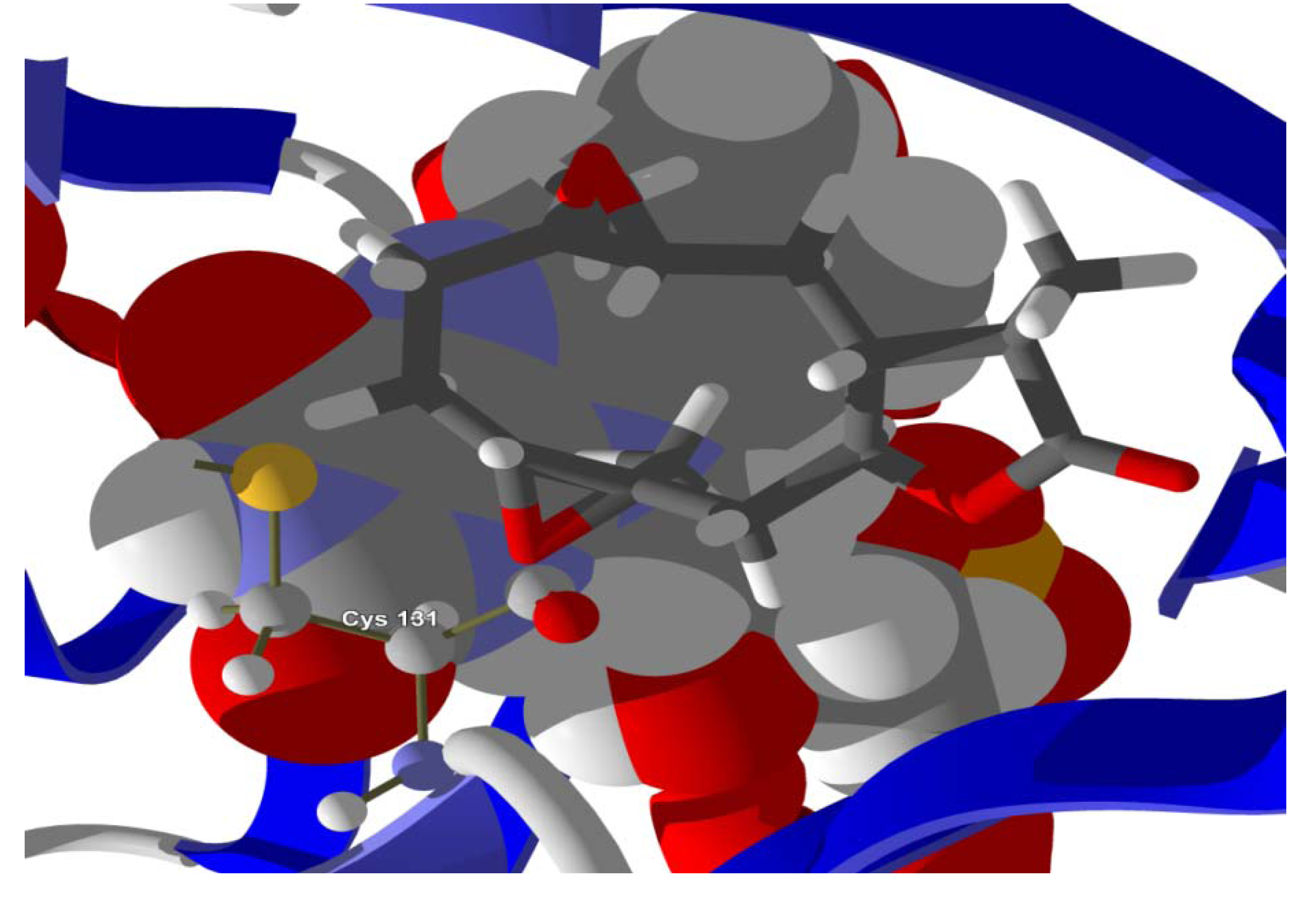
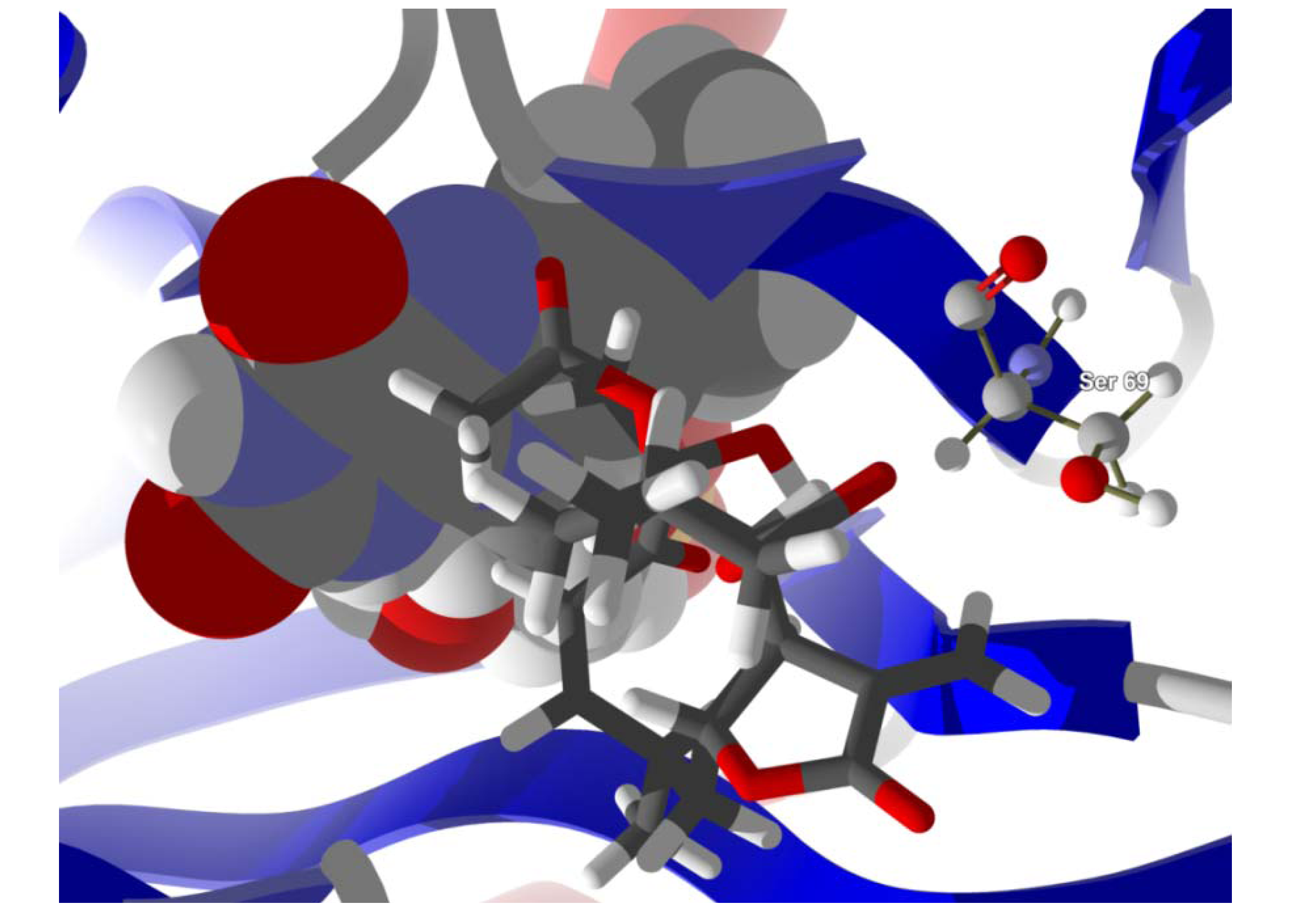
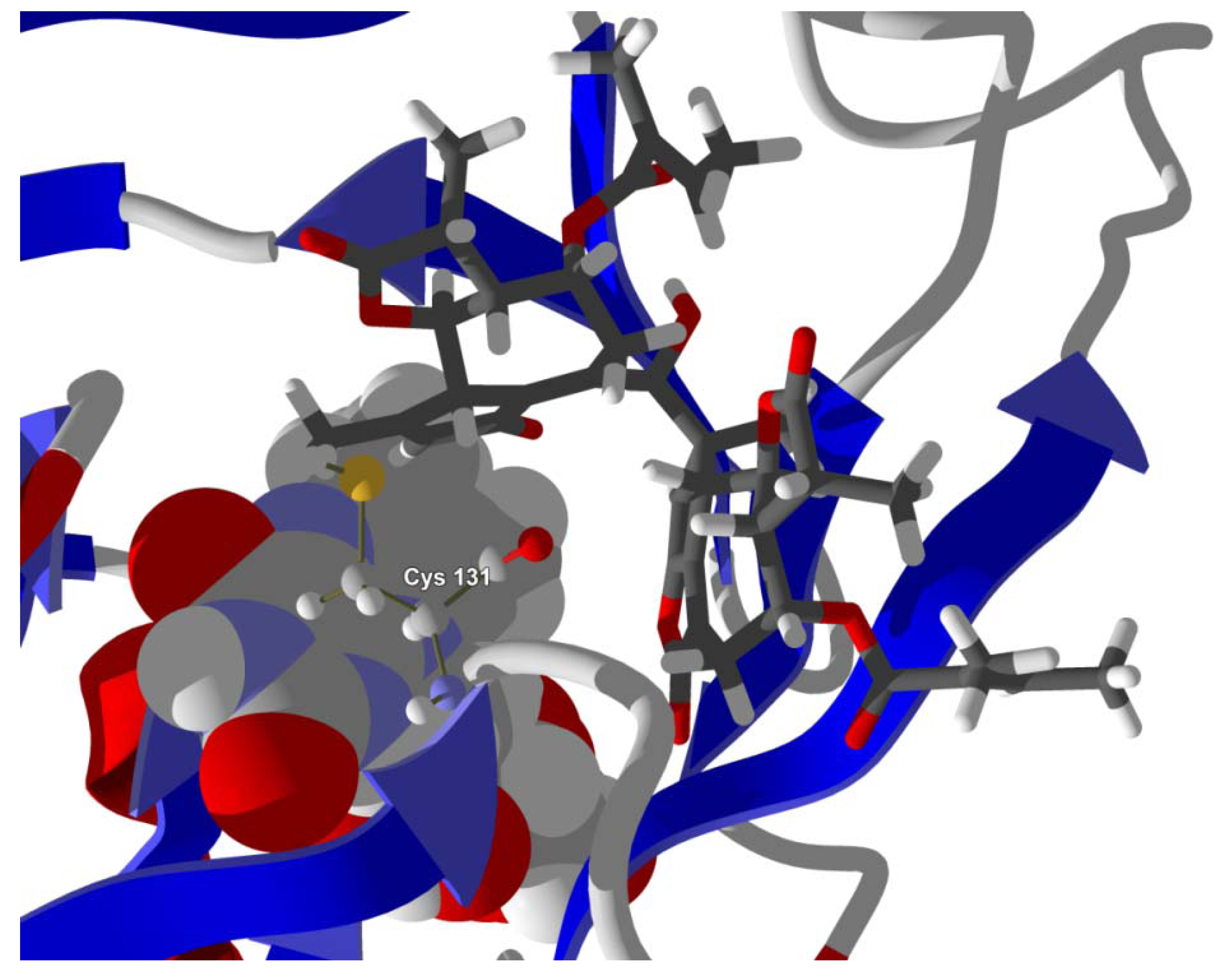
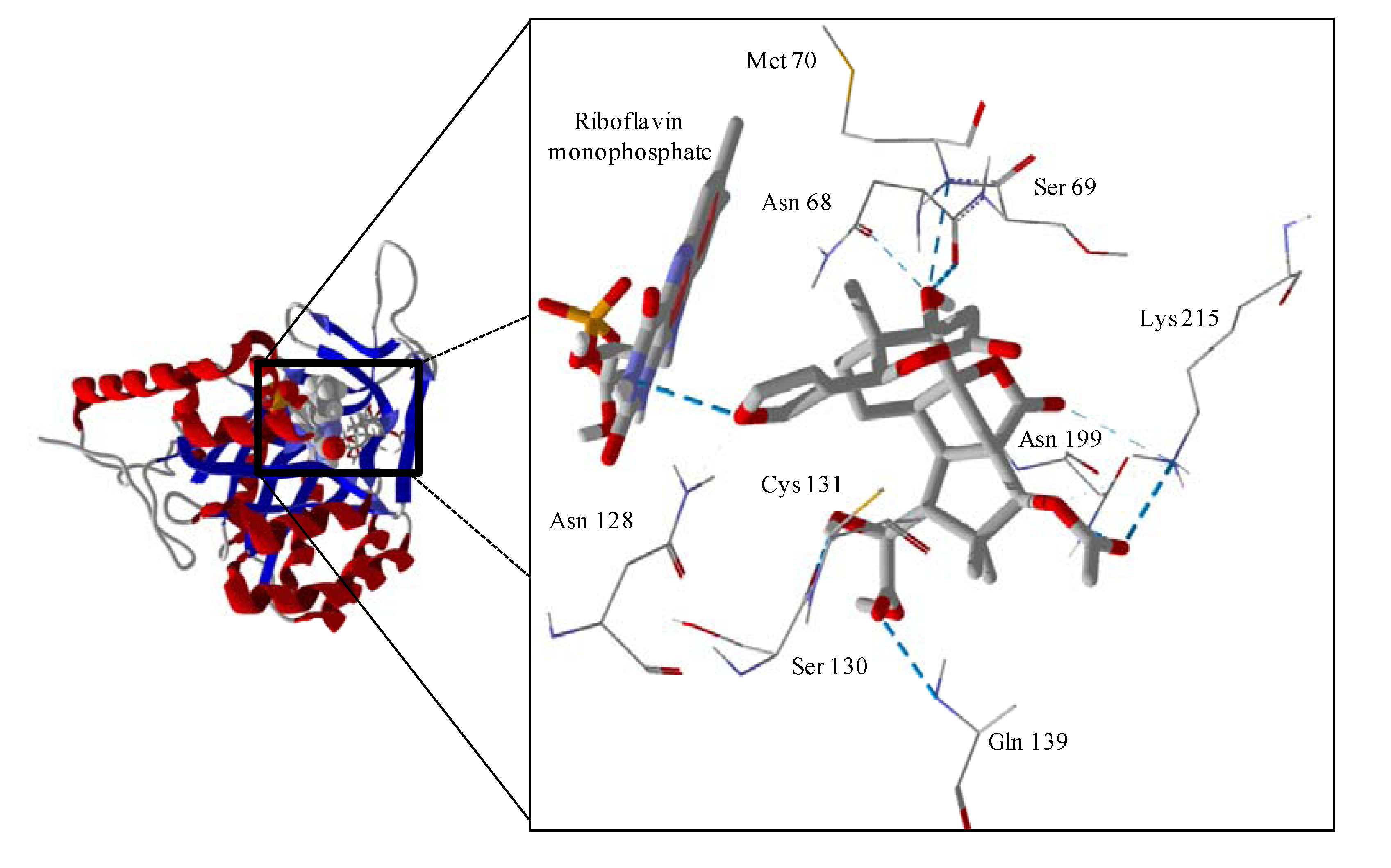
Several electrophilic sesquiterpenoids have exhibited antiprotozoal activity [
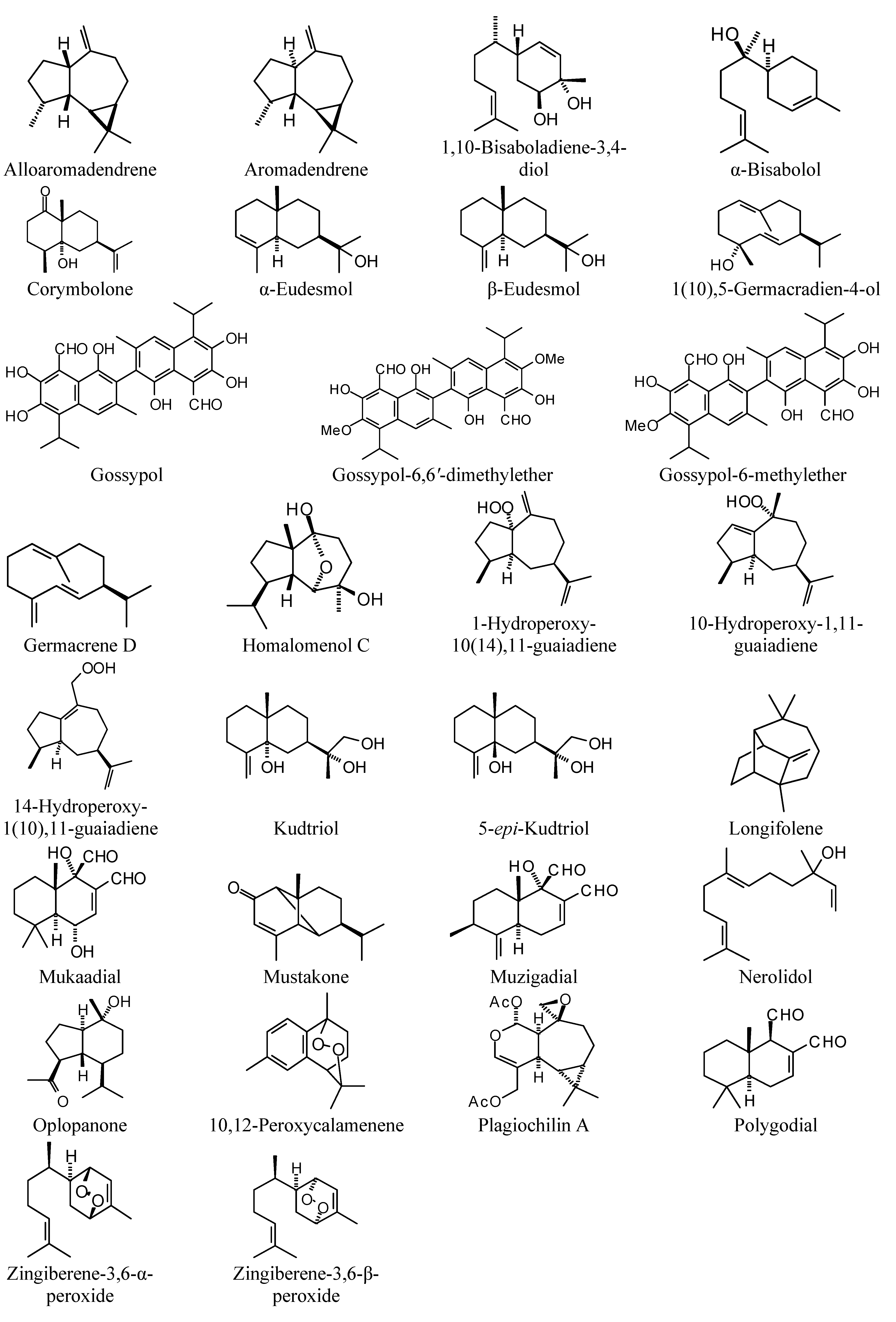
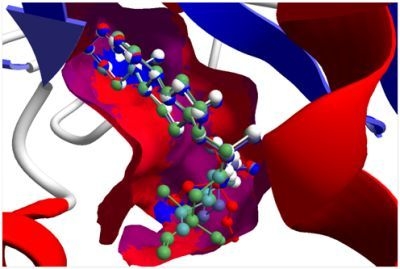
5] and many of these showed docking selectivity to LmajDHODH. The active site of this protein has some potential nucleophilic residues, namely Ser 69, Ser 196, and Cys 131. Suitably oriented electrophilic ligands could form covalent bonds with these nucleophiles and thus inhibit the enzyme. Thus, for example, the germacranolide tatridin A docked preferentially to LmajDHODH, and the lowest-energy docked pose oriented the electrophilic carbon of the α-methylene lactone moiety close to the sulfur atom of Cys 131 (see
Figure 6). Similarly, the lowest-energy docked orientation of 11-
epi-ivaxillin places one of the epoxide groups near to the sulfur atom of Cys 131 (
Figure 7). Conversely, the lowest-energy docked pose of neurolenin B is such that the electrophilic carbon of the α-methylene lactone group of the ligand is near the hydroxyl group of Ser 69 (
Figure 8). The ligand with the lowest docking energy to LmajDHODH was the guaianolide dimer diguaiaperfolin (−154.5 kJ/mol). The lowest-energy pose for this ligand placed the cyclopentenone moiety near the sulfur atom of Cys 131 (
Figure 9).
Figure 6.
Lowest-energy docked pose of tatridin A with L. major dihydroorotate dehydrogenase (LmajDHODH, PDB 3mhu). The cofactor, riboflavin monophosphate, is shown as a space-filling structure.
Figure 6.
Lowest-energy docked pose of tatridin A with L. major dihydroorotate dehydrogenase (LmajDHODH, PDB 3mhu). The cofactor, riboflavin monophosphate, is shown as a space-filling structure.
Figure 7.
Lowest-energy docked pose of 11-epi-ivaxillin with L. major dihydroorotate dehydrogenase (LmajDHODH, PDB 3mhu). The cofactor, riboflavin monophosphate, is shown as a space-filling structure.
Figure 7.
Lowest-energy docked pose of 11-epi-ivaxillin with L. major dihydroorotate dehydrogenase (LmajDHODH, PDB 3mhu). The cofactor, riboflavin monophosphate, is shown as a space-filling structure.
Figure 8.
Lowest-energy docked pose of neurolenin B with L. major dihydroorotate dehydrogenase (LmajDHODH, PDB 3mhu). The cofactor, riboflavin monophosphate, is shown as a space-filling structure.
Figure 8.
Lowest-energy docked pose of neurolenin B with L. major dihydroorotate dehydrogenase (LmajDHODH, PDB 3mhu). The cofactor, riboflavin monophosphate, is shown as a space-filling structure.
Figure 9.
Lowest-energy docked pose of diguaiaperfolin with L. major dihydroorotate dehydrogenase (LmajDHODH, PDB 3mhu). The cofactor, riboflavin monophosphate, is shown as a space-filling structure.
Figure 9.
Lowest-energy docked pose of diguaiaperfolin with L. major dihydroorotate dehydrogenase (LmajDHODH, PDB 3mhu). The cofactor, riboflavin monophosphate, is shown as a space-filling structure.
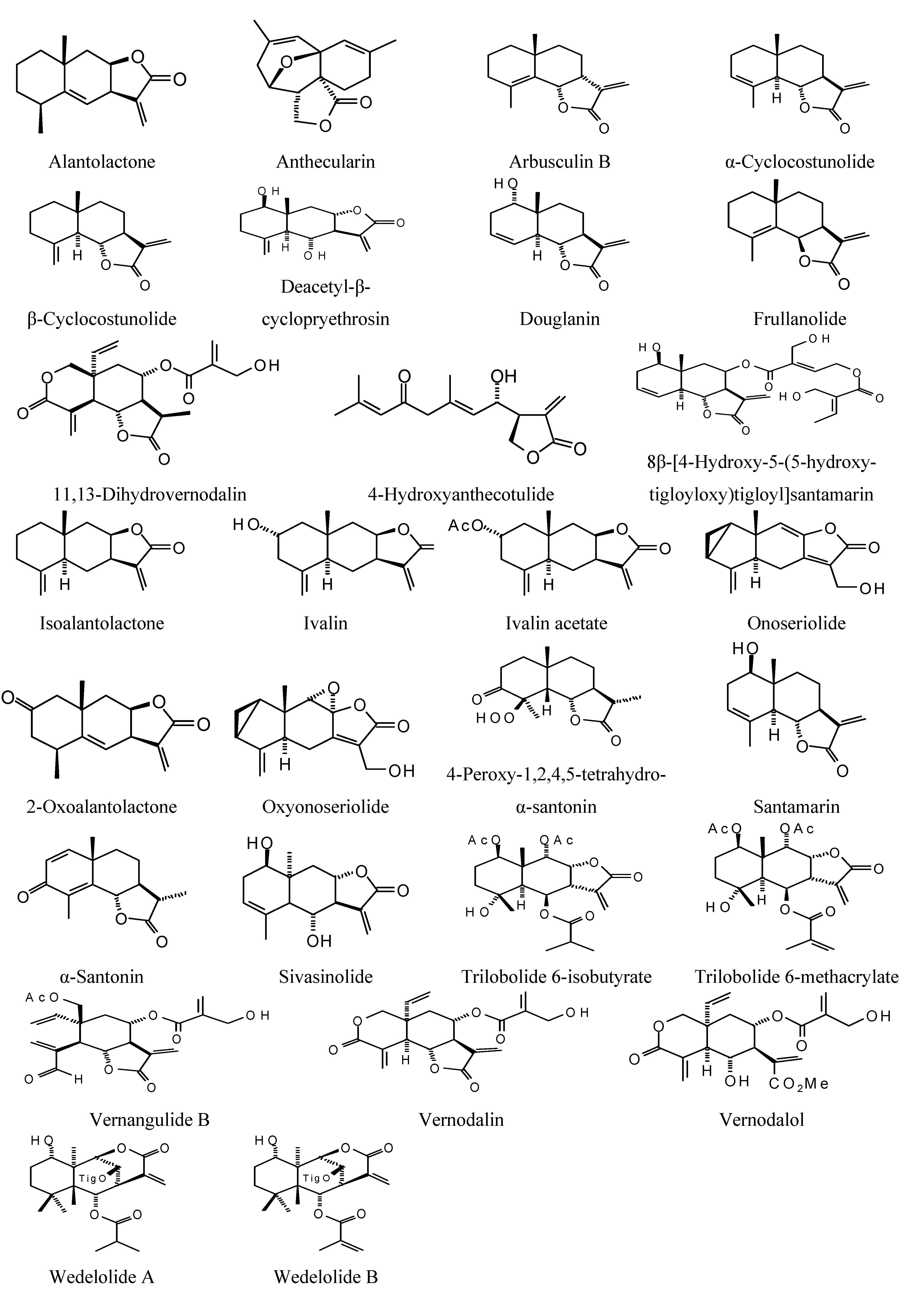
2.3. Diterpenoid Docking
Structures of diterpenoids are shown in
Figure 10,
Figure 11,
Figure 12,
Figure 13,
Figure 14,
Figure 15,
Figure 16,
Figure 17 and
Figure 18. Docking energies of the diterpenoids are assembled in
Table 16,
Table 17,
Table 18,
Table 19,
Table 20,
Table 21,
Table 22,
Table 23,
Table 24,
Table 25,
Table 26,
Table 27 and
Table 28. The diterpenoids ligands generally favored docking to
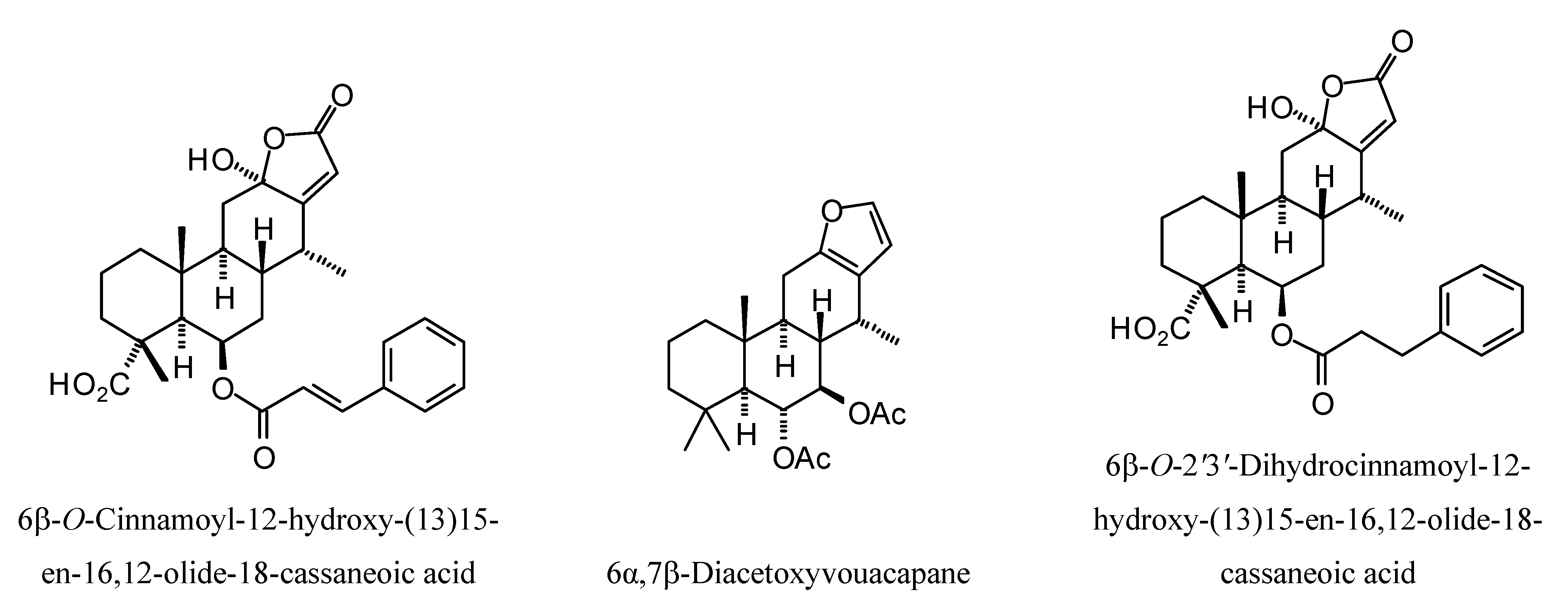
L. mexicana glycerol-3-phosphate dehydrogenase (LmexGPDH). In particular, the kaurane diterpenoids strongly docked to this target. In addition to LmexGPDH, labdane diterpenoids showed docking preferences for LmajMetRS and LmajDHODH. The strongest-docking ligands were the cinnamoyl cassanes 6β-
O-cinnamoyl-12-hydroxy-(13)15-en-16,12-olide-18-cassaneoic acid and 6β-O-2
',3
'-dihydro-cinnamoyl-12-hydroxy-(13)15-en-16,12-olide-18-cassaneoic acid. These two ligands showed significant docking preference to LmajMetRS and LmexPMM.
Figure 10.
Abietane diterpenoids examined in this study.
Figure 10.
Abietane diterpenoids examined in this study.
Figure 11.
Clerodane diterpenoids examined in this study.
Figure 11.
Clerodane diterpenoids examined in this study.
Figure 12.
Labdane diterpenoids examined in this study.
Figure 12.
Labdane diterpenoids examined in this study.
Figure 13.
Kaurane diterpenoids examined in this study.
Figure 13.
Kaurane diterpenoids examined in this study.
Figure 14.
Pimarane diterpenoids examined in this study.
Figure 14.
Pimarane diterpenoids examined in this study.
Figure 15.
Cassane diterpenoids examined in this study.
Figure 15.
Cassane diterpenoids examined in this study.
Figure 16.
Icetaxane diterpenoids examined in this study.
Figure 16.
Icetaxane diterpenoids examined in this study.
Figure 17.
Mulinane diterpenoids examined in this study.
Figure 17.
Mulinane diterpenoids examined in this study.
Figure 18.
Miscellaneoous diterpenoids examined in this study.
Figure 18.
Miscellaneoous diterpenoids examined in this study.
Table 16.
MolDock docking energies (kJ/mol) of abietane diterpenoids with Leishmania major protein targets.
Table 16.
MolDock docking energies (kJ/mol) of abietane diterpenoids with Leishmania major protein targets.
| Abietane diterpenoids | LmajCatB | LmajDHODH | LmajdUTPase | LmajNDKb | LmajNH | LdonNMT | LmajOPB | LmajPDE1 | LmajPTR1 | LmajMetRS | LmajTyrRS | LmajUGPase |
|---|
| Abieta−7,13−diene | −71.5 | −80.8 | −79.3 | −80.3 | −76.3 | −69.7 | −82.2 | −77.0 | −74.7 | −96.9 | −78.1 | −78.3 |
| ar−Abietatriene−12,16−diol−14,16−oxide | −74.0 | −97.8 | −78.9 | −96.2 | −78.0 | −74.8 | −98.2 | −83.2 | −73.8 | −89.0 | −84.6 | −77.8 |
| ar−Abietatrien−12−ol−6,7−dione−14,16−oxide | −66.1 | −91.8 | −86.1 | −79.4 | −72.8 | −74.2 | −98.2 | −83.6 | −84.5 | −92.7 | −91.4 | −83.4 |
| epi−Abietic acid | −75.3 | −93.5 | −81.8 | −74.9 | −84.0 | −78.1 | −86.1 | −80.7 | −82.1 | −99.8 | −87.3 | −84.7 |
| 4−
epi−Abietol | −71.9 | −87.6 | −81.8 | −80.4 | −80.9 | −69.3 | −83.4 | −74.8 | −78.1 | −96.3 | −81.0 | −82.3 |
| Cryptotanshinone | −69.2 | −93.7 | −74.6 | −81.3 | −59.6 | −78.3 | −91.6 | −83.4 | −81.2 | −105.2 | −86.5 | −78.6 |
| 12−
O−Deacetyl−6−O−acetyl−18−acetyloxycoleon Q | −81.2 | −89.0 | −95.0 | −89.7 | −99.9 | −79.5 | −95.5 | −111.8 | −93.9 | −94.2 | −88.9 | −114.2 |
| 12−
O−Deacetyl−6−O−acetyl−19−acetyloxycoleon Q | −76.3 | −104.3 | −82.9 | −93.8 | −101.2 | −75.4 | −100.7 | −107.4 | −84.3 | −99.0 | −92.0 | −96.9 |
| 12−Deoxyroyleanone | −78.2 | −79.2 | −74.3 | −74.8 | −72.9 | −73.1 | −76.5 | −83.6 | −86.4 | −85.9 | −77.6 | −77.4 |
| 9α,13α−
epi−Dioxyabiet−8(14)−en−18−oic acid | −65.5 | −88.2 | −79.9 | −82.1 | −80.1 | −64.0 | −89.6 | −79.4 | −71.2 | −89.5 | −88.2 | −81.6 |
| 9α,13α−
epi−Dioxyabiet−8(14)en−18−ol | −47.3 | −86.6 | −76.2 | −78.7 | −72.3 | −65.3 | −74.4 | −76.8 | −72.7 | −81.5 | −74.0 | −71.2 |
| Dracocephalone A | −66.5 | −82.6 | −70.8 | −76.6 | −71.9 | −66.7 | −93.0 | −83.2 | −87.4 | −83.4 | −72.9 | −78.7 |
| Dracocequinone A | −66.6 | −84.4 | −74.5 | −76.0 | −74.1 | −75.4 | −94.8 | −83.5 | −73.8 | −87.6 | −76.4 | −82.6 |
| Dracocequinone B | −74.5 | −89.1 | −71.5 | −78.4 | −78.4 | −76.6 | −83.4 | −81.9 | −76.0 | −88.7 | −77.3 | −88.2 |
| Ferruginol | −68.7 | −90.3 | −81.0 | −81.7 | −76.0 | −71.3 | −85.3 | −82.2 | −83.7 | −99.3 | −83.4 | −81.2 |
| Hinokiol | −77.9 | −88.2 | −85.2 | −75.8 | −79.1 | −62.3 | −84.5 | −74.9 | −77.9 | −92.4 | −77.7 | −74.0 |
| Hinokiol−1−one | −79.9 | −87.2 | −87.6 | −80.6 | −81.8 | −70.4 | −86.5 | −76.7 | −81.6 | −97.4 | −82.2 | −77.5 |
| 7β−Hydroxyabieta−8,13−diene−11,12−dione | −76.2 | −77.8 | −94.0 | −83.9 | −79.9 | −76.5 | −91.4 | −84.1 | −81.3 | −95.2 | −86.2 | −83.9 |
| 7α−Hydroxyabieta−8,11,13−triene | −76.8 | −85.3 | −84.5 | −75.4 | −69.1 | −66.7 | −83.0 | −81.3 | −77.8 | −90.9 | −78.7 | −82.0 |
| 14−Hydroxy−7,9(11),13−abietatriene−6,12−dione | −74.8 | −83.5 | −71.0 | −82.1 | −68.6 | −76.5 | −88.3 | −76.2 | −78.6 | −94.8 | −84.7 | −85.5 |
| 11−Hydroxy−7,9(11),13−abietatrien−12−one | −72.5 | −82.5 | −80.1 | −82.3 | −75.3 | −72.1 | −83.7 | −73.6 | −82.4 | −95.9 | −74.1 | −80.4 |
| 12−Hydroxy−8,12−abietadiene−3,11,14−trione | −75.9 | −86.0 | −75.2 | −79.9 | −83.7 | −73.9 | −90.3 | −84.7 | −94.2 | −85.3 | −79.6 | −86.0 |
| 1β−Hydroxycryptotanshinone | −74.7 | −96.0 | −70.0 | −80.8 | −70.1 | −79.2 | −99.1 | −83.8 | −84.0 | −104.4 | −90.0 | −78.5 |
| 7−Hydroxy−12−methoxy−20−
nor−abieta−1,5(10),7,9,12−pentaen−6,14−dione | −66.6 | −91.6 | −75.4 | −80.9 | −72.5 | −79.2 | −92.5 | −82.4 | −83.6 | −94.2 | −83.7 | −81.5 |
| 14−Hydroxy−6−oxoferruginol | −56.1 | −91.1 | −72.3 | −85.5 | −75.7 | −73.9 | −87.0 | −79.5 | −83.6 | −90.3 | −75.2 | −79.3 |
| 6−Hydroxysalvinolone | −77.1 | −83.3 | −76.6 | −76.2 | −77.6 | −72.4 | −82.3 | −81.2 | −82.7 | −93.7 | −87.1 | −86.5 |
| Komarovinone A | −82.0 | −82.4 | −79.2 | −73.1 | −72.7 | −67.1 | −96.1 | −83.5 | −84.6 | −100.0 | −77.6 | −84.0 |
| 1−Oxocryptotanshinone | −75.8 | −102.9 | −74.7 | −77.9 | −70.6 | −75.4 | −96.6 | −79.8 | −82.4 | −114.6 | −89.3 | −78.0 |
| 1−Oxomiltirone | −71.6 | −84.7 | −78.4 | −77.8 | −71.6 | −76.8 | −90.3 | −82.8 | −79.8 | −104.9 | −82.4 | −79.1 |
| Royleanone | −73.5 | −84.0 | −72.2 | −81.9 | −79.6 | −72.3 | −90.5 | −82.7 | −91.0 | −87.9 | −80.5 | −83.9 |
| Sugiol | −67.6 | −90.5 | −82.4 | −73.2 | −81.5 | −53.1 | −76.2 | −84.0 | −77.2 | −97.3 | −84.0 | −84.4 |
| Taxodione | −79.0 | −87.2 | −77.8 | −83.5 | −75.7 | −72.3 | −85.6 | −78.3 | −69.0 | −90.7 | −82.0 | −80.4 |
| 6,11,12,16−Tetrahydroxy−5,8,11,13−abietatetraen−7−one | −78.2 | −88.6 | −81.5 | −82.9 | −80.2 | −73.3 | −95.1 | −87.8 | −88.3 | −93.8 | −83.4 | −90.7 |
| 6,12,14−Trihydroxyabieta−5,8,11,13−tetraen−7−one | −69.6 | −86.1 | −70.6 | −78.7 | −73.9 | −80.8 | −89.4 | −84.6 | −81.5 | −97.4 | −85.9 | −83.5 |
| 4−
epi−Triptobenzene L | −68.9 | −94.9 | −89.9 | −81.7 | −79.6 | −76.6 | −87.6 | −85.9 | −83.3 | −88.2 | −82.4 | −82.4 |
| Uncinatone | −84.3 | −81.8 | −81.6 | −85.9 | −62.1 | −74.5 | −86.5 | −86.5 | −108.8 | −107.7 | −95.6 | −77.6 |
Table 17.
MolDock docking energies (kJ/mol) of abietane diterpenoids with Leishmania donovani and L. mexicana protein targets.
Table 17.
MolDock docking energies (kJ/mol) of abietane diterpenoids with Leishmania donovani and L. mexicana protein targets.
| Abietane diterpenoids | LdonCatB | LdonCyp | LdonDHODH | LdonNMT | LmexGAPDH | LmexGPDH | LmexPGI | LmexPMM | LmexPYK | LmexPYK | LmexPYK | LmexTIM |
|---|
| | | | | | | | Site 1 | Site 2 | Site 3 | |
|---|
| Abieta−7,13−diene | −73.0 | −84.2 | −70.8 | −69.7 | −69.1 | −92.6 | −62.7 | −76.6 | −81.3 | −71.3 | −74.2 | −62.9 |
| ar−Abietatriene−12,16−diol−14,16−oxide | −77.9 | −84.2 | −70.5 | −74.8 | −73.7 | −104.7 | −74.6 | −82.6 | −104.4 | −83.5 | −84.9 | −73.1 |
| ar−Abietatrien−12−ol−6,7−dione−14,16−oxide | −76.3 | −90.1 | −75.9 | −74.2 | −69.1 | −102.3 | −68.9 | −93.5 | −99.2 | −87.3 | −90.1 | −72.1 |
| epi−Abietic acid | −77.4 | −86.4 | −67.6 | −78.1 | −60.9 | −97.6 | −67.8 | −81.5 | −84.0 | −74.5 | −91.0 | −68.6 |
| 4−
epi−Abietol | −77.2 | −88.0 | −69.7 | −69.3 | −68.1 | −94.0 | −66.3 | −78.8 | −78.0 | −75.2 | −72.6 | −65.9 |
| Cryptotanshinone | −77.5 | −80.5 | −84.5 | −78.3 | −66.6 | −101.9 | −71.1 | −84.4 | −95.4 | −75.3 | −71.7 | −65.3 |
| 12−
O−Deacetyl−6−O−acetyl−18−acetyloxycoleon Q | −78.6 | −93.4 | −41.3 | −79.5 | −86.6 | −108.4 | −87.4 | −109.1 | −102.6 | −110.1 | −102.8 | −86.7 |
| 12−
O−Deacetyl−6−O−acetyl−19−acetyloxycoleon Q | −99.6 | −80.4 | −45.8 | −75.4 | −90.6 | −105.1 | −87.4 | −109.7 | −101.1 | −96.2 | −88.7 | −73.6 |
| 12−Deoxyroyleanone | −80.4 | −86.7 | −72.1 | −73.1 | −68.2 | −99.6 | −62.7 | −82.7 | −84.0 | −73.0 | −87.4 | −68.4 |
| 9α,13α−
epi−Dioxyabiet−8(14)−en−18−oic acid | −59.0 | −88.1 | −51.5 | −64.0 | −71.2 | −100.3 | −69.3 | −86.9 | −82.4 | −77.1 | −85.3 | −76.1 |
| 9α,13α−
epi−Dioxyabiet−8(14)en−18−ol | −55.8 | −76.8 | −58.2 | −65.3 | −63.2 | −96.6 | −66.9 | −81.2 | −79.7 | −71.6 | −74.1 | −70.9 |
| Dracocephalone A | −74.3 | −81.2 | −73.4 | −66.7 | −69.5 | −103.5 | −64.4 | −82.2 | −84.9 | −74.8 | −81.2 | −68.1 |
| Dracocequinone A | −71.2 | −83.6 | −77.7 | −75.4 | −66.1 | −104.7 | −66.2 | −82.5 | −84.7 | −72.0 | −79.9 | −68.9 |
| Dracocequinone B | −75.7 | −81.6 | −72.0 | −76.6 | −67.4 | −107.2 | −67.7 | −83.7 | −82.8 | −72.2 | −77.3 | −76.7 |
| Ferruginol | −70.5 | −91.1 | −75.4 | −71.3 | −71.2 | −96.7 | −75.1 | −83.7 | −82.9 | −75.3 | −76.1 | −69.8 |
| Hinokiol | −77.1 | −87.8 | −72.0 | −62.3 | −71.1 | −95.9 | −65.9 | −79.3 | −81.4 | −71.9 | −73.5 | −63.7 |
| Hinokiol−1−one | −77.0 | −86.1 | −72.7 | −70.4 | −70.9 | −99.6 | −63.7 | −80.2 | −79.7 | −69.7 | −77.2 | −67.6 |
| 7β−Hydroxyabieta−8,13−diene−11,12−dione | −83.8 | −79.8 | −80.7 | −76.5 | −69.0 | −100.8 | −69.4 | −89.6 | −91.6 | −77.9 | −83.9 | −76.8 |
| 7α−Hydroxyabieta−8,11,13−triene | −75.4 | −76.3 | −73.1 | −66.7 | −68.1 | −96.5 | −70.5 | −78.2 | −82.5 | −74.8 | −82.9 | −67.0 |
| 14−Hydroxy−7,9(11),13−abietatriene−6,12−dione | −74.5 | −89.5 | −81.6 | −76.5 | −72.7 | −96.0 | −68.6 | −86.8 | −85.3 | −76.6 | −75.3 | −67.9 |
| 11−Hydroxy−7,9(11),13−abietatrien−12−one | −79.6 | −87.1 | −76.1 | −72.1 | −61.8 | −97.7 | −68.7 | −83.9 | −78.1 | −76.3 | −78.1 | −69.9 |
| 12−Hydroxy−8,12−abietadiene−3,11,14−trione | −79.7 | −82.0 | −75.6 | −73.9 | −66.2 | −109.4 | −68.9 | −92.5 | −88.8 | −75.3 | −84.8 | −71.0 |
| 1b−Hydroxycryptotanshinone | −78.4 | −84.0 | −80.5 | −79.2 | −68.3 | −98.5 | −71.1 | −87.5 | −94.5 | −77.8 | −74.5 | −70.8 |
| 7−hydroxy−12−methoxy−20−
nor−abieta−1,5(10),7,9,12−pentaen−6,14−dione | −75.6 | −87.7 | −69.1 | −79.2 | −67.5 | −102.0 | −68.2 | −87.1 | −88.6 | −68.1 | −79.9 | −65.2 |
| 14−Hydroxy−6−oxoferruginol | −66.5 | −93.7 | −78.2 | −73.9 | −71.7 | −96.9 | −61.8 | −87.2 | −88.7 | −80.8 | −70.6 | −68.5 |
| 6−Hydroxysalvinolone | −78.8 | −78.9 | −77.6 | −72.4 | −62.2 | −94.7 | −66.0 | −89.9 | −94.9 | −84.3 | −75.8 | −69.0 |
| Komarovinone A | −80.3 | −81.2 | −81.2 | −67.1 | −65.0 | −98.3 | −69.2 | −90.1 | −86.5 | −80.4 | −70.8 | −73.4 |
| 1−Oxocryptotanshinone | −76.1 | −80.4 | −71.9 | −75.4 | −66.9 | −95.0 | −69.1 | −85.7 | −89.6 | −79.8 | −77.1 | −65.4 |
| 1−Oxomiltirone | −74.3 | −71.4 | −78.1 | −76.8 | −61.6 | −92.9 | −65.3 | −81.5 | −84.5 | −76.6 | −71.2 | −66.5 |
| Royleanone | −80.1 | −85.8 | −75.6 | −72.3 | −61.3 | −102.2 | −65.3 | −87.9 | −90.8 | −77.4 | −73.0 | −69.1 |
| Sugiol | −73.9 | −78.4 | −77.3 | −53.1 | −65.1 | −95.8 | −66.2 | −85.3 | −86.3 | −79.1 | −71.1 | −71.8 |
| Taxodione | −80.6 | −94.1 | −77.9 | −72.3 | −68.0 | −97.3 | −70.5 | −89.2 | −81.6 | −79.0 | −75.8 | −49.5 |
| 6,11,12,16−Tetrahydroxy−5,8,11,13−abietatetra−en−7−one | −79.0 | −86.0 | −84.9 | −73.3 | −66.9 | −96.1 | −65.9 | −95.6 | −105.1 | −87.5 | −77.6 | −75.5 |
| 6,12,14−Trihydroxyabieta−5,8,11,13−tetraen−7−one | −72.7 | −84.9 | −81.2 | −80.8 | −72.6 | −97.7 | −62.5 | −89.1 | −89.8 | −89.2 | −77.1 | −62.8 |
| 4−
epi−Triptobenzene L | −76.3 | −89.7 | −76.3 | −76.6 | −72.3 | −101.5 | −70.0 | −85.1 | −85.7 | −77.6 | −77.9 | −72.5 |
| Uncinatone | −85.4 | −73.3 | −72.1 | −74.5 | −69.8 | −89.0 | −65.3 | −84.4 | −94.8 | −84.1 | −89.2 | −64.4 |
Table 18.
MolDock docking energies (kJ/mol) of abietane diterpenoids with Leishmania infantum protein targets.
Table 18.
MolDock docking energies (kJ/mol) of abietane diterpenoids with Leishmania infantum protein targets.
| Abietane diterpenoids | LinfCYP51 | LinfGLO2 | LinfPnC1 | LinfTDR1 | LinfTR |
|---|
| Abieta−7,13−diene | −76.4 | −71.1 | no dock | −65.7 | −72.6 |
| ar−Abietatriene−12,16−diol−14,16−oxide | −87.3 | −76.4 | no dock | −74.3 | −78.4 |
| ar−Abietatrien−12−ol−6,7−dione−14,16−oxide | −84.7 | −72.7 | no dock | −76.8 | −83.5 |
| epi−Abietic acid | −83.8 | −67.7 | no dock | −66.4 | −74.5 |
| 4−
epi−Abietol | −78.5 | −68.9 | no dock | −64.6 | −72.4 |
| Cryptotanshinone | −83.6 | −67.0 | no dock | −71.5 | −84.1 |
| 12−
O−Deacetyl−6−O−acetyl−18−acetyloxycoleon Q | −106.3 | −80.7 | no dock | −88.6 | −103.4 |
| 12−
O−Deacetyl−6−O−acetyl−19−acetyloxycoleon Q | −105.1 | −85.8 | no dock | −87.3 | −98.2 |
| 12−Deoxyroyleanone | −81.7 | −72.8 | no dock | −67.2 | −77.2 |
| 9α,13α−
epi−Dioxyabiet−8(14)−en−18−oic acid | −90.4 | −73.1 | no dock | −68.7 | −77.1 |
| 9α,13α−
epi−Dioxyabiet−8(14)en−18−ol | −80.3 | −70.1 | no dock | −65.9 | −72.3 |
| Dracocephalone A | −83.2 | −64.1 | no dock | −76.6 | −82.5 |
| Dracocequinone A | −82.5 | −63.6 | no dock | −77.7 | −74.9 |
| Dracocequinone B | −85.6 | −69.1 | no dock | −76.4 | −75.5 |
| Ferruginol | −78.9 | −71.0 | no dock | −73.2 | −79.0 |
| Hinokiol | −77.7 | −68.5 | no dock | −62.4 | −70.0 |
| Hinokiol−1−one | −84.6 | −69.6 | no dock | −72.9 | −72.4 |
| 7β−Hydroxyabieta−8,13−diene−11,12−dione | −85.1 | −71.6 | no dock | −73.0 | −85.6 |
| 7α−Hydroxyabieta−8,11,13−triene | −79.5 | −68.9 | no dock | −67.6 | −77.1 |
| 14−Hydroxy−7,9(11),13−abietatriene−6,12−dione | −82.9 | −71.7 | no dock | −71.5 | −84.5 |
| 11−Hydroxy−7,9(11),13−abietatrien−12−one | −81.1 | −68.0 | no dock | −69.6 | −73.3 |
| 12−Hydroxy−8,12−abietadiene−3,11,14−trione | −83.4 | −76.4 | no dock | −73.8 | −86.9 |
| 1β−Hydroxycryptotanshinone | −84.1 | −66.5 | no dock | −75.5 | −88.2 |
| 7−Hydroxy−12−methoxy−20−
nor−abieta−1,5(10),7,9,12−pentaen−6,14−dione | −84.6 | −67.7 | no dock | −73.0 | −78.3 |
| 14−Hydroxy−6−oxoferruginol | −85.2 | −73.0 | no dock | −71.7 | −79.3 |
| 6−Hydroxysalvinolone | −81.2 | −68.6 | no dock | −80.7 | −85.5 |
| Komarovinone A | −86.9 | −66.3 | no dock | −74.9 | −84.7 |
| 1−Oxocryptotanshinone | −83.9 | −66.9 | no dock | −72.6 | −84.1 |
| 1−Oxomiltirone | −85.5 | −64.4 | no dock | −69.7 | −78.6 |
| Royleanone | −82.6 | −72.0 | no dock | −72.8 | −82.1 |
| Sugiol | −81.8 | −70.4 | no dock | −72.6 | −78.7 |
| Taxodione | −81.2 | −68.2 | no dock | −71.1 | −75.8 |
| 6,11,12,16−Tetrahydroxy−5,8,11,13−abietatetraen−7−one | −88.5 | −66.5 | no dock | −85.7 | −93.8 |
| 6,12,14−Trihydroxyabieta−5,8,11,13−tetraen−7−one | −83.5 | −73.4 | no dock | −75.9 | −81.9 |
| 4−
epi−Triptobenzene L | −83.0 | −72.3 | no dock | −65.2 | −76.3 |
| Uncinatone | −93.0 | −73.3 | no dock | −82.5 | −85.5 |
Table 19.
MolDock docking energies (kJ/mol) of clerodane diterpenoids with Leishmania major protein targets.
Table 19.
MolDock docking energies (kJ/mol) of clerodane diterpenoids with Leishmania major protein targets.
| Clerodane diterpenoids | LmajCatB | LmajDHODH | LmajdUTPase | LmajNDKb | LmajNH | LdonNMT | LmajOPB | LmajPDE1 | LmajPTR1 | LmajMetRS | LmajTyrRS | LmajUGPase |
|---|
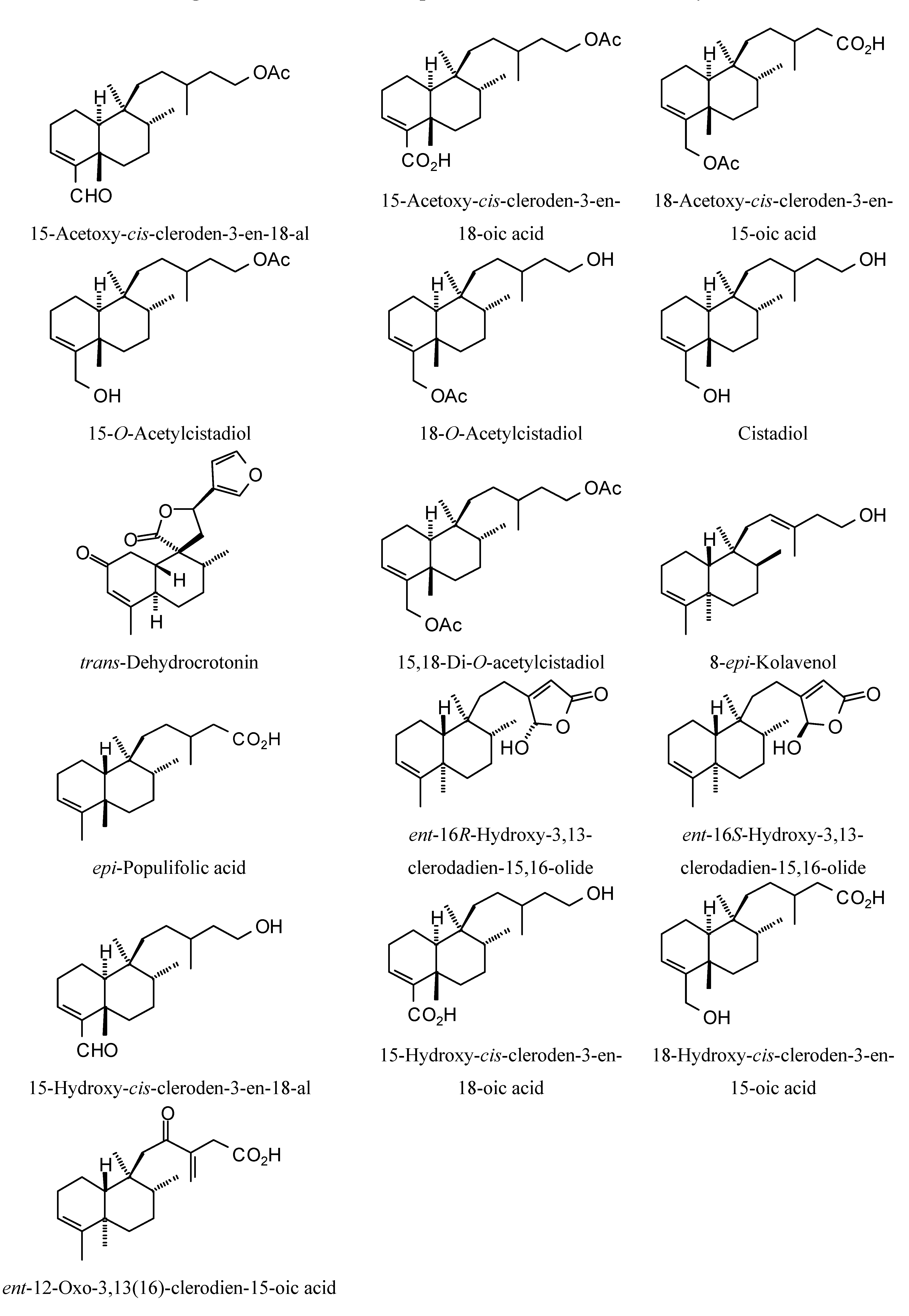
| 15−Acetoxy−cis−cleroden−3−en−18−al | −68.6 | −94.1 | −85.0 | −97.9 | −96.0 | −87.3 | −90.4 | −94.9 | −94.7 | −107.1 | −92.6 | −106.9 |
| 15−Acetoxy−cis−cleroden−3−en−18−oic acid | −75.0 | −103.1 | −88.3 | −97.2 | −92.3 | −87.0 | −91.6 | −95.3 | −93.2 | −97.7 | −94.4 | −97.3 |
| 18−Acetoxy−cis−cleroden−3−en−15−oic acid | −76.9 | −103.7 | −82.7 | −96.7 | −88.8 | −80.7 | −97.3 | −96.8 | −89.3 | −104.2 | −97.5 | −98.2 |
| 15−O−Acetylcistadiol | −68.0 | −94.7 | −87.4 | −97.3 | −94.7 | −80.4 | −96.8 | −89.2 | −92.9 | −106.7 | −95.0 | −95.3 |
| 18−O−Acetylcistadiol | −70.5 | −103.4 | −88.1 | −91.7 | −93.2 | −85.0 | −94.0 | −93.8 | −82.3 | −101.2 | −95.9 | −100.3 |
| Cistadiol | −64.1 | −93.4 | −79.7 | −81.6 | −81.9 | −69.0 | −77.1 | −85.2 | −79.2 | −78.9 | −80.7 | −92.5 |
| trans−Dehydrocrotonin | −78.6 | −114.9 | −90.2 | −91.8 | −84.5 | −75.9 | −89.0 | −89.1 | −99.2 | −115.1 | −97.7 | −92.1 |
| 15,18−Di−O−acetylcistadiol | −77.8 | −100.5 | −92.8 | −109.4 | −94.8 | −84.2 | −90.5 | −107.0 | −91.6 | −117.5 | −100.6 | −110.4 |
| 8−epi−Kolavenol | −71.5 | −94.5 | −79.2 | −80.4 | −85.0 | −72.3 | −78.7 | −80.6 | −82.8 | −92.9 | −85.3 | −95.3 |
| epi−Populifolic acid | −69.9 | −85.5 | −72.7 | −82.3 | −83.4 | −71.7 | −76.3 | −82.2 | −78.3 | −85.0 | −78.2 | −89.9 |
| ent−16R−Hydroxy−3,13−clerodadien−15,16−olide | −77.7 | −108.7 | −86.4 | −88.2 | −87.7 | −88.2 | −90.8 | −87.5 | −101.9 | −105.9 | −104.9 | −97.4 |
| ent−16S−Hydroxy−3,13−clerodadien−15,16−olide | −81.9 | −101.9 | −84.6 | −91.0 | −83.9 | −78.4 | −93.4 | −82.2 | −100.3 | −110.7 | −99.4 | −105.1 |
| 15−Hydroxy−cis−cleroden−3−en−18−al | −66.5 | −91.8 | −77.4 | −81.8 | −86.8 | −76.7 | −76.8 | −83.7 | −77.5 | −92.8 | −80.1 | −91.4 |
| 15−Hydroxy−cis−cleroden−3−en−18−oic acid | −69.0 | −95.8 | −90.1 | −83.5 | −86.9 | −80.5 | −82.1 | −86.0 | −80.4 | −95.9 | −84.3 | −95.2 |
| 18−Hydroxy−cis−cleroden−3−en−15−oic acid | −70.0 | −93.4 | −76.1 | −86.3 | −86.8 | −76.2 | −79.7 | −87.2 | −81.1 | −96.1 | −82.9 | −97.4 |
| ent−12−Oxo−3,13(16)−clerodien−15−oic acid | −75.0 | −112.7 | −86.3 | −90.0 | −93.2 | −88.2 | −87.7 | −83.0 | −96.5 | −107.2 | −87.6 | −110.5 |
Table 20.
MolDock docking energies (kJ/mol) of clerodane diterpenoids with Leishmania donovani and L. mexicana protein targets.
Table 20.
MolDock docking energies (kJ/mol) of clerodane diterpenoids with Leishmania donovani and L. mexicana protein targets.
| Clerodane diterpenoids | LdonCatB | LdonCyp | LdonDHODH | LdonNMT | LmexGAPDH | LmexGPDH | LmexPGI | LmexPMM | LmexPYK | LmexPYK | LmexPYK | LmexTIM |
|---|
| | | | | | | | Site 1 | Site 2 | Site 3 | |
|---|
| 15−Acetoxy−cis−cleroden−3−en−18−al | −80.4 | −84.1 | −75.1 | −87.3 | −76.7 | −107.2 | −75.4 | −101.2 | −96.3 | −96.5 | −89.3 | −84.6 |
| 15−Acetoxy−cis−cleroden−3−en−18−oic acid | −83.5 | −85.0 | −75.2 | −87.0 | −86.3 | −93.6 | −84.4 | −104.8 | −100.2 | −92.9 | −90.4 | −80.9 |
| 18−Acetoxy−cis−cleroden−3−en−15−oic acid | −82.9 | −90.6 | −85.1 | −80.7 | −77.3 | −109.2 | −77.6 | −97.4 | −95.9 | −90.8 | −89.5 | −95.5 |
| 15−O−Acetylcistadiol | −82.8 | −79.6 | −72.7 | −80.4 | −77.4 | −102.7 | −77.8 | −98.9 | −96.3 | −96.8 | −87.7 | −89.7 |
| 18−O−Acetylcistadiol | −79.8 | −92.7 | −66.9 | −85.0 | −73.2 | −105.6 | −78.9 | −97.1 | −92.5 | −96.8 | −92.6 | −98.6 |
| Cistadiol | −66.6 | −81.0 | −58.3 | −69.0 | −70.5 | −91.1 | −70.4 | −88.0 | −90.4 | −82.8 | −82.5 | −80.2 |
| trans−Dehydrocrotonin | −92.8 | −98.7 | −51.3 | −75.9 | −73.0 | −100.5 | −77.1 | −94.2 | −107.8 | −87.6 | −92.4 | −95.2 |
| 15,18−Di−O−acetylcistadiol | −82.2 | −82.4 | −94.8 | −84.2 | −85.2 | −122.2 | −86.7 | −104.1 | −102.9 | −101.7 | −97.4 | −99.1 |
| 8−epi−Kolavenol | −75.5 | −83.0 | −77.8 | −72.3 | −71.7 | −100.8 | −70.6 | −85.6 | −92.1 | −84.2 | −94.6 | −78.9 |
| epi−Populifolic acid | −68.3 | −75.8 | −63.4 | −71.7 | −73.8 | −89.4 | −69.7 | −88.7 | −91.1 | −79.3 | −85.0 | −82.4 |
| ent−16R−Hydroxy−3,13−clerodadien−15,16−olide | −74.7 | −89.1 | −82.9 | −88.2 | −85.4 | −105.8 | −79.8 | −96.5 | −103.1 | −89.0 | −95.5 | −83.1 |
| ent−16S−Hydroxy−3,13−clerodadien−15,16−olide | −88.6 | −91.8 | −82.7 | −78.4 | −83.9 | −100.1 | −81.3 | −97.2 | −102.0 | −92.3 | −90.3 | −85.8 |
| 15−Hydroxy−cis−cleroden−3−en−18−al | −77.5 | −87.9 | −67.4 | −76.7 | −74.1 | −97.2 | −70.9 | −87.6 | −96.8 | −86.4 | −85.4 | −82.4 |
| 15−Hydroxy−cis−cleroden−3−en−18−oic acid | −71.4 | −86.4 | −66.4 | −80.5 | −74.9 | −99.5 | −73.3 | −85.9 | −99.3 | −88.1 | −88.5 | −80.5 |
| 18−Hydroxy−cis−cleroden−3−en−15−oic acid | −72.5 | −84.3 | −59.0 | −76.2 | −78.2 | −95.4 | −73.2 | −90.2 | −89.2 | −83.9 | −90.5 | −88.0 |
| ent−12−Oxo−3,13(16)−clerodien−15−oic acid | −84.3 | −95.2 | −72.0 | −88.2 | −85.3 | −88.0 | −76.6 | −95.9 | −108.8 | −88.3 | −91.6 | −86.9 |
Table 21.
MolDock docking energies (kJ/mol) of clerodane and labdane diterpenoids with Leishmania infantum protein targets.
Table 21.
MolDock docking energies (kJ/mol) of clerodane and labdane diterpenoids with Leishmania infantum protein targets.
| Clerodane diterpenoids | LinfCYP51 | LinfGLO2 | LinfPnC1 | LinfTDR1 | LinfTR |
|---|
| 15−Acetoxy−cis−cleroden−3−en−18−al | −96.8 | −78.2 | no dock | −90.4 | −91.8 |
| 15−Acetoxy−cis−cleroden−3−en−18−oic acid | −93.0 | −82.7 | no dock | −87.6 | −91.7 |
| 18−Acetoxy−cis−cleroden−3−en−15−oic acid | −98.2 | −78.1 | no dock | −87.6 | −97.6 |
| 15−O−Acetylcistadiol | −101.7 | −89.8 | no dock | −88.4 | −94.3 |
| 18−O−Acetylcistadiol | −95.8 | −78.3 | no dock | −88.4 | −93.8 |
| Cistadiol | −86.8 | −68.7 | no dock | −78.3 | −82.2 |
| trans−Dehydrocrotonin | −89.6 | −74.5 | no dock | −77.6 | −88.9 |
| 15,18−Di−O−acetylcistadiol | −95.6 | −85.8 | no dock | −89.2 | −103.5 |
| 8−epi−Kolavenol | −83.6 | −72.3 | −18.2 | −80.5 | −78.8 |
| epi−Populifolic acid | −83.8 | −71.4 | no dock | −75.9 | −77.0 |
| ent−16R−Hydroxy−3,13−clerodadien−15,16−olide | −87.9 | −75.1 | −22.6 | −83.9 | −87.5 |
| ent−16S−Hydroxy−3,13−clerodadien−15,16−olide | −88.4 | −67.5 | no dock | −79.6 | −78.8 |
| 15−Hydroxy−cis−cleroden−3−en−18−al | −83.0 | −68.9 | no dock | −81.0 | −81.2 |
| 15−Hydroxy−cis−cleroden−3−en−18−oic acid | −89.4 | −66.8 | no dock | −78.4 | −78.1 |
| 18−Hydroxy−cis−cleroden−3−en−15−oic acid | −87.5 | −69.6 | −31.7 | −82.8 | −81.3 |
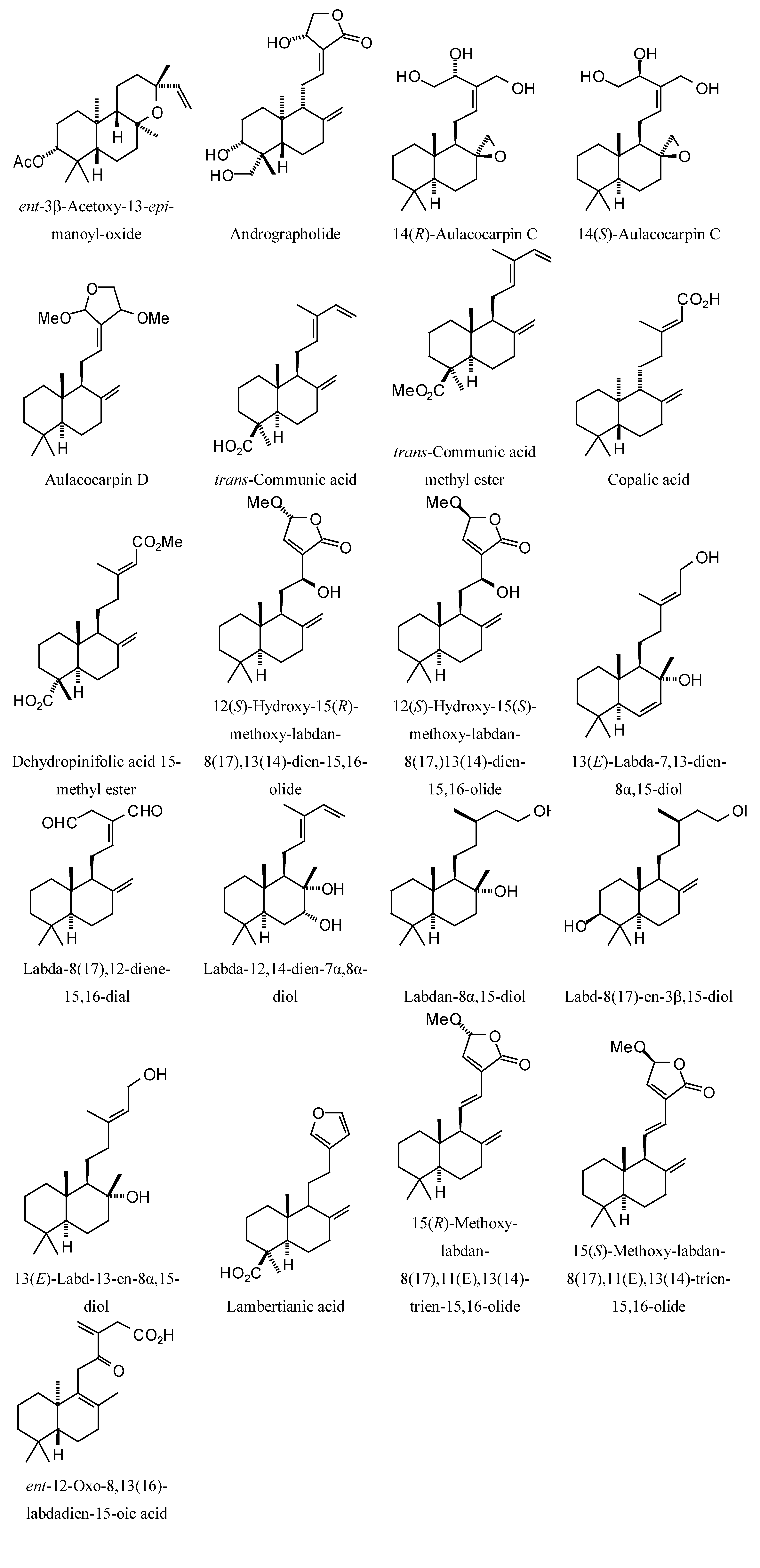
| Labdane diterpenoids | | | | | |
| ent−3β−Acetoxy−13−epi−manoyl−oxide | −94.3 | −72.4 | no dock | −67.4 | −88.0 |
| Andrographolide | −97.9 | −80.4 | no dock | −81.1 | −91.9 |
| 14(R)−Aulacocarpin C | | | | | |
| 14(S)−Aulacocarpin C | −93.1 | −81.7 | no dock | −81.7 | −92.8 |
| Aulacocarpin D | −92.7 | −74.5 | no dock | −85.8 | −93.5 |
| trans−Communic acid | −90.8 | −75.0 | no dock | −82.6 | −91.6 |
| trans−Communic acid methyl ester | −84.3 | −77.2 | no dock | −76.0 | −82.7 |
| Copalic acid | −89.8 | −74.8 | no dock | −72.8 | −86.8 |
| Dehydropinifolic acid 15−methyl ester | −93.9 | −84.1 | no dock | −78.1 | −83.6 |
| 12(S)−Hydroxy−15(R)−methoxy−labdan−8(17),13(14)−dien−15,16−olide | −93.3 | −84.8 | no dock | −89.8 | −93.5 |
| 12(S)−Hydroxy−15(S)−methoxy−labdan−8(17,)13(14)−dien−15,16−olide | −99.3 | −88.5 | no dock | −88.8 | −95.1 |
| Labda−8(17),12−diene−15,16−dial | −94.2 | −84.8 | no dock | −89.2 | −100.2 |
| 13(E)−Labda−7,13−dien−8α,15−diol | −87.7 | −76.3 | no dock | −81.7 | −86.6 |
| Labda−12,14−dien−7α,8α−diol | −89.1 | −77.8 | no dock | −79.8 | −88.4 |
| Labdan−8α,15−diol | −85.9 | −77.1 | no dock | −76.1 | −87.2 |
| Labd−8(17)−en−3β,15−diol | −85.8 | −77.4 | no dock | −82.0 | −90.2 |
| 13(E)−Labd−13−en−8α,15−diol | −84.2 | −76.9 | no dock | −79.7 | −87.2 |
| Lambertianic acid | −87.0 | −75.4 | no dock | −78.4 | −83.4 |
| 15(R)−Methoxy−labdan−8(17),11(E),13(14)−trien−15,16−olide | −90.0 | −73.6 | no dock | −72.5 | −89.3 |
| 15(S)−Methoxy−labdan−8(17),11(E),13(14)−trien−15,16−olide | −94.2 | −87.8 | no dock | −88.3 | −92.3 |
| ent−12−Oxo−8,13(16)−labdadien−15−oic acid | −96.0 | −86.3 | no dock | −85.3 | −96.8 |
| ent−3β−Acetoxy−13−epi−manoyl−oxide | −94.1 | −77.8 | no dock | −78.5 | −88.9 |
Table 22.
MolDock docking energies (kJ/mol) of labdane diterpenoids with Leishmania major protein targets.
Table 22.
MolDock docking energies (kJ/mol) of labdane diterpenoids with Leishmania major protein targets.
| Labdane diterpenoids | LmajCatB | LmajDHODH | LmajdUTPase | LmajNDKb | LmajNH | LdonNMT | LmajOPB | LmajPDE1 | LmajPTR1 | LmajMetRS | LmajTyrRS | LmajUGPase |
|---|
| ent−3β−Acetoxy−13−epi−manoyl−oxide | −74.9 | −96.4 | −76.3 | −82.2 | −82.7 | −78.7 | −90.7 | −82.0 | −89.0 | −85.9 | −80.1 | −91.2 |
| Andrographolide | −86.7 | −114.6 | −90.5 | −92.1 | −96.5 | −101.8 | −98.5 | −94.4 | −101.6 | −114.1 | −97.7 | −95.9 |
| 14(R)−Aulacocarpin C | −74.4 | −116.1 | −85.2 | −91.7 | −97.7 | −99.7 | −95.7 | −85.2 | −98.1 | −107.0 | −91.8 | −103.3 |
| 14(S)−Aulacocarpin C | −83.6 | −114.7 | −88.3 | −90.3 | −91.7 | −97.4 | −90.6 | −83.1 | −99.9 | −107.9 | −94.0 | −100.3 |
| Aulacocarpin D | −80.8 | −108.3 | −89.0 | −87.3 | −87.3 | −86.0 | −91.8 | −98.6 | −92.9 | −108.3 | −93.8 | −98.9 |
| trans−Communic acid | −69.5 | −109.4 | −86.3 | −86.7 | −87.5 | −89.7 | −83.7 | −84.0 | −88.4 | −97.1 | −88.2 | −88.3 |
| trans−Communic acid methyl ester | −81.3 | −103.5 | −84.7 | −86.3 | −85.6 | −84.1 | −85.7 | −83.0 | −92.3 | −99.6 | −89.8 | −88.7 |
| Copalic acid | −69.5 | −102.7 | −88.4 | −89.5 | −83.2 | −83.9 | −86.0 | −96.2 | −90.1 | −105.1 | −93.9 | −104.6 |
| Dehydropinifolic acid 15−methyl ester | −87.3 | −116.3 | −102.2 | −90.1 | −87.9 | −91.3 | −101.0 | −89.8 | −89.7 | −104.1 | −99.8 | −103.3 |
| 12(S)−Hydroxy−15(R)−methoxy−labdan−8(17),13(14)−dien−15,16−olide | −85.8 | −114.5 | −87.2 | −97.5 | −88.7 | −93.8 | −96.4 | −88.0 | −96.1 | −108.9 | −103.0 | −110.1 |
| 12(S)−Hydroxy−15(S)−methoxy−labdan−8(17,)13(14)−dien−15,16−olide | −90.2 | −112.6 | −89.4 | −98.7 | −98.8 | −84.0 | −107.3 | −89.1 | −91.2 | −110.4 | −103.6 | −111.4 |
| Labda−8(17),12−diene−15,16−dial | −77.5 | −107.2 | −87.2 | −82.9 | −93.0 | −95.8 | −82.8 | −89.0 | −97.5 | −103.2 | −95.5 | −102.8 |
| 13(E)−Labda−7,13−dien−8α,15−diol | −76.9 | −100.6 | −85.8 | −87.0 | −86.3 | −89.7 | −100.7 | −85.8 | −87.9 | −107.5 | −91.0 | −90.8 |
| Labda−12,14−dien−7α,8α−diol | −71.9 | −99.4 | −76.2 | −75.8 | −89.8 | −89.8 | −90.5 | −85.1 | −81.5 | −101.3 | −87.8 | −94.7 |
| Labdan−8α,15−diol | −78.9 | −101.4 | −84.6 | −81.5 | −80.3 | −92.1 | −93.7 | −82.7 | −86.0 | −108.6 | −83.9 | −92.3 |
| Labd−8(17)−en−3β,15−diol | −75.5 | −106.7 | −82.2 | −82.9 | −80.6 | −71.0 | −88.0 | −81.0 | −93.9 | −104.9 | −85.9 | −90.8 |
| 13(E)−Labd−13−en−8α,15−diol | −69.4 | −105.0 | −82.1 | −83.3 | −85.1 | −89.1 | −88.5 | −85.8 | −88.9 | −107.8 | −89.8 | −92.2 |
| Lambertianic acid | −69.8 | −110.5 | −86.3 | −82.0 | −84.0 | −79.8 | −81.8 | −83.9 | −93.5 | −100.7 | −87.7 | −88.9 |
| 15(R)−Methoxy−labdan−8(17),11(E),13(14)−trien−15,16−olide | −88.3 | −111.4 | −84.5 | −97.3 | −91.3 | −91.7 | −104.9 | −94.4 | −91.8 | −101.5 | −95.0 | −98.9 |
| 15(S)−Methoxy−labdan−8(17),11(E),13(14)−trien−15,16−olide | −83.1 | −104.7 | −84.8 | −89.0 | −95.4 | −88.5 | −109.2 | −98.3 | −99.0 | −106.2 | −98.3 | −97.3 |
| ent−12−Oxo−8,13(16)−labdadien−15−oic acid | −78.0 | −107.7 | −93.4 | −92.3 | −88.2 | −92.1 | −90.6 | −90.0 | −94.1 | −100.0 | −97.0 | −100.4 |
Table 23.
MolDock docking energies (kJ/mol) of labdane diterpenoids with Leishmania donovani and L. mexicana protein targets.
Table 23.
MolDock docking energies (kJ/mol) of labdane diterpenoids with Leishmania donovani and L. mexicana protein targets.
| LdonCatB | LdonCyp | LdonDHODH | LdonNMT | LmexGAPDH | LmexGPDH | LmexPGI | LmexPMM | LmexPYK | LmexPYK | LmexPYK | LmexTIM |
|---|
| Labdane diterpenoids | | | | | | | | | Site 1 | Site 2 | Site 3 | |
|---|
| ent−3β−Acetoxy−13−epi−manoyl−oxide | −83.3 | −77.6 | −60.4 | −78.7 | −67.9 | −94.4 | −69.1 | −86.9 | −90.2 | −84.0 | −77.7 | −78.3 |
| Andrographolide | −88.9 | −93.2 | −94.9 | −101.8 | −86.1 | −114.0 | −79.6 | −100.7 | −110.3 | −100.3 | −107.0 | −95.5 |
| 14(R)−Aulacocarpin C | −87.0 | −98.3 | −92.5 | −99.7 | −78.4 | −103.3 | −76.8 | −95.7 | −99.6 | −91.7 | −114.0 | −84.2 |
| 14(S)−Aulacocarpin C | −85.6 | −95.6 | −90.3 | −97.4 | −75.0 | −100.7 | −77.8 | −90.6 | −97.1 | −95.8 | −109.0 | −86.7 |
| Aulacocarpin D | −96.6 | −88.1 | −75.2 | −86.0 | −87.6 | −99.9 | −73.6 | −87.5 | −95.9 | −89.8 | −88.3 | −87.8 |
| trans−Communic acid | −77.9 | −87.9 | −79.6 | −89.7 | −70.6 | −100.8 | −75.8 | −88.3 | −92.3 | −82.9 | −98.3 | −83.1 |
| trans−Communic acid methyl ester | −85.8 | −90.8 | −81.9 | −84.1 | −73.5 | −104.8 | −72.2 | −88.1 | −95.4 | −82.4 | −90.0 | −85.2 |
| Copalic acid | −89.0 | −92.6 | −85.1 | −83.9 | −73.8 | −90.3 | −76.9 | −92.0 | −94.8 | −88.0 | −92.4 | −97.5 |
| Dehydropinifolic acid 15−methyl ester | −85.8 | −95.2 | −88.4 | −91.3 | −84.5 | −123.2 | −85.7 | −100.6 | −111.6 | −93.2 | −104.2 | −99.0 |
| 12(S)−Hydroxy−15(R)−methoxy−labdan−8(17),13(14)−dien−15,16−olide | −92.1 | −93.8 | −90.8 | −93.8 | −75.2 | −117.2 | −79.4 | −101.2 | −117.2 | −96.5 | −104.7 | −93.8 |
| 12(S)−Hydroxy−15(S)−methoxy−labdan−8(17,)13(14)−dien−15,16−olide | −95.4 | −92.4 | −82.7 | −84.0 | −88.1 | −116.2 | −91.2 | −103.1 | −117.1 | −92.4 | −109.7 | −93.1 |
| Labda−8(17),12−diene−15,16−dial | −90.6 | −93.4 | −85.7 | −95.8 | −75.4 | −97.3 | −72.3 | −93.0 | −97.0 | −87.4 | −99.5 | −85.4 |
| 13(E)−Labda−7,13−dien−8α,15−diol | −88.4 | −82.3 | −80.9 | −89.7 | −76.3 | −109.7 | −73.5 | −87.1 | −100.2 | −82.1 | −93.7 | −88.8 |
| Labda−12,14−dien−7α,8α−diol | −86.9 | −83.8 | −80.5 | −89.8 | −77.9 | −86.3 | −78.7 | −89.2 | −100.4 | −85.5 | −91.2 | −91.6 |
| Labdan−8α,15−diol | −86.8 | −82.0 | −82.5 | −92.1 | −73.2 | −94.9 | −76.4 | −95.9 | −93.7 | −86.0 | −90.8 | −88.8 |
| Labd−8(17)−en−3β,15−diol | −78.5 | −85.4 | −86.9 | −71.0 | −71.0 | −87.8 | −72.0 | −93.5 | −97.3 | −81.6 | −93.8 | −86.2 |
| 13(E)−Labd−13−en−8α,15−diol | −81.4 | −82.1 | −77.1 | −89.1 | −71.9 | −104.1 | −83.4 | −88.7 | −99.2 | −84.5 | −92.7 | −92.0 |
| Lambertianic acid | −82.2 | −82.9 | −81.6 | −79.8 | −72.9 | −109.5 | −75.1 | −95.1 | −96.2 | −85.6 | −98.4 | −80.3 |
| 15(R)−Methoxy−labdan−8(17),11(E),13(14)−trien−15,16−olide | −106.6 | −91.2 | −92.5 | −91.7 | −79.3 | −121.2 | −76.7 | −97.1 | −105.9 | −94.3 | −95.6 | −91.2 |
| 15(S)−Methoxy−labdan−8(17),11(E),13(14)−trien−15,16−olide | −92.3 | −92.2 | −93.3 | −88.5 | −87.3 | −115.8 | −78.5 | −94.5 | −101.8 | −97.0 | −89.2 | −91.5 |
| ent−12−Oxo−8,13(16)−labdadien−15−oic acid | −76.2 | −84.3 | −84.0 | −92.1 | −89.2 | −99.0 | −72.6 | −102.2 | −105.2 | −91.1 | −103.5 | −91.8 |
Table 24.
MolDock docking energies (kJ/mol) of kaurane and pimarane diterpenoids with Leishmania major protein targets.
Table 24.
MolDock docking energies (kJ/mol) of kaurane and pimarane diterpenoids with Leishmania major protein targets.
| Kaurane diterpenoids | LmajCatB | LmajDHODH | LmajdUTPase | LmajNDKb | LmajNH | LdonNMT | LmajOPB | LmajPDE1 | LmajPTR1 | LmajMetRS | LmajTyrRS | LmajUGPase |
|---|
| 15−Angeloyl−4α,15β−kaur−16−en−18−oic acid | −88.6 | −116.3 | −75.8 | −97.5 | −97.3 | −78.2 | −85.5 | −98.6 | −97.4 | −116.0 | −91.4 | −106.3 |
| ent−11α−Hydroxy−16−kauren−15−one | −64.3 | −98.8 | −69.7 | −76.7 | −83.3 | −64.7 | −87.8 | −83.0 | −74.0 | −85.7 | −75.1 | −81.0 |
| Kaurenoic acid | −70.4 | −95.2 | −77.8 | −76.3 | −81.4 | −58.4 | −86.5 | −80.6 | −89.7 | −97.5 | −75.5 | −83.2 |
| Perymenic acid | −97.6 | −115.4 | −75.2 | −95.4 | −103.0 | −81.2 | −88.9 | −99.6 | −98.5 | −118.4 | −91.2 | −103.7 |
| ent−15β−Senecioyloxy−16,17−epoxy−kauran−18−oic acid | −97.3 | −120.5 | −76.6 | −93.6 | −102.4 | −87.8 | −100.7 | −100.8 | −86.6 | −111.7 | −93.3 | −104.6 |
| Pimarane diterpenoids | | | | | | | | | | | | |
| Acanthoic acid | −66.0 | −97.0 | −73.1 | −84.0 | −86.6 | −72.3 | −87.9 | −81.1 | −78.7 | −87.9 | −72.7 | −82.5 |
| 7β−Hydroxy−
ent−pimara−8(14),15−dien−19−oic acid | −74.0 | −93.7 | −79.3 | −84.3 | −87.3 | −71.7 | −92.6 | −87.7 | −76.7 | −87.6 | −78.9 | −81.2 |
| ent−15−Pimarene−8β,19−diol | −72.1 | −97.1 | −69.0 | −78.0 | −86.3 | −66.1 | −87.4 | −79.1 | −77.5 | −84.4 | −74.3 | −85.5 |
| ent−8(14),15−Pimaradien−3β−acetoxy | −72.5 | −85.3 | −83.9 | −90.3 | −89.4 | −71.7 | −88.6 | −82.1 | −83.7 | −99.7 | −88.9 | −92.1 |
| ent−8(14),15−Pimaradien−3β,19−diol | −66.9 | −90.8 | −66.2 | −78.3 | −91.3 | −71.5 | −83.3 | −80.0 | −75.0 | −86.6 | −80.9 | −81.6 |
| ent−Pimara−8(14),15−dien−19−oic acid | −65.8 | −91.7 | −69.9 | −81.3 | −87.1 | −70.6 | −89.3 | −80.2 | −81.5 | −85.7 | −75.7 | −77.4 |
| ent−8(14),15−Pimaradien−3β−ol | −64.7 | −89.0 | −66.5 | −77.7 | −86.2 | −66.6 | −82.3 | −76.4 | −73.8 | −79.4 | −81.8 | −76.4 |
Table 25.
MolDock docking energies (kJ/mol) of kaurane and pimarane diterpenoids with Leishmania donovani and L. mexicana protein targets.
Table 25.
MolDock docking energies (kJ/mol) of kaurane and pimarane diterpenoids with Leishmania donovani and L. mexicana protein targets.
| LdonCatB | LdonCyp | LdonDHODH | LdonNMT | LmexGAPDH | LmexGPDH | LmexPGI | LmexPMM | LmexPYK | LmexPYK | LmexPYK | LmexTIM |
|---|
| Kaurane diterpenoids | | | | | | | | | Site 1 | Site 2 | Site 3 | |
|---|
| 15−Angeloyl−4α,15β−kaur−16−en−18−oic acid | −97.4 | −78.7 | −27.8 | −78.2 | −91.8 | −121.4 | −75.9 | −96.9 | −104.0 | −89.2 | −92.8 | −77.3 |
| ent−11α−Hydroxy−16−kauren−15−one | −66.2 | −82.3 | no dock | −64.7 | −61.9 | −84.4 | −63.0 | −81.1 | −79.6 | −73.8 | −70.1 | −80.5 |
| Kaurenoic acid | −69.6 | −73.9 | −21.6 | −58.4 | −64.6 | −88.8 | −64.5 | −80.3 | −93.9 | −77.4 | −81.3 | −78.1 |
| Perymenic acid | −100.6 | −92.3 | −45.0 | −81.2 | −85.7 | −128.3 | −76.8 | −100.4 | −99.7 | −95.7 | −91.9 | −80.3 |
| ent−15β−Senecioyloxy−16,17−epoxy−kauran−18−oic acid | −99.1 | −88.1 | −38.0 | −87.8 | −90.1 | −128.9 | −77.2 | −96.5 | −99.3 | −92.9 | −98.0 | −81.1 |
| Pimarane diterpenoids | | | | | | | | | | | | |
| Acanthoic_acid | −69.3 | −79.7 | −62.5 | −72.3 | −59.8 | −90.8 | −69.0 | −81.5 | −92.4 | −79.2 | −78.3 | −79.4 |
| 7β−Hydroxy−
ent−pimara−8(14),15−dien−19−oic acid | −71.2 | −78.8 | −13.1 | −71.7 | −71.7 | −101.0 | −68.7 | −88.0 | −96.6 | −83.5 | −76.9 | −71.3 |
| ent−15−Pimarene−8β,19−diol | −75.9 | −77.9 | −63.4 | −66.1 | −72.6 | −85.2 | −68.6 | −79.0 | −79.7 | −80.1 | −71.6 | −79.0 |
| ent−8(14),15−Pimaradien−3β−acetoxy | −79.6 | −80.5 | −36.8 | −71.7 | −71.4 | −90.2 | −63.2 | −83.7 | −88.7 | −84.4 | −73.5 | −76.4 |
| ent−8(14),15−Pimaradien−3β,19−diol | −67.6 | −73.6 | −31.7 | −71.5 | −65.3 | −90.2 | −69.2 | −83.6 | −95.0 | −79.6 | −73.6 | −82.7 |
| ent−Pimara−8(14),15−dien−19−oic acid | −66.6 | −77.7 | −53.0 | −70.6 | −64.0 | −94.0 | −66.0 | −85.4 | −88.0 | −79.7 | −70.8 | −77.1 |
| ent−8(14),15−Pimaradien−3b−ol | −69.3 | −71.6 | −26.5 | −66.6 | −63.1 | −88.7 | −64.5 | −80.7 | −94.3 | −77.1 | −72.6 | −79.9 |
Table 26.
MolDock docking energies (kJ/mol) of miscellaneous diterpenoids with Leishmania major protein targets.
Table 26.
MolDock docking energies (kJ/mol) of miscellaneous diterpenoids with Leishmania major protein targets.
| LmajCatB | LmajDHODH | LmajdUTPase | LmajNDKb | LmajNH | LdonNMT | LmajOPB | LmajPDE1 | LmajPTR1 | LmajMetRS | LmajTyrRS | LmajUGPase |
|---|
| Cassane diterpenoids | | | | | | | | | | | | |
|---|
| 6β−
O−Cinnamoyl−12−hydroxy−(13)15−en−16,12−olide−18−cassaneoic acid | −78.8 | −123.0 | −110.4 | −111.0 | −119.5 | −108.1 | −107.4 | −108.1 | −120.0 | −134.4 | −115.8 | −116.4 |
| 6α,7β−Diacetoxyvouacapane | −77.8 | −118.7 | −80.7 | −91.0 | −92.6 | −74.7 | −85.6 | −99.8 | −86.5 | −90.4 | −88.3 | −88.0 |
| 6β−
O−2'3'−Dihydrocinnamoyl−12−hydroxy−(13)15−en−16,12−olide−18−cassaneoic acid | −107.0 | −125.0 | −97.1 | −107.3 | −121.6 | −101.3 | −109.0 | −109.0 | −117.3 | −135.2 | −115.1 | −114.3 |
| Icetaxane diterpenoids | | | | | | | | | | | | |
| Cyclocoulterone | −70.3 | −92.1 | −83.7 | −88.6 | −78.7 | −81.3 | −86.7 | −96.1 | −91.6 | −100.2 | −91.8 | −92.0 |
| 5−
epi−Icetexone | −80.9 | −91.0 | −74.2 | −81.6 | −88.3 | −27.2 | −95.7 | −88.5 | −88.5 | −96.9 | −98.8 | −85.9 |
| Komaroviquinone | −75.7 | −83.0 | −87.6 | −84.9 | −75.6 | −76.5 | −90.9 | −98.1 | −97.8 | −105.2 | −87.8 | −87.0 |
| Mulinane diterpenoids | | | | | | | | | | | | |
| Azorellanol | −64.7 | −107.8 | −74.6 | −79.1 | −98.7 | −80.0 | −94.5 | −99.7 | −85.3 | −98.6 | −88.5 | −91.9 |
| 7−Deacetylazorellanol | −72.5 | −98.4 | −72.3 | −80.2 | −79.0 | −72.4 | −88.9 | −91.8 | −76.2 | −94.0 | −90.1 | −78.7 |
| Mulin−11,13−dien−20−oic acid | −83.1 | −103.0 | −78.0 | −84.3 | −80.7 | −71.5 | −87.5 | −83.5 | −87.0 | −100.5 | −88.1 | −88.3 |
| Mulinic acid | −69.4 | −98.9 | −71.9 | −82.4 | −80.7 | −82.3 | −83.6 | −85.7 | −74.6 | −93.7 | −78.3 | −97.0 |
| Mulinolic acid | −75.6 | −94.5 | −87.3 | −82.5 | −84.5 | −85.1 | −84.9 | −90.2 | −81.1 | −102.3 | −92.2 | −88.7 |
| Miscellaneous diterpenoids | | | | | | | | | | | | |
| Hypoestoxide | −74.1 | −117.7 | −88.3 | −80.1 | −87.8 | −85.2 | −94.0 | −97.0 | −78.1 | −81.1 | −99.5 | −99.2 |
| Komarovispirone | −74.1 | −92.9 | −75.8 | −88.9 | −90.5 | −75.5 | −85.4 | −86.4 | −75.3 | −79.9 | −94.0 | −91.4 |
| Sacculatal | −69.5 | −112.7 | −85.2 | −87.1 | −86.1 | −82.4 | −88.0 | −85.9 | −104.4 | −103.5 | −95.2 | −100.4 |
| Serratol | −69.2 | −90.5 | −75.7 | −77.2 | −78.8 | −77.4 | −82.1 | −89.7 | −79.1 | −93.2 | −90.3 | −88.6 |
| Totarol | −62.7 | −96.6 | −64.9 | −85.1 | −76.4 | −69.8 | −89.2 | −80.7 | −74.6 | −97.7 | −77.8 | −93.1 |
Table 27.
MolDock docking energies (kJ/mol) of miscellaneous diterpenoids with Leishmania donovani and L. mexicana protein targets.
Table 27.
MolDock docking energies (kJ/mol) of miscellaneous diterpenoids with Leishmania donovani and L. mexicana protein targets.
| Cassane diterpenoids | LdonCatB | LdonCyp | LdonDHODH | LdonNMT | LmexGAPDH | LmexGPDH | LmexPGI | LmexPMM | LmexPYK | LmexPYK | LmexPYK | LmexTIM |
|---|
| | | | | | | | Site 1 | Site 2 | Site 3 | |
|---|
| 6β−
O−Cinnamoyl−12−hydroxy−(13)15−en−16,12−olide−18−cassaneoic acid | −107.7 | −94.7 | −104.3 | −108.1 | −103.7 | −126.5 | −103.9 | −136.7 | −133.4 | −117.8 | −110.2 | −91.8 |
| 6α,7β−Diacetoxyvouacapane | −83.2 | −82.4 | −78.8 | −74.7 | −77.3 | −99.7 | −86.4 | −103.6 | −102.4 | −86.7 | −85.6 | −81.5 |
| 6β−
O−2'3'−Dihydrocinnamoyl−12−hydroxy−(13)15−en−16,12−olide−18−cassaneoic acid | −108.6 | −100.4 | −52.6 | −101.3 | −97.7 | −125.8 | −92.8 | −140.1 | −130.8 | −118.5 | −109.0 | −102.6 |
| Icetaxane diterpenoids | | | | | | | | | | | | |
| Cyclocoulterone | −89.0 | −82.5 | −71.2 | −81.3 | −78.1 | −117.3 | −70.7 | −87.7 | −98.4 | −87.3 | −83.5 | −79.3 |
| 5−
epi−Icetexone | −90.5 | −80.3 | −85.1 | −27.2 | −75.6 | −99.1 | −68.4 | −89.5 | −88.3 | −78.6 | −80.3 | −80.3 |
| Komaroviquinone | −75.5 | −81.9 | −77.6 | −76.5 | −72.6 | −105.7 | −73.9 | −90.4 | −92.8 | −82.4 | −81.2 | −69.5 |
| Mulinane diterpenoids | | | | | | | | | | | | |
| Azorellanol | −80.6 | −82.6 | −87.2 | −80.0 | −71.4 | −100.2 | −77.7 | −95.1 | −100.2 | −89.9 | −94.0 | −84.8 |
| 7−Deacetylazorellanol | −73.7 | −90.1 | −81.0 | −72.4 | −75.4 | −103.4 | −72.2 | −91.8 | −88.7 | −82.8 | −89.1 | −72.3 |
| Mulin−11,13−dien−20−oic acid | −84.7 | −95.2 | −85.0 | −71.5 | −71.3 | −97.5 | −72.5 | −98.2 | −99.2 | −80.6 | −92.2 | −86.8 |
| Mulinic acid | −63.1 | −93.4 | −72.3 | −82.3 | −70.2 | −98.9 | −63.7 | −85.5 | −94.4 | −77.2 | −87.1 | −83.0 |
| Mulinolic acid | −76.7 | −95.1 | −77.5 | −85.1 | −78.7 | −105.7 | −68.8 | −94.1 | −94.7 | −80.4 | −87.4 | −82.6 |
| Miscellaneous diterpenoids | | | | | | | | | | | | |
| Hypoestoxide | −80.6 | −92.1 | −67.1 | −85.2 | −78.1 | −94.9 | −85.7 | −114.8 | −102.4 | −79.6 | −87.9 | −72.7 |
| Komarovispirone | −77.7 | −80.5 | −67.4 | −75.5 | −70.3 | −101.0 | −77.5 | −90.5 | −92.3 | −86.1 | −80.8 | −79.0 |
| Sacculatal | −86.4 | −84.1 | −84.0 | −82.4 | −77.2 | −103.5 | −78.1 | −92.5 | −104.3 | −87.9 | −98.7 | −79.1 |
| Serratol | −67.7 | −78.7 | −59.5 | −77.4 | −67.0 | −89.9 | −71.4 | −94.3 | −92.3 | −82.9 | −90.1 | −70.1 |
| Totarol | −63.9 | −70.3 | −59.4 | −69.8 | −60.5 | −88.0 | −68.6 | −84.0 | −81.5 | −72.2 | −90.4 | −65.7 |
Table 28.
MolDock docking energies (kJ/mol) of miscellaneous diterpenoids with Leishmania infantum protein targets.
Table 28.
MolDock docking energies (kJ/mol) of miscellaneous diterpenoids with Leishmania infantum protein targets.
| Kaurane diterpenoids | LinfCYP51 | LinfGLO2 | LinfPnC1 | LinfTDR1 | LinfTR |
|---|
| 15−Angeloyl−4α,15β−kaur−16−en−18−oic acid | −97.4 | −64.6 | no dock | −82.4 | −95.6 |
| ent−11α−Hydroxy−16−kauren−15−one | −82.8 | −66.5 | no dock | −66.5 | −76.6 |
| Kaurenoic acid | −79.9 | −65.8 | no dock | −77.4 | −82.6 |
| Perymenic acid | −98.3 | −77.0 | no dock | −83.2 | −97.0 |
| ent−15β−Senecioyloxy−16,17−epoxy−kauran−18−oic acid | −99.6 | −77.9 | no dock | −85.4 | −102.2 |
| Pimarane diterpenoids | | | | | |
| Acanthoic_acid | −83.6 | −64.6 | no dock | −72.4 | −82.6 |
| 7β−Hydroxy−
ent−pimara−8(14),15−dien−19−oic acid | −90.4 | −67.2 | no dock | −75.0 | −79.2 |
| ent−15−Pimarene−8β,19−diol | −84.0 | −63.8 | no dock | −69.9 | −77.4 |
| ent−8(14),15−Pimaradien−3β−acetoxy | −90.6 | −73.3 | no dock | −70.5 | −92.1 |
| ent−8(14),15−Pimaradien−3β,19−diol | −82.2 | −69.2 | no dock | −67.0 | −78.9 |
| ent−Pimara−8(14),15−dien−19−oic acid | −84.2 | −69.0 | no dock | −70.8 | −80.3 |
| ent−8(14),15−Pimaradien−3b−ol | −78.3 | −67.2 | no dock | −66.8 | −79.1 |
| Cassane diterpenoids | | | | | |
| 6β−
O−Cinnamoyl−12−hydroxy−(13)15−en−16,12−olide−18−cassaneoic acid | −125.4 | −91.1 | no dock | −105.3 | −119.8 |
| 6α,7β−Diacetoxyvouacapane | −93.8 | −77.7 | no dock | −83.7 | −93.9 |
| 6β−
O−2'3'−Dihydrocinnamoyl−12−hydroxy−(13)15−en−16,12−olide−18−cassaneoic acid | −124.7 | −88.7 | no dock | −99.0 | −114.1 |
| Icetaxane diterpenoids | | | | | |
| Cyclocoulterone | −90.3 | −68.7 | no dock | −76.9 | −90.0 |
| 5−
epi−Icetexone | −99.1 | −75.8 | no dock | −81.0 | −81.6 |
| Komaroviquinone | −96.8 | −72.4 | no dock | −75.6 | −88.7 |
| Mulinane diterpenoids | | | | | |
| Azorellanol | −99.1 | −72.4 | no dock | −78.2 | −91.2 |
| 7−Deacetylazorellanol | −88.5 | −71.4 | no dock | −76.5 | −85.9 |
| Mulin−11,13−dien−20−oic acid | −87.5 | −72.9 | no dock | −79.1 | −78.6 |
| Mulinic acid | −88.3 | −70.6 | no dock | −73.8 | −84.3 |
| Mulinolic acid | −89.0 | −78.1 | no dock | −78.6 | −85.8 |
| Miscellaneous diterpenoids | | | | | |
| Hypoestoxide | −97.2 | −82.2 | no dock | −73.6 | −91.2 |
| Komarovispirone | −83.1 | −72.0 | no dock | −74.4 | −79.9 |
| Sacculatal | −93.5 | −77.7 | no dock | −84.4 | −88.1 |
| Serratol | −81.7 | −77.1 | no dock | −78.5 | −78.5 |
| Totarol | −76.4 | −67.7 | no dock | −67.9 | −73.4 |
Over 100 antiparasitic diterpenoids were docked into the protein targets in this study. These include abietane-, clerodane-, kaurane-, labdane-, pimarane-, cassane- and mulinane-like diterpenoids. In general, diterpenoids preferentially dock to LmajMetRS, LmajDHODH and LmexGPDH. 12-
O-deacetyl-6-
O-acetyl-18-acetyloxycoleon Q, the antiplasmodial diterpene isolated from the tropical Asia plant
Anisochilus harmandii (Lamiaceae) [
64] has a stronger docking energy (−111.8 kJ/mol) for LmajPDE1 than the co-crystallized competitive phosphodiesterase inhibitor, 3-isobutyl-1-methylxanthine (docking energy = −78.1 kJ/mol). 14(
R)-Aulacocarpin C and 15(
S)-methoxy-labdan-8(17),11(
E),13(14)-trien-15,16-olide are also selective for LmexPYK and LmajOPB, respectively. The cassane-like antileishmanial diterpenes 6β-
O-2
'3
'-dihydrocinnamoyl-12-hydroxy-(13)15-en-16,12-olide-18-cassaneoic acid and 6β-
O-cinnamoyl-12-hydroxy-(13)15-en-16,12-olide-18-cassaneoic acid have stronger docking energies than any of the other docked diterpenoids for most of the protein targets. These higher docking energies correlate with the fact they have higher molecular weights that the other diterpenoids. Despite the molecular weight correlation, 6β-
O-2
'3
'-dihydrocinnamoyl-12-hydroxy-(13)15-en-16,12-olide-18-cassaneoic acid (MW: 496.592) is selective for LmajNMT. The compound’s docking score for LmajNMT is comparable to that of the protein’s co-crystallized pyrazole sulfonamide inhibitor (PDB ID: 4a30; MW: 495.425; docking energy = −121.1 kJ/mol). The lowest energy pose of 6β-
O-2
'3
'-dihydrocinnamoyl-12-hydroxy-(13)15-en-16,12-olide-18-cassaneoic acid is predicted to have extensive interactions with residues Ala 204, Asp 83, Asp 84, Glu 82, Gly 205, Phe 88, Phe 90, Tyr 217, Tyr 345, Val 81 and Val 206. The cassane diterpenoid is predicted to have hydrogen bonding interactions through its carboxylic acid moiety and the ester bond of its cinnamoyl substituent to the backbone carbonyl group of Gly 205, and the side chain phenolic residue of Tyr 217, respectively (
Figure 19). The icetaxane diterpenoids 5-
epi-icetaxone showed preferential docking to
L. infantum sterol 14α-demethylase (LinfCYP51) with a lower docking energy than the co-crystallized ligand, fluconazole (−99.1 and −91.3 kJ/mol, respectively).
Figure 19.
The lowest-energy pose of 6β-O-2'3'-dihydrocinnamoyl-12-hydroxy-(13)15-en-16,12-olide-18-cassaneoic acid and LmajNMT. The hydrogen bonding interactions between the ligand and the protein (Gly 205 and Tyr 217) is shown as blue dash lines. Val 81, Asp 83 and Phe 88 were also predicted to have very strong steric interactions with the ligand.
Figure 19.
The lowest-energy pose of 6β-O-2'3'-dihydrocinnamoyl-12-hydroxy-(13)15-en-16,12-olide-18-cassaneoic acid and LmajNMT. The hydrogen bonding interactions between the ligand and the protein (Gly 205 and Tyr 217) is shown as blue dash lines. Val 81, Asp 83 and Phe 88 were also predicted to have very strong steric interactions with the ligand.
2.4. Triterpenoid Docking
Triterpenoids, including limonoids, withanolides, quassinoids, and steroids are shown in
Figure 20,
Figure 21,
Figure 22,
Figure 23, and
Figure 24. The docking energies for the triterpenoid-based ligands are compiled in
Table 29,
Table 30,
Table 31,
Table 32,
Table 33,
Table 34,
Table 35,
Table 36,
Table 37,
Table 38,
Table 39 and
Table 40. The triterpenoids ligands with the strongest docking energies were the limonoids carapolide A and khayanolide A, and the withanolides 24,25-epoxywithanolide D and withangulatin A. Carapolide A showed significant docking to LmajDHODH (docking energy = −140.0 kJ/mol). Khayanolide A preferentially docked with LmajMetRS and LmexGPDH (docking energies = −138.5 and −137.8 kJ/mol, respectively). The limonoid with the strongest docking was, however, grandifotane with LmajDHODH with a docking energy of −154.4 kJ/mol. 6-
O-Acetylswietenolide docked strongly to LmexGPDH (docking energy = −142.7 kJ/mol). The withanolide with the strongest docking was physagulin F with LmajMetRS, which had a docking energy of −133.2 kJ/mol. As a class, the steroids showed the most target selectivity with six of the seven steroids significantly docking more strongly to LmajMetRS. In general, limonoids showed some selectivity for LmexGPDH and LmajDHODH, while withanolides docked more selectively with LmajUGPase.
Figure 20.
Triterpenoids examined in this work.
Figure 20.
Triterpenoids examined in this work.
Figure 21.
Steroids examined in this work.
Figure 21.
Steroids examined in this work.
Figure 22.
Quassinoids examined in this work.
Figure 22.
Quassinoids examined in this work.
Figure 23.
Limonoids examined in this work.
Figure 23.
Limonoids examined in this work.
Figure 24.
Withanolides examined in this work.
Figure 24.
Withanolides examined in this work.
Grandifotane showed selective docking to LmajDHODH with a docking energy of −154.4 kJ/mol. The lowest-energy pose of the ligand placed the compound at the binding site of the co-crystallized ligand, 5-nitroorotic acid (
Figure 25). The furan ring of the ligand is sandwiched between the riboflavin monophosphate cofactor and Cys 131. There are hydrogen-bonding interactions between the docked ligand and residues Asn 199, Asn 68, Ser 69, and Ser 130. Grandifotane, isolated from
Khaya grandifoliola [
65], has apparently not been reported to be antiparasitic. The bark and seeds of
K. grandifoliola, however, have shown significant antimalarial activity [
66]. 6-
O-Acetylswietenolide, also isolated from
K. grandifoliola, has shown antiplasmodial activity [
67], and this compound showed preferential docking to LmexGPDH. The lowest-energy docking pose of 6-
O-acetylswietenolide with LmexGPDH (
Figure 26) lies in a cavity surrounded by Arg 274, Phe 26, Lys 125, Phe 156, Val 298, Lys 210, and Ala 157. Lys 125 and Lys 210 have been identified as critical to catalytic activity of this enzyme [
15]. With the exception of lanosterol, the steroid ligands showed preferential docking to LmajMetRS. The lowest-energy poses for these steroids show them all occupying the methionyl adenylate binding site (
Figure 27). The tetracyclic steroidal structures all occupy the same position in a hydrophobic pocket surrounded by Asp 486, Trp 515, Lys 522, His 228, Gly 227, His 513, and Tyr 218, and hydrogen-bonded by way of the 3-hydroxyl group of the steroid to Trp 516 and Gly 514. Clerosterol has shown
in vitro antileishmanial activity [
68], while saringosterol, stigmasterol, and 24-hydroperoxy-24-vinylcholesterol, in addition to clerosterol, have shown
in vitro antitrypanosomal activity [
69]. β-Sitosterol has shown modest antitrypanosomal activity [
70].
Table 29.
MolDock docking energies (kJ/mol) of limonoids with Leishmania major protein targets.
Table 29.
MolDock docking energies (kJ/mol) of limonoids with Leishmania major protein targets.
| Limonoids | LmajCatB | LmajDHODH | LmajdUTPase | LmajNDKb | LmajNH | LmajNMT | LmajOPB | LmajPDE1 | LmajPTR1 | LmajMetRS | LmajTyrRS | LmajUGPase |
|---|
| 11α−Acetoxy−2α−hydroxy−6−deoxyswietenine_acetate | −77.4 | −100.5 | −112.6 | −84.0 | −112.9 | −113.7 | −79.4 | −90.7 | −99.9 | −85.3 | −106.7 | −103.7 |
| 3−
O−Acetylanthothecanolide | −102.6 | −129.9 | −114.6 | −91.2 | −111.4 | −121.1 | −103.5 | −96.4 | −102.1 | −103.7 | −106.8 | −107.6 |
| 3−
O−Acetylkhayalactone | −70.4 | −103.4 | −96.4 | −98.9 | −115.9 | −123.4 | −96.9 | −109.5 | −109.5 | −114.5 | −106.6 | −123.7 |
| 1−
O−Acetylkhayanolide A | −87.7 | −103.2 | −85.4 | −86.3 | −96.5 | −114.1 | −92.2 | −93.2 | −88.7 | −81.1 | −105.0 | −106.8 |
| 1−
O−Acetylkhayanolide B | −80.0 | −86.6 | −87.0 | −87.1 | −80.7 | −105.0 | −105.5 | −74.5 | −72.8 | −91.8 | −95.5 | −94.5 |
| 3−
O−Acetylswietenine | −79.2 | −90.6 | −95.8 | −69.9 | −102.9 | −104.3 | −82.6 | −84.6 | −82.4 | −92.3 | −95.5 | −121.8 |
| 3−
O−Acetylswietenolide | −91.9 | −115.0 | −92.7 | −82.9 | −98.6 | −115.0 | −74.1 | −81.5 | −86.8 | −80.1 | −93.6 | −100.2 |
| 6−
O−Acetylswietenolide | −87.9 | −107.6 | −80.0 | −86.6 | −97.4 | −102.0 | −81.6 | −98.4 | −85.7 | −96.3 | −107.5 | −108.7 |
| Anthothecanolide | −103.7 | −133.3 | −85.5 | −101.6 | −93.7 | −112.3 | −98.4 | −94.8 | −104.4 | −97.4 | −96.8 | −114.1 |
| Carapa spirolactone | −92.7 | −91.4 | −87.0 | −89.7 | −99.1 | −90.6 | −93.9 | −104.0 | −71.8 | −92.1 | −95.7 | −86.5 |
| Carapin | −79.9 | −109.2 | −89.9 | −95.0 | −92.8 | −95.0 | −84.8 | −92.6 | −86.4 | −88.3 | −98.9 | −110.5 |
| Carapolide A | −107.8 | −140.0 | −101.1 | −114.9 | −111.7 | −119.2 | −98.5 | −113.5 | −92.4 | −125.5 | −107.1 | −123.5 |
| Carapolide B | −104.9 | −128.3 | −88.1 | −117.3 | −111.7 | −117.4 | −124.7 | −106.0 | −82.3 | −98.6 | −107.9 | −107.4 |
| Carapolide C | −94.5 | −105.1 | −82.7 | −106.2 | −99.2 | −110.1 | −118.5 | −109.8 | −76.3 | −95.5 | −104.4 | −107.8 |
| 7−Deacetoxy−7−oxogedunin | −51.6 | −95.8 | −82.2 | −90.1 | −86.0 | −93.6 | −100.1 | −100.2 | −91.2 | −79.0 | −91.3 | −86.5 |
| 1−
O−Deacetyl−6−deoxykhayanolide E | −104.2 | −81.4 | −75.8 | −79.7 | −96.9 | −94.3 | −88.0 | −72.9 | −85.6 | −96.6 | −94.1 | −102.8 |
| 7−Deacetylgedunin | −84.7 | −86.9 | −78.5 | −84.8 | −85.2 | −87.5 | −96.9 | −99.7 | −89.9 | −83.2 | −91.5 | −90.3 |
| 1−
O−Deacetyl−2α−hydroxykhayanolide E | −96.4 | −105.3 | −77.1 | −74.1 | −91.7 | −101.1 | −93.7 | −81.9 | −85.9 | −97.6 | −109.8 | −108.7 |
| Deacetylkhayanolide E | −104.7 | −85.6 | −76.5 | −80.6 | −88.0 | −97.2 | −87.0 | −72.6 | −83.1 | −98.8 | −110.8 | −102.9 |
| 1−Deacetylkhivorin | −60.5 | −92.7 | −96.1 | −78.3 | −96.3 | −107.5 | −58.1 | −84.0 | −81.8 | −97.0 | −98.5 | −97.7 |
| 3−Deacetylkhivorin | −73.2 | −95.7 | −90.4 | −71.8 | −96.0 | −110.6 | −99.1 | −99.4 | −81.4 | −82.4 | −98.7 | −108.2 |
| 7−Deacetylkhivorin | −77.8 | −94.0 | −99.2 | −85.0 | −88.2 | −96.3 | −93.5 | −79.1 | −87.1 | −83.7 | −89.9 | −95.6 |
| 6−Deoxyswietenolide | −84.5 | −100.2 | −79.3 | −84.0 | −97.3 | −108.8 | −90.1 | −84.6 | −93.8 | −90.9 | −98.2 | −100.5 |
| 3,7−Dideacetylkhivorin | −81.5 | −93.0 | −83.4 | −64.8 | −105.3 | −96.3 | −93.9 | −109.3 | −108.1 | −76.6 | −104.4 | −99.6 |
| Evodulone | −83.0 | −113.4 | −87.5 | −83.7 | −103.1 | −103.3 | −96.8 | −114.6 | −87.5 | −80.9 | −96.0 | −101.9 |
| Fissinolide | −89.4 | −112.8 | −93.5 | −80.4 | −99.0 | −121.6 | −99.6 | −85.5 | −94.3 | −110.6 | −88.8 | −97.1 |
| Gedunin | −74.5 | −106.8 | −81.4 | −70.2 | −89.5 | −102.1 | −93.5 | −102.8 | −96.4 | −87.1 | −93.1 | −97.7 |
| Grandifolide A | −75.5 | −113.8 | −93.3 | −101.7 | −115.1 | −128.4 | −96.9 | −94.9 | −95.7 | −98.4 | −108.3 | −102.9 |
| Grandifolin | −91.5 | −110.0 | −99.7 | −92.9 | −96.2 | −100.3 | −94.8 | −94.1 | −114.7 | −95.8 | −108.9 | −100.0 |
| Grandifoliolenone | −84.6 | −88.5 | −89.0 | −88.7 | −103.9 | −106.0 | −102.8 | −97.8 | −89.6 | −86.1 | −94.2 | −119.8 |
| Grandifotane | −102.5 | −154.5 | −94.6 | −80.1 | −99.5 | −106.1 | −90.4 | −107.9 | −96.4 | −96.5 | −104.0 | −112.7 |
| 6−Hydroxykhayalactone | −112.4 | −105.3 | −89.5 | −111.0 | −106.7 | −116.6 | −92.8 | −88.3 | −98.9 | −101.2 | −111.5 | −126.4 |
| 3β−Isobutyryloxy−1−oxomeliac−8(30)−enate | −95.6 | −119.9 | −109.6 | −89.9 | −101.4 | −111.4 | −105.5 | −81.3 | −98.8 | −86.9 | −109.1 | −110.0 |
| Khayalactone | −107.6 | −110.0 | −90.3 | −103.4 | −102.6 | −115.8 | −92.1 | −96.1 | −104.7 | −101.0 | −113.2 | −126.4 |
| Khayanolide A | −110.8 | −123.5 | −101.3 | −104.4 | −111.1 | −115.5 | −98.4 | −105.9 | −107.9 | −138.5 | −127.4 | −123.9 |
| Khayanolide B | −104.1 | −93.6 | −77.9 | −86.4 | −93.6 | −99.7 | −94.8 | −59.4 | −86.1 | −97.9 | −99.8 | −105.4 |
| Khivorin | −75.6 | −98.7 | −99.2 | −91.1 | −99.1 | −101.7 | −97.5 | −77.8 | −92.1 | −89.3 | −101.3 | −109.6 |
| Methyl acetoxyangolensate | −90.2 | −102.1 | −75.7 | −99.5 | −102.2 | −107.2 | −82.9 | −93.5 | −80.3 | −108.2 | −98.2 | −94.5 |
| Methyl angolensate | −89.2 | −104.2 | −82.9 | −100.1 | −92.6 | −109.9 | −85.1 | −90.8 | −79.7 | −110.5 | −113.2 | −108.6 |
| Methyl hydroxyangolensate | −82.9 | −91.4 | −73.0 | −102.8 | −87.6 | −109.2 | −73.0 | −84.1 | −79.5 | −100.2 | −108.8 | −101.4 |
| Methyl ivorensate | −79.5 | −106.7 | −95.7 | −102.4 | −98.9 | −99.8 | −84.7 | −76.1 | −87.2 | −95.8 | −106.5 | −108.2 |
| Mexicanolide | −86.9 | −103.0 | −82.7 | −79.6 | −95.4 | −111.6 | −90.8 | −90.2 | −95.6 | −100.5 | −93.9 | −103.4 |
| Proceranolide | −86.5 | −110.3 | −77.8 | −96.1 | −97.0 | −108.7 | −76.0 | −91.5 | −94.3 | −91.7 | −102.2 | −102.0 |
| Proceranolide butanoate | −94.0 | −119.7 | −87.9 | −93.8 | −97.5 | −119.2 | −95.9 | −103.9 | −89.2 | −90.9 | −107.1 | −106.6 |
| Proceranone | −84.4 | −103.0 | −93.9 | −97.8 | −101.8 | −106.0 | −88.2 | −115.6 | −91.4 | −82.7 | −89.5 | −113.2 |
| Procerin | −71.9 | −98.4 | −75.8 | −19.8 | −76.4 | −96.8 | −67.9 | −46.1 | −70.5 | −58.8 | −88.9 | −78.6 |
| Seneganolide | −101.7 | −122.0 | −94.5 | −106.6 | −96.4 | −110.3 | −92.6 | −92.4 | −104.9 | −106.9 | −105.8 | −104.6 |
| Swiemahogin A | −92.4 | −125.6 | −91.5 | −115.5 | −116.8 | −119.1 | −103.7 | −122.0 | −81.1 | −111.1 | −106.4 | −123.7 |
| Swietenine | −93.7 | −114.2 | −99.4 | −84.6 | −113.1 | −107.1 | −88.8 | −87.7 | −87.6 | −91.1 | −106.8 | −110.9 |
| Swietenolide | −85.7 | −108.1 | −80.4 | −83.7 | −97.4 | −109.0 | −71.8 | −82.8 | −84.3 | −102.3 | −97.4 | −98.9 |
| 1,3,7−Trideacetylkhivorin | −86.0 | −95.7 | −79.2 | −76.7 | −94.8 | −88.5 | −101.1 | −97.0 | −61.9 | −81.2 | −97.1 | −84.1 |
Table 30.
MolDock docking energies (kJ/mol) of limonoids with Leishmania donovani and L. mexicana protein targets.
Table 30.
MolDock docking energies (kJ/mol) of limonoids with Leishmania donovani and L. mexicana protein targets.
| Limonoids | LdonCatB | LdonCyp | LdonDHODH | LdonNMT | LmexGAPDH | LmexGPDH | LmexPGI | LmexPMM | LmexPYK | LmexPYK | LmexPYK | LmexTIM |
|---|
| | | | | | | | Site 1 | Site 2 | Site 3 | |
|---|
| 11α−Acetoxy−2α−hydroxy−6−deoxyswietenine acetate | −92.2 | −85.4 | −46.3 | −82.9 | −94.0 | −99.6 | −88.0 | −121.3 | −95.5 | −103.8 | −103.9 | −81.9 |
| 3−O−Acetylanthothecanolide | −100.8 | −85.9 | −54.7 | −70.9 | −108.1 | −100.9 | −94.3 | −117.9 | −110.2 | −97.0 | −94.7 | −97.5 |
| 3−O−Acetylkhayalactone | −77.2 | −66.9 | −90.3 | −100.7 | −108.8 | −113.1 | −104.3 | −110.2 | −104.4 | −111.4 | −107.4 | −107.8 |
| 1−O−Acetylkhayanolide A | −80.2 | −47.6 | −38.3 | −92.4 | −96.6 | −115.5 | −103.5 | −110.9 | −116.4 | −101.2 | −90.2 | −84.7 |
| 1−O−Acetylkhayanolide B | −81.3 | −68.4 | −65.9 | −72.5 | −83.2 | −106.6 | −82.6 | −88.0 | −94.9 | −84.9 | −91.4 | −82.7 |
| 3−O−Acetylswietenine | −75.2 | −78.2 | −57.4 | −106.0 | −74.8 | −118.6 | −81.1 | −106.8 | −101.8 | −108.3 | −103.5 | −61.4 |
| 3−O−Acetylswietenolide | −88.0 | −81.2 | −37.9 | −106.8 | −72.1 | −103.8 | −83.5 | −89.0 | −98.3 | −95.5 | −96.0 | −68.2 |
| 6−O−Acetylswietenolide | −86.6 | −69.6 | −58.4 | −91.1 | −82.4 | −142.7 | −92.8 | −85.9 | −110.1 | −97.9 | −95.8 | −67.7 |
| Anthothecanolide | −97.5 | −66.6 | −69.0 | −92.0 | −95.0 | −99.1 | −86.8 | −118.1 | −100.7 | −98.3 | −91.0 | −91.5 |
| Carapa spirolactone | −89.9 | −75.5 | −65.2 | −61.7 | −88.6 | −94.8 | −78.3 | −96.5 | −91.4 | −94.8 | −85.9 | −78.6 |
| Carapin | −71.7 | −102.8 | no dock | −94.8 | −84.6 | −106.0 | −78.4 | −99.7 | −104.6 | −94.8 | −82.9 | −78.9 |
| Carapolide A | −113.0 | −101.2 | −88.5 | −97.5 | −88.6 | −118.9 | −94.1 | −112.6 | −114.2 | −115.6 | −106.5 | −100.6 |
| Carapolide B | −96.9 | −82.5 | −21.9 | −93.1 | −90.7 | −107.7 | −90.1 | −98.7 | −107.0 | −109.9 | −99.4 | −91.4 |
| Carapolide C | −102.9 | −99.2 | −58.0 | −78.9 | −83.1 | −112.1 | −88.5 | −115.3 | −107.4 | −113.3 | −98.4 | −94.4 |
| 7−Deacetoxy−7−oxogedunin | −68.4 | −66.9 | −14.8 | −79.7 | −79.6 | −92.1 | −73.6 | −83.5 | −111.5 | −101.0 | −95.5 | −83.9 |
| 1−O−Deacetyl−6−deoxykhayanolide E | −101.8 | −87.2 | −70.1 | −73.5 | −82.3 | −99.9 | −82.4 | −75.6 | −97.4 | −99.5 | −91.7 | −71.8 |
| 7−Deacetylgedunin | −52.7 | −81.4 | −11.3 | −82.6 | −84.8 | −91.5 | −74.1 | −82.5 | −98.3 | −93.9 | −93.3 | −80.6 |
| 1−O−Deacetyl−2α−hydroxykhayanolide E | −98.1 | −58.4 | −48.5 | −64.2 | −79.0 | −111.8 | −87.0 | −84.9 | −100.4 | −104.5 | −86.2 | −66.2 |
| Deacetylkhayanolide E | −96.2 | −69.1 | −56.0 | −58.8 | −84.2 | −92.6 | −85.0 | −77.8 | −97.5 | −101.8 | −88.6 | −64.2 |
| 1−Deacetylkhivorin | −52.5 | −89.2 | no dock | −74.9 | −76.6 | −99.4 | −83.8 | −84.7 | −98.4 | −106.4 | −93.5 | −66.4 |
| 3−Deacetylkhivorin | −68.8 | −92.2 | −66.2 | −84.2 | −96.9 | −100.5 | −99.7 | −85.4 | −98.4 | −91.3 | −79.4 | −71.6 |
| 7−Deacetylkhivorin | −68.1 | −68.6 | −36.1 | −94.8 | −88.0 | −99.8 | −83.1 | −85.5 | −105.3 | −104.6 | −94.1 | −75.9 |
| 6−Deoxyswietenolide | −90.3 | −75.0 | no dock | −94.3 | −74.2 | −114.6 | −84.8 | −105.7 | −90.4 | −86.9 | −94.5 | −73.0 |
| 3,7−Dideacetylkhivorin | −62.6 | −69.3 | −63.2 | −87.3 | −92.3 | −96.2 | −88.0 | −78.6 | −113.5 | −85.2 | −98.5 | −73.1 |
| Evodulone | −80.2 | −84.8 | −74.3 | −90.6 | −79.0 | −98.7 | −86.6 | −103.2 | −103.4 | −105.3 | −93.2 | −88.4 |
| Fissinolide | −83.6 | −80.3 | −54.6 | −102.4 | −64.2 | −95.1 | −85.1 | −111.9 | −90.4 | −95.2 | −92.9 | −78.1 |
| Gedunin | −84.0 | −88.2 | no dock | −88.3 | −85.0 | −98.3 | −80.4 | −82.0 | −88.4 | −99.1 | −95.6 | −88.9 |
| Grandifolide A | −77.1 | −96.3 | no dock | −98.1 | −91.2 | −103.8 | −90.9 | −93.4 | −104.5 | −115.5 | −87.3 | −85.8 |
| Grandifolin | −53.7 | −94.9 | −67.0 | −73.2 | −83.7 | −109.1 | −89.4 | −95.4 | −105.1 | −98.9 | −104.0 | −99.3 |
| Grandifoliolenone | −93.7 | −93.8 | −68.9 | −77.3 | −78.7 | −115.1 | −80.0 | −101.4 | −115.4 | −103.6 | −100.1 | −87.3 |
| Grandifotane | −99.7 | −70.8 | −40.6 | −99.4 | −107.8 | −113.7 | −87.5 | −117.2 | −100.5 | −100.6 | −106.8 | −29.1 |
| 6−Hydroxykhayalactone | −106.7 | −80.2 | −88.2 | −96.4 | −94.0 | −106.1 | −97.3 | −98.6 | −103.4 | −110.1 | −91.2 | −95.9 |
| 3β−Isobutyryloxy−1−oxomeliac−8(30)−enate | −92.4 | −82.3 | no dock | −99.8 | −87.6 | −99.2 | −87.8 | −89.7 | −95.3 | −117.6 | −92.1 | −60.0 |
| Khayalactone | −107.0 | −68.1 | −55.1 | −97.7 | −94.6 | −116.0 | −95.9 | −102.0 | −100.2 | −100.8 | −95.6 | −94.2 |
| Khayanolide A | −110.9 | −98.7 | −18.3 | −97.8 | −92.5 | −137.8 | −98.6 | −112.1 | −115.3 | −107.8 | −107.3 | −89.9 |
| Khayanolide B | −102.5 | −72.5 | −39.5 | −70.4 | −90.8 | −94.6 | −88.3 | −77.6 | −88.2 | −105.2 | −94.7 | −71.2 |
| Khivorin | −64.6 | −89.4 | −72.0 | −30.2 | −92.4 | −107.4 | −82.0 | −84.0 | −93.9 | −108.2 | −85.6 | −78.8 |
| Methyl acetoxyangolensate | −89.1 | −58.8 | −59.5 | −88.8 | −98.0 | −94.6 | −72.7 | −81.8 | −108.9 | −98.4 | −94.8 | −41.3 |
| Methyl angolensate | −89.5 | −52.0 | −74.5 | −27.42 | −98.6 | −112.9 | −75.3 | −109.7 | −99.1 | −95.9 | −87.2 | −79.6 |
| Methyl hydroxyangolensate | −91.0 | −73.9 | −74.7 | −75.6 | −94.5 | −115.7 | −76.6 | −112.5 | −93.1 | −93.7 | −87.6 | −78.9 |
| Methyl ivorensate | −86.9 | −75.4 | −63.9 | −82.2 | −102.0 | −119.7 | −77.1 | −80.8 | −104.0 | −91.3 | −96.2 | −70.8 |
| Mexicanolide | −85.4 | −67.6 | −33.9 | −84.3 | −67.9 | −112.8 | −92.2 | −105.5 | −97.3 | −90.4 | −98.3 | −59.4 |
| Proceranolide | −90.2 | −77.6 | no dock | −94.7 | −66.8 | −121.6 | −87.2 | −106.0 | −90.0 | −88.8 | −94.6 | −73.9 |
| Proceranolide butanoate | −94.7 | −81.0 | −70.3 | −111.8 | −76.2 | −92.9 | −89.1 | −95.1 | −100.9 | −106.8 | −107.4 | −63.0 |
| Proceranone | −87.7 | −89.0 | no dock | −84.6 | −75.0 | −99.3 | −86.0 | −109.9 | −98.0 | −108.4 | −87.7 | −98.6 |
| Procerin | −77.5 | −22.4 | −59.7 | −107.5 | −94.8 | −111.3 | −108.8 | −84.5 | −93.9 | −86.6 | −70.7 | −64.8 |
| Seneganolide | −99.5 | −92.0 | −51.9 | −88.4 | −89.9 | −98.8 | −86.6 | −109.8 | −95.9 | −92.5 | −92.5 | −83.5 |
| Swiemahogin A | −69.7 | −94.9 | −52.2 | −81.4 | −79.9 | −125.4 | −92.8 | −117.4 | −120.1 | −106.2 | −110.0 | −91.4 |
| Swietenine | −90.7 | −67.4 | no dock | −99.3 | −85.0 | −104.1 | −86.3 | −91.6 | −99.6 | −122.3 | −100.7 | −78.0 |
| Swietenolide | −87.3 | −72.3 | no dock | −93.6 | −72.3 | −121.0 | −88.9 | −94.4 | −99.2 | −91.2 | −87.7 | −68.3 |
| 1,3,7−Trideacetylkhivorin | −53.7 | −80.5 | no dock | −76.2 | −87.1 | −97.2 | −81.2 | −71.5 | −96.9 | −93.0 | −82.8 | −83.2 |
Table 31.
MolDock docking energies (kJ/mol) of limonoids with Leishmania infantum protein targets.
Table 31.
MolDock docking energies (kJ/mol) of limonoids with Leishmania infantum protein targets.
| Limonoids | LinfCYP51 | LinfGLO2 | LinfPnC1 | LinfTDR1 | LinfTR |
|---|
| 11α−Acetoxy−2α−hydroxy−6−deoxyswietenine acetate | −98.0 | −75.1 | no dock | −93.6 | −89.4 |
| 3−O−Acetylanthothecanolide | −102.5 | −77.7 | no dock | −82.5 | −101.6 |
| 3−O−Acetylkhayalactone | −116.2 | −99.5 | no dock | −103.7 | −113.2 |
| 1−O−Acetylkhayanolide A | −102.4 | −89.1 | no dock | −87.9 | −108.3 |
| 1−O−Acetylkhayanolide B | −114.3 | −82.0 | no dock | −80.6 | −84.0 |
| 3−O−Acetylswietenine | −95.3 | −74.9 | no dock | −76.4 | −84.2 |
| 3−O−Acetylswietenolide | −105.4 | −80.3 | no dock | −79.5 | −95.4 |
| 6−O−Acetylswietenolide | −103.1 | −77.4 | no dock | −91.1 | −92.6 |
| Anthothecanolide | −107.9 | −74.6 | −57.2 | −82.9 | −94.3 |
| Carapa spirolactone | −108.8 | −69.7 | no dock | −68.3 | −87.3 |
| Carapin | −94.3 | −80.2 | no dock | −80.9 | −86.3 |
| Carapolide A | −111.8 | −89.1 | no dock | −98.1 | −98.3 |
| Carapolide B | −103.1 | −71.6 | no dock | −91.8 | −92.9 |
| Carapolide C | −106.9 | −80.4 | no dock | −79.4 | −92.5 |
| 7−Deacetoxy−7−oxogedunin | −104.3 | −71.9 | no dock | −70.0 | −84.6 |
| 1−
O−Deacetyl−6−deoxykhayanolide E | −98.0 | −77.3 | no dock | −78.6 | −90.9 |
| 7−Deacetylgedunin | −91.2 | −72.8 | no dock | −72.6 | −80.4 |
| 1−
O−Deacetyl−2α−hydroxykhayanolide E | −110.6 | −75.1 | no dock | −83.8 | −88.6 |
| Deacetylkhayanolide E | −103.4 | −81.0 | −45.4 | −83.0 | −84.9 |
| 1−Deacetylkhivorin | −101.2 | −72.0 | no dock | −75.8 | −87.7 |
| 3−Deacetylkhivorin | −106.0 | −76.8 | no dock | −76.0 | −98.3 |
| 7−Deacetylkhivorin | −94.5 | −83.8 | no dock | −87.0 | −73.7 |
| 6−Deoxyswietenolide | −95.9 | −76.5 | no dock | −83.0 | −92.4 |
| 3,7−Dideacetylkhivorin | −95.9 | −74.8 | no dock | −76.3 | −103.3 |
| Evodulone | −105.3 | −81.6 | no dock | −88.6 | −87.5 |
| Fissinolide | −99.2 | −78.0 | no dock | −78.8 | −99.0 |
| Gedunin | −107.4 | −75.9 | no dock | −82.2 | −78.4 |
| Grandifolide A | −109.3 | −89.6 | no dock | −90.2 | −99.8 |
| Grandifolin | −105.8 | −76.1 | no dock | −75.8 | −96.8 |
| Grandifoliolenone | −98.7 | −90.7 | −43.1 | −75.9 | −92.7 |
| Grandifotane | −111.5 | −80.8 | no dock | −89.8 | −90.0 |
| 6−Hydroxykhayalactone | −116.0 | −83.1 | no dock | −100.2 | −98.6 |
| 3β−Isobutyryloxy−1−oxomeliac−8(30)−enate | −102.7 | −78.4 | no dock | −82.5 | −103.4 |
| Khayalactone | −101.0 | −90.6 | no dock | −100.3 | −97.5 |
| Khayanolide A | −104.5 | −83.4 | no dock | −84.3 | −97.4 |
| Khayanolide B | −110.0 | −77.4 | −44.1 | −97.2 | −103.1 |
| Khivorin | −100.9 | −71.4 | no dock | −82.0 | −93.8 |
| Methyl acetoxyangolensate | −99.6 | −66.4 | no dock | −84.9 | −82.8 |
| Methyl angolensate | −90.5 | −71.0 | no dock | −77.8 | −85.6 |
| Methyl hydroxyangolensate | −95.8 | −65.2 | −48.5 | −78.2 | −85.4 |
| Methyl ivorensate | −95.5 | −77.8 | no dock | −85.2 | −92.5 |
| Mexicanolide | −90.4 | −78.8 | −27.5 | −75.8 | −92.3 |
| Proceranolide | −95.7 | −78.9 | no dock | −83.1 | −91.5 |
| Proceranolide butanoate | −100.6 | −82.1 | no dock | −89.5 | −93.5 |
| Proceranone | −107.9 | −89.7 | no dock | −90.7 | −94.6 |
| Procerin | −94.9 | −70.1 | no dock | −68.8 | −81.8 |
| Seneganolide | −101.8 | −76.9 | no dock | −82.0 | −86.4 |
| Swiemahogin A | −113.3 | −84.7 | no dock | −93.4 | −97.4 |
| Swietenine | −108.3 | −79.6 | no dock | −83.2 | −87.1 |
| Swietenolide | −97.0 | −72.7 | no dock | −86.3 | −93.3 |
| 1,3,7−Trideacetylkhivorin | −92.0 | −73.6 | no dock | −62.4 | −86.6 |
Table 32.
MolDock docking energies (kJ/mol) of withanolides with Leishmania major protein targets.
Table 32.
MolDock docking energies (kJ/mol) of withanolides with Leishmania major protein targets.
| Withanolides | LmajCatB | LmajDHODH | LmajdUTPase | LmajNDKb | LmajNH | LmajNMT | LmajOPB | LmajPDE1 | LmajPTR1 | LmajMetRS | LmajTyrRS | LmajUGPase |
|---|
| 24,25−Epoxywithanolide D | −92.9 | −121.0 | −109.3 | −104.7 | −104.2 | −124.1 | −102.7 | −114.1 | −114.2 | −98.2 | −119.5 | −108.2 |
| 14−Hydroxyixocarpanolide | −77.9 | −104.9 | −89.2 | −77.9 | −100.9 | −98.2 | −90.0 | −113.0 | −81.8 | −99.0 | −99.6 | −106.6 |
| Physagulin A | −102.7 | −123.7 | −98.3 | −103.1 | −108.2 | −113.7 | −100.6 | −115.8 | −110.0 | −98.4 | −104.0 | −110.6 |
| Physagulin B | −97.0 | −107.6 | −89.8 | −105.1 | −104.5 | −119.6 | −95.6 | −103.5 | −93.8 | −114.8 | −106.0 | −111.0 |
| Physagulin C | −93.4 | −99.5 | −89.6 | −105.5 | −106.5 | −113.8 | −95.9 | −118.3 | −93.8 | −127.3 | −105.0 | −122.7 |
| Physagulin F | −92.8 | −88.2 | −88.0 | −96.5 | −106.1 | −102.6 | −96.9 | −107.5 | −90.9 | −133.2 | −98.8 | −98.5 |
| Physagulin H | −91.8 | −112.0 | −88.3 | −92.9 | −105.4 | −111.2 | −103.2 | −111.8 | −95.5 | −126.1 | −104.1 | −112.8 |
| Physagulin I | −94.2 | −107.1 | −92.3 | −99.1 | −107.6 | −103.0 | −97.3 | −110.6 | −92.1 | −123.8 | −103.8 | −106.2 |
| Physagulin J | −96.8 | −105.4 | −87.2 | −90.6 | −96.5 | −106.7 | −94.5 | −117.6 | −91.2 | −105.6 | −100.1 | −110.1 |
| Physagulin K | −97.5 | −118.5 | −81.7 | −69.4 | −111.2 | −112.6 | −108.9 | −108.9 | −95.5 | −101.7 | −97.3 | −104.1 |
| Physagulin L | −98.8 | −114.8 | −88.0 | −69.9 | −111.2 | −112.1 | −110.3 | −97.3 | −106.0 | −103.0 | −99.2 | −107.6 |
| Physagulin L' | −107.0 | −109.0 | −104.2 | −99.2 | −99.7 | −114.2 | −108.2 | −111.9 | −108.2 | −115.0 | −103.5 | −101.3 |
| Physagulin M | −72.5 | −95.0 | −90.9 | −83.2 | −104.8 | −110.0 | −88.3 | −95.8 | −89.1 | −92.9 | −103.3 | −115.4 |
| Physagulin M' | −88.5 | −115.0 | −93.6 | −99.3 | −98.2 | −112.8 | −92.4 | −106.9 | −97.1 | −120.2 | −100.5 | −101.0 |
| Physagulin N | −93.1 | −102.8 | −92.3 | −93.1 | −101.9 | −115.2 | −94.7 | −108.5 | −84.4 | −110.6 | −106.6 | −118.1 |
| Physagulin N' | −94.4 | −102.5 | −72.7 | −94.6 | −99.7 | −99.0 | −96.1 | −116.1 | −90.0 | −110.4 | −90.2 | −91.9 |
| Physagulin O | −87.3 | −94.3 | −81.5 | −91.4 | −100.8 | −103.6 | −95.0 | −98.0 | −87.0 | −88.9 | −95.5 | −108.1 |
| Physalin A | −86.5 | −92.6 | −71.2 | −86.8 | −87.3 | −92.0 | −110.7 | −70.2 | −75.0 | −84.2 | −90.0 | −101.2 |
| Physalin B | −76.9 | −92.3 | −80.8 | −62.9 | −90.0 | −100.2 | −86.9 | −95.8 | −76.7 | −78.1 | −88.3 | −101.7 |
| Physalin D | −83.9 | −87.6 | −85.1 | −65.4 | −101.7 | −99.7 | −86.9 | −85.4 | −68.4 | −83.2 | −83.8 | −96.5 |
| Physalin E | −82.1 | −91.5 | −85.3 | −65.5 | −88.5 | −100.6 | −94.1 | −76.3 | −66.7 | −83.2 | −82.2 | −99.1 |
| Physalin F | −81.2 | −90.1 | −80.7 | −66.0 | −93.0 | −101.4 | −83.5 | −96.3 | −73.2 | −78.0 | −96.7 | −101.7 |
| Physalin G | −95.6 | −103.9 | −80.1 | −82.3 | −96.3 | −89.6 | −91.7 | −91.6 | −88.0 | −89.8 | −103.5 | −103.8 |
| Physalin H | −82.8 | −89.5 | −80.4 | −65.0 | −98.4 | −95.8 | −86.2 | −86.6 | −67.2 | −80.8 | −87.1 | −93.0 |
| Physalin I | −78.8 | −85.4 | −82.6 | −71.4 | −85.9 | −93.0 | −87.3 | −75.2 | −60.5 | −79.5 | −82.5 | −98.4 |
| Physalin J | −80.2 | −91.0 | −85.1 | −74.9 | −96.4 | −103.1 | −86.6 | −91.8 | −73.9 | −77.7 | −84.1 | −94.1 |
| Physalin K | −83.1 | −106.9 | −73.7 | −81.7 | −87.5 | −88.7 | −66.7 | −68.5 | −71.5 | −77.6 | −81.3 | −105.3 |
| Physalin U | −81.2 | −96.3 | −83.0 | −50.7 | −93.4 | −93.0 | −90.4 | −99.2 | −74.5 | −85.2 | −91.9 | −103.1 |
| Physalin V | −88.4 | −101.1 | −83.0 | −83.5 | −103.5 | −100.9 | −96.4 | −95.5 | −79.4 | −84.8 | −86.9 | −107.5 |
| Physalin W | −93.4 | −95.0 | −85.1 | −75.9 | −91.0 | −92.0 | −78.6 | −100.4 | −80.2 | −84.0 | −91.7 | −110.3 |
| Physangulide | −101.9 | −88.9 | −104.0 | −97.8 | −101.8 | −109.1 | −92.9 | −117.4 | −110.6 | −104.4 | −96.4 | −101.8 |
| Physanolide A | −85.7 | −102.9 | −84.6 | −69.0 | −97.6 | −107.9 | −93.8 | −103.5 | −100.5 | −94.5 | −88.4 | −101.1 |
| Vamonolide | −83.4 | −96.7 | −87.8 | −109.9 | −95.7 | −103.8 | −96.5 | −109.0 | −84.4 | −92.2 | −106.7 | −97.4 |
| Withangulatin A | −92.4 | −121.9 | −97.8 | −108.1 | −110.6 | −124.2 | −97.1 | −112.9 | −110.1 | −99.4 | −105.1 | −112.2 |
| Withangulatin B | −105.4 | −89.8 | −96.1 | −96.6 | −95.7 | −99.9 | −94.3 | −107.2 | −96.2 | −117.9 | −104.7 | −91.0 |
| Withangulatin C | −85.7 | −93.6 | −86.6 | −100.7 | −90.1 | −109.7 | −106.3 | −89.2 | −94.1 | −125.9 | −104.6 | −98.5 |
| Withangulatin D | −88.8 | −80.3 | −89.4 | −102.6 | −96.6 | −107.2 | −92.0 | −113.3 | −86.9 | −126.2 | −105.2 | −104.8 |
| Withangulatin E | −57.3 | −94.8 | −36.3 | −92.5 | −85.7 | −102.4 | −89.1 | −95.7 | −88.9 | −96.0 | −112.7 | −97.9 |
| Withangulatin F | −85.1 | −96.0 | −83.8 | −103.9 | −97.7 | −98.9 | −87.9 | −120.8 | −85.8 | −96.0 | −102.4 | −96.9 |
| Withangulatin G | −91.5 | −122.3 | −76.0 | −100.4 | −95.2 | −100.1 | −93.7 | −99.7 | −87.6 | −97.7 | −105.8 | −92.5 |
| Withangulatin H | −91.6 | −105.5 | −89.1 | −96.8 | −81.0 | −94.8 | −105.1 | −88.8 | −71.0 | −115.3 | −102.2 | −106.9 |
| Withangulatin I | −98.8 | −112.4 | −90.0 | −91.3 | −106.6 | −113.1 | −88.1 | −105.6 | −106.0 | −98.2 | −102.3 | −110.7 |
Table 33.
MolDock docking energies (kJ/mol) of withanolides with Leishmania donovani and L. mexicana protein targets.
Table 33.
MolDock docking energies (kJ/mol) of withanolides with Leishmania donovani and L. mexicana protein targets.
| LdonCatB | LdonCyp | LdonDHODH | LdonNMT | LmexGAPDH | LmexGPDH | LmexPGI | LmexPMM | LmexPYK | LmexPYK | LmexPYK | LmexTIM |
|---|
| Withanolides | | | | | | | | | Site 1 | Site 2 | Site 3 | |
|---|
| 24,25−Epoxywithanolide D | −95.7 | −101.5 | −63.4 | −103.0 | −97.7 | −118.1 | −110.4 | −125.7 | −119.7 | −106.2 | −113.6 | −95.4 |
| 14−Hydroxyixocarpanolide | −88.0 | −92.8 | −30.7 | −52.2 | −80.7 | −102.3 | −77.5 | −95.6 | −103.6 | −92.2 | −100.3 | −74.3 |
| Physagulin A | −97.4 | −103.3 | −82.8 | −91.9 | −91.4 | −116.2 | −100.5 | −119.2 | −111.6 | −104.4 | −106.5 | −85.0 |
| Physagulin B | −105.4 | −104.2 | −85.9 | −89.9 | −94.2 | −107.6 | −96.3 | −114.4 | −110.3 | −109.3 | −103.8 | −82.9 |
| Physagulin C | −98.7 | −86.0 | −80.6 | −104.7 | −91.6 | −108.7 | −98.2 | −124.7 | −103.4 | −100.7 | −103.8 | −85.9 |
| Physagulin F | −94.8 | −88.7 | −84.6 | −93.6 | −88.7 | −106.0 | −87.9 | −107.8 | −105.8 | −106.3 | −91.6 | −82.3 |
| Physagulin H | −96.5 | −85.2 | −83.6 | −98.8 | −86.0 | −101.4 | −97.2 | −113.0 | −106.2 | −104.6 | −101.7 | −79.3 |
| Physagulin I | −95.9 | −87.5 | −77.2 | −94.7 | −90.2 | −99.9 | −91.5 | −102.3 | −101.2 | −103.5 | −95.0 | −77.7 |
| Physagulin J | −94.4 | −84.6 | −60.7 | −92.5 | −88.5 | −108.7 | −91.2 | −100.6 | −98.5 | −111.2 | −100.6 | −86.6 |
| Physagulin K | −85.1 | −95.8 | no dock | −92.5 | −94.2 | −109.1 | −90.1 | −92.6 | −112.2 | −106.4 | −97.1 | −76.0 |
| Physagulin L | −90.6 | −83.3 | −67.6 | −97.6 | −96.2 | −112.3 | −98.2 | −104.5 | −117.3 | −103.6 | −92.0 | −68.1 |
| Physagulin L' | −108.8 | −104.3 | −102.2 | −94.7 | −94.2 | −110.7 | −87.0 | −109.4 | −111.5 | −98.2 | −105.4 | −101.2 |
| Physagulin M | −91.2 | −91.3 | −70.6 | −81.4 | −96.1 | −101.8 | −78.6 | −87.4 | −105.6 | −111.5 | −101.9 | −75.9 |
| Physagulin M' | −84.3 | −94.8 | −30.7 | −90.9 | −89.8 | −106.2 | −88.9 | −88.5 | −110.5 | −103.7 | −96.3 | −106.7 |
| Physagulin N | −94.9 | −99.8 | −89.5 | −95.8 | −95.0 | −106.2 | −80.7 | −115.1 | −105.1 | −112.2 | −95.4 | −82.5 |
| Physagulin N' | −93.2 | −83.6 | −65.4 | −85.2 | −86.2 | −112.3 | −83.4 | −103.2 | −107.5 | −103.8 | −85.1 | −55.3 |
| Physagulin O | −90.3 | −88.8 | −88.3 | −93.9 | −93.4 | −103.5 | −84.5 | −94.6 | −99.4 | −102.4 | −106.3 | −72.0 |
| Physalin A | −85.6 | −71.4 | −41.9 | −82.3 | −83.6 | −93.8 | −82.0 | −102.9 | −88.1 | −100.4 | −91.4 | −81.7 |
| Physalin B | −84.9 | −71.7 | −48.5 | −81.8 | −78.4 | −88.5 | −84.8 | −92.6 | −92.0 | −89.4 | −84.8 | −73.9 |
| Physalin D | −82.6 | −79.1 | −54.4 | −80.5 | −79.6 | −91.8 | −84.5 | −85.0 | −89.8 | −95.0 | −79.9 | −71.7 |
| Physalin E | −83.4 | −63.3 | −30.0 | −75.9 | −81.3 | −83.5 | −74.5 | −83.7 | −96.2 | −93.5 | −81.5 | −76.0 |
| Physalin F | −83.7 | −73.8 | −57.4 | −83.2 | −80.7 | −92.8 | −92.4 | −85.1 | −90.9 | −91.0 | −76.6 | −74.9 |
| Physalin G | −93.6 | −71.0 | −81.2 | −92.5 | −76.9 | −78.4 | −87.9 | −97.9 | −100.6 | −90.4 | −93.0 | −69.2 |
| Physalin H | −85.2 | −60.7 | no dock | −73.0 | −81.0 | −92.5 | −86.0 | −81.7 | −92.6 | −89.6 | −81.2 | −70.1 |
| Physalin I | −84.2 | −51.6 | no dock | −68.3 | −82.5 | −96.2 | −87.4 | −79.0 | −84.0 | −86.5 | −82.9 | −61.6 |
| Physalin J | −83.1 | −80.1 | −55.5 | −79.5 | −82.2 | −92.4 | −78.2 | −97.8 | −97.0 | −94.9 | −88.0 | −72.1 |
| Physalin K | −82.6 | −74.7 | −65.5 | −84.2 | −70.4 | −90.2 | −77.6 | −76.5 | −96.7 | −87.4 | −94.2 | −60.7 |
| Physalin U | −83.0 | −65.1 | −68.5 | −94.0 | −85.9 | −97.6 | −93.2 | −85.0 | −97.0 | −93.2 | −85.9 | −81.6 |
| Physalin V | −83.2 | −83.6 | −27.1 | −84.3 | −81.2 | −93.9 | −93.1 | −82.8 | −95.0 | −92.7 | −90.5 | −52.8 |
| Physalin W | −93.9 | −67.5 | −77.2 | −89.7 | −84.0 | −101.5 | −92.1 | −91.4 | −98.1 | −99.0 | −86.9 | −80.2 |
| Physangulide | −103.2 | −99.4 | −71.3 | −106.0 | −89.9 | −105.2 | −83.7 | −111.5 | −114.6 | −108.0 | −103.3 | −82.8 |
| Physanolide A | −93.4 | −96.3 | −22.9 | −103.0 | −84.0 | −99.6 | −81.4 | −106.3 | −97.4 | −96.3 | −86.2 | −44.5 |
| Vamonolide | −83.4 | −89.7 | −52.6 | −90.2 | −75.4 | −96.2 | −81.5 | −107.1 | −101.6 | −100.9 | −90.8 | −77.4 |
| Withangulatin A | −106.1 | −102.6 | −39.9 | −98.6 | −96.7 | −119.8 | −105.4 | −122.0 | −109.2 | −102.9 | −107.5 | −87.8 |
| Withangulatin B | −95.7 | −90.0 | no dock | −94.6 | −87.5 | −99.4 | −85.5 | −94.2 | −99.6 | −106.1 | −112.4 | −92.4 |
| Withangulatin C | −91.7 | −87.1 | −27.3 | −90.6 | −88.3 | −110.5 | −87.0 | −99.0 | −106.2 | −101.2 | −113.8 | −78.8 |
| Withangulatin D | −101.2 | −92.4 | no dock | −93.6 | −84.6 | −105.1 | −86.5 | −96.2 | −92.3 | −104.8 | −108.2 | −86.5 |
| Withangulatin E | −81.0 | −77.9 | −25.9 | −95.5 | −76.9 | −105.7 | −85.8 | −99.9 | −102.7 | −101.4 | −100.8 | −87.8 |
| Withangulatin F | −54.3 | −94.3 | −58.6 | −83.3 | −88.7 | −108.4 | −88.3 | −100.2 | −106.2 | −97.7 | −104.7 | −99.5 |
| Withangulatin G | −91.5 | −87.6 | no dock | −96.1 | −74.2 | −100.9 | −85.5 | −100.5 | −99.8 | −103.2 | −95.7 | −64.3 |
| Withangulatin H | −95.2 | −85.5 | −65.1 | −85.9 | −80.3 | −102.5 | −75.5 | −98.8 | −95.8 | −106.9 | −97.6 | −80.7 |
| Withangulatin I | −93.5 | −95.3 | −82.6 | −93.6 | −95.5 | −113.7 | −89.3 | −114.2 | −116.1 | −105.4 | −94.5 | −71.1 |
Table 34.
MolDock docking energies (kJ/mol) of withanolides with Leishmania infantum protein targets.
Table 34.
MolDock docking energies (kJ/mol) of withanolides with Leishmania infantum protein targets.
| Withanolides | LinfCYP51 | LinfGLO2 | LinfPnC1 | LinfTDR1 | LinfTR |
|---|
| 24,25−Epoxywithanolide D | −128.1 | −102.8 | no dock | −103.8 | −98.4 |
| 14−Hydroxyixocarpanolide | −111.7 | −81.1 | no dock | −85.3 | −94.4 |
| Physagulin A | −106.7 | −88.5 | no dock | −103.6 | −102.7 |
| Physagulin B | −107.7 | −82.8 | no dock | −99.0 | −95.1 |
| Physagulin C | −122.5 | −98.8 | no dock | −99.7 | −94.2 |
| Physagulin F | −108.2 | −70.6 | no dock | −80.8 | −96.1 |
| Physagulin H | −114.9 | −92.6 | no dock | −96.8 | −91.9 |
| Physagulin I | −102.9 | −75.1 | no dock | −84.5 | −97.7 |
| Physagulin J | −115.4 | −79.4 | no dock | −87.5 | −93.2 |
| Physagulin K | −105.4 | −76.4 | no dock | −88.5 | −95.0 |
| Physagulin L | −109.4 | −83.5 | no dock | −90.1 | −94.7 |
| Physagulin L
' | −107.4 | −90.3 | no dock | −102.4 | −101.4 |
| Physagulin M | −106.5 | −78.5 | no dock | −95.7 | −94.4 |
| Physagulin M
' | −113.3 | −81.6 | no dock | −94.8 | −91.5 |
| Physagulin N | −114.0 | −91.4 | no dock | −92.6 | −95.2 |
| Physagulin N
' | −100.5 | −87.7 | no dock | −85.7 | −106.2 |
| Physagulin O | −108.2 | −77.9 | no dock | −94.4 | −92.2 |
| Physalin A | −105.0 | −75.2 | no dock | −81.4 | −73.9 |
| Physalin B | −110.4 | −72.6 | no dock | −86.0 | −93.8 |
| Physalin D | −113.5 | −86.0 | no dock | −69.0 | −93.0 |
| Physalin E | −115.4 | −72.5 | no dock | −82.9 | −92.6 |
| Physalin F | −111.9 | −73.6 | no dock | −76.2 | −93.0 |
| Physalin G | −99.1 | −69.1 | no dock | −77.5 | −91.0 |
| Physalin H | −112.4 | −83.7 | no dock | −70.2 | −94.5 |
| Physalin I | −101.3 | −72.0 | no dock | −72.8 | −94.2 |
| Physalin J | −111.1 | −83.9 | −34.5 | −91.4 | −93.0 |
| Physalin K | −93.9 | −57.4 | no dock | −70.4 | −89.5 |
| Physalin U | −99.8 | −80.0 | −37.3 | −78.6 | −94.1 |
| Physalin V | −114.5 | −67.3 | −50.4 | −76.0 | −102.3 |
| Physalin W | −98.6 | −76.3 | no dock | −71.1 | −81.1 |
| Physangulide | −116.5 | −94.1 | no dock | −103.3 | −103.2 |
| Physanolide A | −104.8 | −90.5 | no dock | −88.1 | −95.2 |
| Vamonolide | −105.8 | −78.1 | no dock | −88.6 | −97.3 |
| Withangulatin A | −114.5 | −92.1 | no dock | −98.6 | −94.2 |
| Withangulatin B | −112.3 | −83.6 | no dock | −93.2 | −90.6 |
| Withangulatin C | −112.1 | −77.3 | no dock | −89.5 | −87.7 |
| Withangulatin D | −106.6 | −75.7 | no dock | −79.5 | −96.3 |
| Withangulatin E | −120.7 | −82.1 | no dock | −89.9 | −85.6 |
| Withangulatin F | −111.9 | −80.4 | no dock | −93.8 | −92.6 |
| Withangulatin G | −115.5 | −75.1 | no dock | −77.0 | −97.7 |
| Withangulatin H | −119.1 | −74.0 | −40.1 | −103.9 | −89.9 |
| Withangulatin I | −118.5 | −92.6 | no dock | −98.5 | −97.6 |
Table 35.
MolDock docking energies (kJ/mol) of triterpenoids with Leishmania major protein targets.
Table 35.
MolDock docking energies (kJ/mol) of triterpenoids with Leishmania major protein targets.
| Triterpenoids | LmajCatB | LmajDHODH | LmajdUTPase | LmajNDKb | LmajNH | LmajNMT | LmajOPB | LmajPDE1 | LmajPTR1 | LmajMetRS | LmajTyrRS | LmajUGPase |
|---|
| α−Amyrin | −74.4 | −81.7 | −52.9 | −55.1 | −73.4 | −84.8 | −75.6 | −100.9 | −72.3 | −71.8 | −84.5 | −82.7 |
| β−Amyrin | −43.0 | −78.8 | −66.1 | −59.1 | −76.8 | −87.7 | −84.4 | −83.8 | −65.7 | −72.6 | −84.0 | −82.4 |
| Betulin | −71.9 | −85.3 | −75.8 | −61.5 | −99.0 | −93.6 | −94.4 | −83.0 | −71.2 | −80.8 | −99.1 | −85.5 |
| Betulinaldehyde | −74.6 | −81.7 | −75.7 | −71.9 | −98.9 | −93.5 | −94.2 | −88.4 | −76.0 | −78.5 | −99.1 | −89.4 |
| Betulinic acid | −74.2 | −88.5 | −66.7 | −61.3 | −96.3 | −104.6 | −89.1 | −82.6 | −73.5 | −82.0 | −103.3 | −88.7 |
| Corosolic acid | −80.2 | −74.7 | −81.7 | −63.1 | −91.3 | −107.7 | −84.5 | −106.4 | −81.6 | −79.3 | −86.4 | −81.4 |
| Erythrodiol | −34.6 | −84.6 | −64.6 | −64.9 | −79.5 | −92.7 | −87.0 | −84.9 | −67.2 | −74.7 | −87.6 | −83.5 |
| Friedelin | −57.3 | −72.5 | −74.6 | −53.9 | −84.9 | −75.9 | −78.0 | −79.6 | −75.9 | −71.3 | −102.4 | −76.5 |
| Isoiguesterin | −67.3 | −79.4 | −64.7 | −73.5 | −80.5 | −95.5 | −81.0 | −92.2 | −77.3 | −77.2 | −88.2 | −85.2 |
| 20−
epi−Isoiguesterinol | −69.7 | −74.0 | −64.7 | −83.9 | −85.3 | −96.4 | −98.2 | −96.2 | −84.4 | −81.4 | −83.6 | −78.6 |
| Lawnermis acid methyl ester | −75.8 | −95.9 | −81.6 | −70.3 | −76.5 | −91.4 | −81.6 | −98.3 | −66.7 | −78.8 | −92.2 | −88.3 |
| Lupeol | −73.2 | −84.6 | −76.7 | −53.1 | −99.0 | −87.5 | −91.5 | −85.5 | −69.9 | −74.3 | −96.2 | −75.9 |
| Methyl
seco−3,4−betulonic acid | −87.2 | −93.8 | −84.8 | −76.1 | −101.2 | −113.5 | −96.5 | −106.3 | −97.0 | −79.2 | −90.1 | −110.1 |
| 3−
O−Methyl−6−oxopristimerol | −91.4 | −92.8 | −75.9 | −77.1 | −99.5 | −102.5 | −88.4 | −95.2 | −79.1 | −94.0 | −99.9 | −96.1 |
| Oleanolic acid | −82.5 | −90.6 | −80.2 | −66.3 | −72.7 | −97.7 | −79.8 | −95.3 | −66.6 | −69.7 | −88.4 | −76.2 |
| epi−Oleanolic acid | −75.9 | −76.7 | −77.2 | −54.2 | −72.4 | −83.7 | −88.7 | −98.5 | −70.3 | −84.2 | −82.3 | −95.6 |
| 6−Oxopristimerol | −87.2 | −91.5 | −72.9 | −76.4 | −98.8 | −103.6 | −90.2 | −91.8 | −82.5 | −93.2 | −93.8 | −101.2 |
| (24
Z)−3−Oxotirucalla−7,24−dien−26−oic acid | −96.4 | −95.4 | −86.1 | −129.2 | −104.4 | −95.7 | −99.7 | −106.1 | −90.3 | −91.9 | −102.2 | −98.9 |
| Pristimerin | −69.3 | −87.8 | −72.3 | −77.6 | −88.1 | −112.9 | −86.3 | −86.4 | −87.8 | −86.6 | −102.0 | −88.6 |
| Rotundic acid | −78.5 | −85.1 | −74.2 | −49.1 | −55.9 | −85.7 | −86.8 | −98.4 | −65.6 | −80.8 | −78.5 | −83.4 |
| Taraxerol | −22.5 | −83.4 | −60.2 | −48.0 | −70.1 | −82.4 | −88.0 | −78.8 | −85.5 | −85.1 | −84.9 | −88.1 |
| Ursolic acid | −70.8 | −80.0 | −74.0 | −59.1 | −72.0 | −92.4 | −80.4 | −73.7 | −71.9 | −82.7 | −84.3 | −89.9 |
| Uvaol | −74.9 | −82.4 | −67.9 | −67.8 | −75.2 | −88.3 | −85.4 | −89.9 | −75.2 | −74.9 | −86.4 | −87.0 |
| Wallichianol | −81.9 | −93.2 | −71.1 | −74.6 | −87.2 | −102.7 | −92.0 | −75.2 | −86.2 | −80.1 | −80.6 | −89.4 |
Table 36.
MolDock docking energies (kJ/mol) of triterpenoids with Leishmania donovani and L. mexicana protein targets.
Table 36.
MolDock docking energies (kJ/mol) of triterpenoids with Leishmania donovani and L. mexicana protein targets.
| LdonCatB | LdonCyp | LdonDHODH | LdonNMT | LmexGAPDH | LmexGPDH | LmexPGI | LmexPMM | LmexPYK | LmexPYK | LmexPYK | LmexTIM |
|---|
| Triterpenoids | | | | | | | | | Site 1 | Site 2 | Site 3 | |
|---|
| α−Amyrin | −79.7 | −79.4 | −19.0 | −53.4 | −73.7 | −88.0 | −73.5 | −71.7 | −70.4 | −87.4 | −85.4 | −54.4 |
| β−Amyrin | −65.3 | −75.6 | no dock | −64.7 | −73.9 | −77.2 | −65.3 | −71.0 | −79.9 | −94.4 | −75.2 | −35.5 |
| Betulin | −80.1 | −78.2 | no dock | −77.8 | −81.6 | −101.1 | −79.1 | −82.6 | −87.1 | −98.0 | −83.7 | −70.3 |
| Betulinaldehyde | −82.8 | −78.6 | no dock | −67.2 | −75.9 | −96.9 | −83.5 | −84.5 | −87.5 | −96.6 | −82.5 | −75.0 |
| Betulinic acid | −86.3 | −65.2 | no dock | −80.7 | −77.7 | −97.3 | −81.0 | −87.8 | −89.5 | −97.8 | −85.4 | −76.3 |
| Corosolic acid | −80.8 | −80.2 | no dock | −78.2 | −78.0 | −93.2 | −81.9 | −75.8 | −80.7 | −89.6 | −73.0 | −60.9 |
| Erythrodiol | −57.7 | −75.8 | −30.0 | −69.5 | −76.1 | −80.8 | −69.9 | −73.0 | −79.2 | −96.6 | −75.4 | −39.3 |
| Friedelin | −65.9 | −53.9 | −48.8 | −63.2 | −72.7 | −83.8 | −64.8 | −77.1 | −73.6 | −86.1 | −76.7 | −70.1 |
| Isoiguesterin | −69.9 | −70.0 | −48.8 | −70.4 | −76.6 | −80.9 | −68.8 | −83.0 | −80.7 | −87.9 | −77.0 | −65.9 |
| 20−
epi−Isoiguesterinol | −70.5 | −83.0 | −55.8 | −74.8 | −75.9 | −93.0 | −73.1 | −91.0 | −88.9 | −88.7 | −76.3 | −58.3 |
| Lawnermis acid methyl ester | −73.7 | −77.4 | −44.5 | −73.4 | −84.7 | −97.0 | −80.1 | −77.6 | −81.5 | −96.3 | −98.1 | −57.7 |
| Lupeol | −80.7 | −76.5 | no dock | −71.9 | −88.5 | −93.0 | −72.1 | −81.3 | −82.0 | −96.0 | −79.6 | −69.2 |
| Methyl
seco−3,4−betulonic acid | −91.6 | −83.9 | no dock | −92.0 | −87.8 | −102.4 | −89.7 | −98.0 | −109.4 | −102.6 | −86.7 | −82.2 |
| 3−
O−Methyl−6−oxopristimerol | −90.8 | −83.7 | no dock | −85.0 | −76.0 | −90.2 | −78.6 | −92.9 | −109.5 | −96.4 | −83.2 | −61.4 |
| Oleanolic acid | −82.4 | −74.2 | no dock | −69.7 | −79.0 | −86.9 | −69.9 | −80.8 | −77.5 | −90.4 | −79.9 | −74.9 |
| epi−Oleanolic acid | −61.1 | −73.7 | −50.7 | −69.1 | −81.2 | −88.8 | −66.3 | −78.6 | −79.6 | −91.9 | −74.8 | −65.7 |
| 6−Oxopristimerol | −90.6 | −84.3 | −60.9 | −82.9 | −80.8 | −90.0 | −79.1 | −85.5 | −98.4 | −95.2 | −94.1 | −68.0 |
| (24
Z)−3−Oxotirucalla−7,24−dien−26−oic acid | −100.0 | −88.1 | −82.2 | −89.0 | −94.3 | −108.5 | −99.2 | −104.0 | −122.1 | −104.4 | −100.1 | −91.2 |
| Pristimerin | −71.3 | −78.4 | −46.7 | −85.9 | −93.1 | −90.7 | −74.4 | −102.4 | −96.4 | −97.4 | −80.4 | −63.0 |
| Rotundic acid | −71.6 | −78.2 | −36.3 | −77.0 | −75.8 | −100.3 | −71.4 | −75.5 | −80.8 | −93.7 | −82.5 | −58.0 |
| Taraxerol | −51.7 | −78.0 | −29.6 | −61.4 | −69.4 | −82.8 | −68.2 | −90.4 | −81.4 | −90.1 | −69.8 | −81.4 |
| Ursolic acid | −69.6 | −78.7 | no dock | −71.7 | −76.5 | −97.3 | −80.3 | −75.5 | −71.8 | −93.0 | −75.4 | −46.5 |
| Uvaol | −81.4 | −79.1 | no dock | −51.7 | −75.9 | −95.0 | −78.0 | −74.3 | −76.4 | −92.0 | −82.8 | −64.9 |
| Wallichianol | −81.5 | −70.6 | −58.4 | −85.4 | −83.8 | −87.6 | −80.4 | −91.9 | −96.8 | −93.8 | −82.3 | −77.4 |
Table 37.
MolDock docking energies (kJ/mol) of triterpenoids with Leishmania infantum protein targets.
Table 37.
MolDock docking energies (kJ/mol) of triterpenoids with Leishmania infantum protein targets.
| Triterpenoids | LinfCYP51 | LinfGLO2 | LinfPnC1 | LinfTDR1 | LinfTR |
|---|
| α−Amyrin | −99.3 | −66.0 | no dock | −66.7 | −71.9 |
| β−Amyrin | −98.9 | −72.6 | −43.9 | −73.5 | −68.4 |
| Betulin | −101.6 | −73.8 | −30.4 | −79.1 | −79.0 |
| Betulinaldehyde | −104.8 | −74.8 | no dock | −74.1 | −85.8 |
| Betulinic acid | −95.5 | −75.2 | no dock | −80.5 | −85.8 |
| Corosolic acid | −111.8 | −64.5 | no dock | −74.2 | −82.2 |
| Erythrodiol | −100.4 | −71.2 | no dock | −76.7 | −78.9 |
| Friedelin | −92.7 | −64.4 | −40.3 | −71.0 | −75.4 |
| Isoiguesterin | −83.7 | −70.6 | no dock | −81.9 | −77.2 |
| 20−
epi−Isoiguesterinol | −81.1 | −71.1 | no dock | −80.4 | −77.1 |
| Lawnermis acid methyl ester | −97.9 | −72.9 | −33.4 | −72.3 | −81.2 |
| Lupeol | −97.9 | −70.7 | −35.1 | −77.1 | −81.0 |
| Methyl
seco−3,4−betulonic acid | −94.7 | −67.1 | no dock | −88.7 | −91.9 |
| 3−
O−Methyl−6−oxopristimerol | −98.6 | −73.3 | −12.0 | −84.9 | −92.2 |
| Oleanolic acid | −105.6 | −70.7 | no dock | −79.2 | −74.9 |
| epi−Oleanolic acid | −105.5 | −74.4 | no dock | −81.6 | −80.5 |
| 6−Oxopristimerol | −95.8 | −66.4 | no dock | −77.6 | −94.9 |
| (24
Z)−3−Oxotirucalla−7,24−dien−26−oic acid | −112.4 | −99.4 | no dock | −83.9 | −97.0 |
| Pristimerin | −97.5 | −78.3 | no dock | −82.4 | −84.9 |
| Rotundic acid | −107.8 | −60.5 | −33.1 | −76.5 | −78.6 |
| Taraxerol | −91.5 | −74.2 | −39.1 | −74.8 | −76.9 |
| Ursolic acid | −102.3 | −62.5 | no dock | −69.4 | −76.3 |
| Uvaol | −101.1 | −65.3 | no dock | −69.4 | −77.4 |
Table 38.
MolDock docking energies (kJ/mol) of quassinoids and steroids with Leishmania major protein targets.
Table 38.
MolDock docking energies (kJ/mol) of quassinoids and steroids with Leishmania major protein targets.
| LmajCatB | LmajDHODH | LmajdUTPase | LmajNDKb | LmajNH | LmajNMT | LmajOPB | LmajPDE1 | LmajPTR1 | LmajMetRS | LmajTyrRS | LmajUGPase |
|---|
| Quassinoids | | | | | | | | | | | | |
|---|
| 15−β−Heptylchaparrinone | −92.7 | −121.0 | −89.4 | −118.4 | −101.9 | −112.4 | −98.2 | −109.3 | −96.2 | −120.5 | −106.5 | −101.0 |
| Simalikalactone D | −88.4 | −106.2 | −80.5 | −99.3 | −84.9 | −98.5 | −63.6 | −105.2 | −91.8 | −111.7 | −105.5 | −111.1 |
| Steroids | | | | | | | | | | | | |
| Cholesterol | −90.5 | −95.2 | −93.3 | −106.4 | −101.7 | −98.6 | −111.4 | −109.2 | −109.5 | −115.2 | −99.3 | −97.7 |
| Clerosterol | −97.9 | −100.6 | −95.5 | −110.6 | −104.8 | −102.4 | −91.6 | −113.3 | −110.8 | −121.3 | −101.0 | −101.9 |
| 24−Hydroperoxy−24−vinylcholesterol | −81.0 | −99.9 | −92.1 | −111.0 | −103.4 | −105.0 | −115.9 | −117.7 | −109.8 | −127.0 | −100.2 | −112.4 |
| Lanosterol | −85.0 | −85.4 | −81.3 | −94.1 | −103.2 | −96.3 | −111.4 | −108.4 | −87.2 | −88.8 | −97.2 | −99.3 |
| Saringosterol | −88.4 | −103.8 | −89.8 | −108.1 | −105.9 | −101.6 | −109.5 | −115.5 | −107.1 | −126.4 | −99.0 | −106.1 |
| β−Sitosterol | −94.8 | −98.7 | −94.8 | −106.4 | −105.6 | −102.1 | −101.7 | −111.9 | −110.0 | −121.5 | −102.2 | −111.6 |
| Stigmasterol | −87.8 | −102.3 | −94.6 | −105.6 | −108.1 | −101.1 | −101.0 | −109.8 | −109.3 | −121.4 | −106.4 | −102.1 |
Table 39.
MolDock docking energies (kJ/mol) of quassinoids and steroids with Leishmania donovani and L. mexicana protein targets.
Table 39.
MolDock docking energies (kJ/mol) of quassinoids and steroids with Leishmania donovani and L. mexicana protein targets.
| LdonCatB | LdonCyp | LdonDHODH | LdonNMT | LmexGAPDH | LmexGPDH | LmexPGI | LmexPMM | LmexPYK | LmexPYK | LmexPYK | LmexTIM |
|---|
| Quassinoids | | | | | | | | | Site 1 | Site 2 | Site 3 | |
|---|
| 15−β−Heptylchaparrinone | −93.0 | −90.0 | −88.8 | −100.5 | −84.9 | −111.8 | −90.5 | −110.0 | −123.2 | −108.0 | −98.8 | −92.0 |
| Simalikalactone D | −98.1 | −102.9 | no dock | −90.9 | −89.5 | −115.2 | −70.9 | −103.9 | −93.4 | −99.6 | −107.4 | −75.8 |
| Steroids | | | | | | | | | | | | |
| Cholesterol | −95.4 | −99.4 | −78.2 | −90.9 | −91.6 | −102.8 | −93.8 | −104.3 | −102.0 | −95.5 | −100.2 | −87.3 |
| Clerosterol | −82.5 | −99.9 | −89.1 | −99.9 | −92.8 | −104.0 | −80.0 | −108.3 | −106.8 | −104.2 | −100.1 | −96.0 |
| 24−Hydroperoxy−24−vinylcholesterol | −97.8 | −98.0 | −101.7 | −89.9 | −93.5 | −97.2 | −77.0 | −111.9 | −103.8 | −104.4 | −89.5 | −91.6 |
| Lanosterol | −84.1 | −88.4 | −55.0 | −81.9 | −81.1 | −99.7 | −91.5 | −98.8 | −103.6 | −93.5 | −103.8 | −82.4 |
| Saringosterol | −97.9 | −106.5 | −98.9 | −99.1 | −86.7 | −113.8 | −91.1 | −111.0 | −106.5 | −102.3 | −88.7 | −95.8 |
| β−Sitosterol | −100.9 | −104.5 | −86.7 | −88.8 | −92.5 | −100.0 | −96.7 | −107.8 | −105.5 | −97.3 | −92.6 | −95.0 |
| Stigmasterol | −97.0 | −98.5 | −93.7 | −92.3 | −90.2 | −112.3 | −88.5 | −107.6 | −106.0 | −104.3 | −97.8 | −96.5 |
Table 40.
MolDock docking energies (kJ/mol) of quassinoids and steroids with Leishmania infantum protein targets.
Table 40.
MolDock docking energies (kJ/mol) of quassinoids and steroids with Leishmania infantum protein targets.
| Quassinoids | LinfCYP51 | LinfGLO2 | LinfPnC1 | LinfTDR1 | LinfTR |
|---|
| 15−β−Heptylchaparrinone | −107.6 | −100.4 | no dock | −92.2 | −96.8 |
| Simalikalactone D | −98.5 | −85.8 | no dock | −83.7 | −80.2 |
| Steroids | | | | | |
| Cholesterol | −110.9 | −86.9 | no dock | −87.4 | −93.0 |
| Clerosterol | −116.3 | −94.5 | no dock | −91.4 | −95.8 |
| 24−Hydroperoxy−24−vinylcholesterol | −121.5 | −89.9 | no dock | −91.4 | −96.6 |
| Lanosterol | −116.0 | −85.7 | no dock | −81.5 | −90.4 |
| Saringosterol | −117.2 | −90.0 | no dock | −97.9 | −99.8 |
| β−Sitosterol | −113.8 | −87.6 | no dock | −94.1 | −103.0 |
| Stigmasterol | −116.1 | −86.4 | no dock | −96.6 | −98.4 |
Not surprisingly, all of the steroids and many of the triterpenoids examined in this study showed significant docking preference for
L. infantum sterol 14α-demethylase (LinfCYP51). This had been noted previously with
Trypanosoma brucei sterol 14α-demethylase [
71]. In particular, 24-hydroperoxy-24-vinylcholesterol (docking energy = −121.5 kJ/mol) and 24,25-epoxywithanolide D (docking energy = −128.1 kJ/mol) were strongly docking with LinfCYP51. The lowest-energy pose of 24-hydroperoxy-24,25-vinylcholesterol with LinfCYP51 places the hydroperoxy group of the ligand adjacent to the heme Fe (
Figure 28); this ligand, then, can presumably oxidize the Fe and render the enzyme inactive.
Figure 25.
Lowest-energy docked pose of grandifontane with L. major dihydroorotate dehydrogenase (LmajDHODH, PDB 3mhu) showing key interactions with Cys 131, Asn 199, Asn 68, Ser 69, and Gln 139. Hydrogen-bonds are shown as blue dashed lines.
Figure 25.
Lowest-energy docked pose of grandifontane with L. major dihydroorotate dehydrogenase (LmajDHODH, PDB 3mhu) showing key interactions with Cys 131, Asn 199, Asn 68, Ser 69, and Gln 139. Hydrogen-bonds are shown as blue dashed lines.
Figure 26.
Lowest-energy docked pose of 6-O-acetylswietenolide with L. mexicana glycerol-3-phosphate dehydrogenase (LmexGPDH, PDB 1n1e) showing key interactions with Arg 274, Ser 293, Phe 26 and Ala 157, and Gln 139. Hydrogen-bonds are shown as blue dashed lines.
Figure 26.
Lowest-energy docked pose of 6-O-acetylswietenolide with L. mexicana glycerol-3-phosphate dehydrogenase (LmexGPDH, PDB 1n1e) showing key interactions with Arg 274, Ser 293, Phe 26 and Ala 157, and Gln 139. Hydrogen-bonds are shown as blue dashed lines.
Figure 27.
Lowest-energy poses of steroids, cholesterol (white), clerosterol (purple), saringosterol (green), stigmasterol (cyan), β-sitosterol (blue), and 24-hydroperoxy-24-vinylcholesterol (red), in the hydropobic pocket of L. major methionyl t-RNA synthetase (LmajMetRS, PDB 3kfl).
Figure 27.
Lowest-energy poses of steroids, cholesterol (white), clerosterol (purple), saringosterol (green), stigmasterol (cyan), β-sitosterol (blue), and 24-hydroperoxy-24-vinylcholesterol (red), in the hydropobic pocket of L. major methionyl t-RNA synthetase (LmajMetRS, PDB 3kfl).
Figure 28.
Lowest-energy poses of 24-hydroperoxy-24-vinylcholesterol with L. infantum sterol 14α-demethylase (LinfCYP51, PDB 3l4d). Note the proximity of the hydroperoxy group of the ligand with the Fe atom of the heme cofactor.
Figure 28.
Lowest-energy poses of 24-hydroperoxy-24-vinylcholesterol with L. infantum sterol 14α-demethylase (LinfCYP51, PDB 3l4d). Note the proximity of the hydroperoxy group of the ligand with the Fe atom of the heme cofactor.
An examination of docking energies with respect to ligand molecular size suggests that for terpenoid ligands there is a threshold where larger size does not correspond to stronger binding to the protein target. A plot of molecular weights of representative terpenoids (monoterpenoids, germacranolide sesquiterpenoids, labdane diterpenoids, and triterpenoids) and docking energies to three different protein targets (LmajMetRS, LmexGPDH, and LdonCyp) (
Figure 29) shows that strongest docking energies are terpenoids with molecular weights around 360–430 amu.