Solid-, Solution-, and Gas-state NMR Monitoring of 13C-Cellulose Degradation in an Anaerobic Microbial Ecosystem
Abstract
:1. Introduction
2. Results and Discussion
2.1. Solid-State NMR Spectroscopy
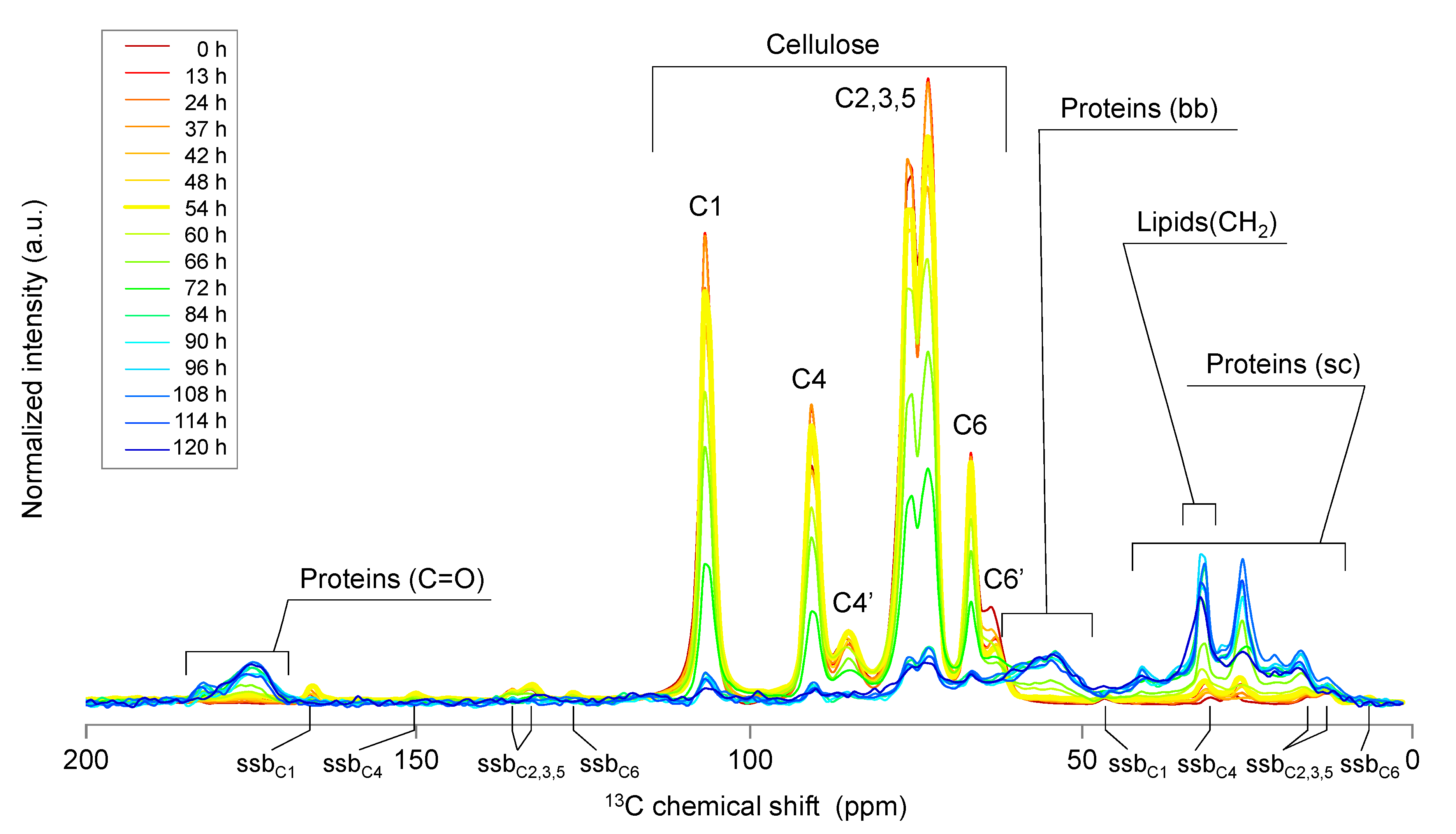

2.2. Solution-State NMR Spectroscopy
2.3. Relationship between 13C-BC Degradation and Production of 13C-Labeled Metabolites
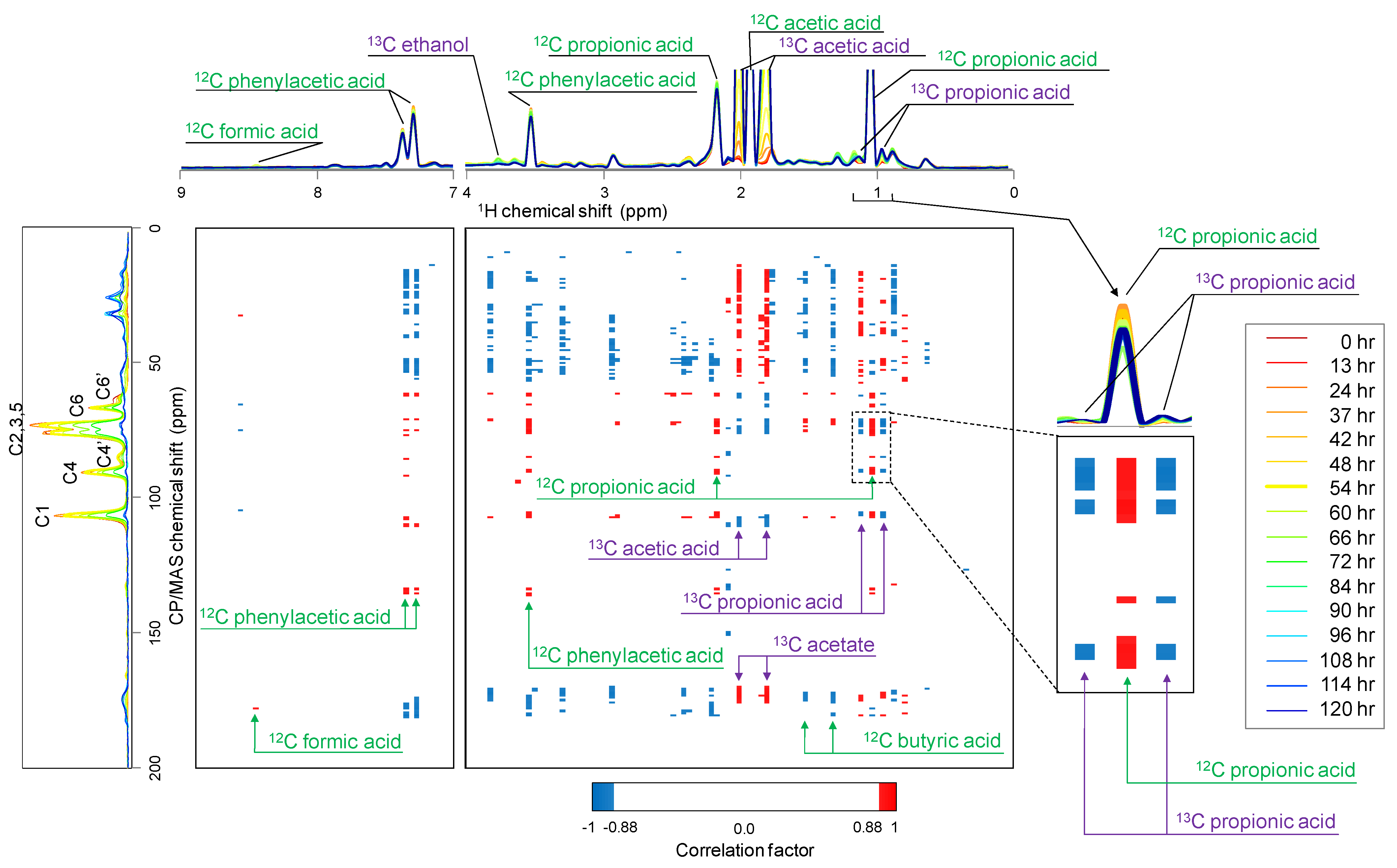
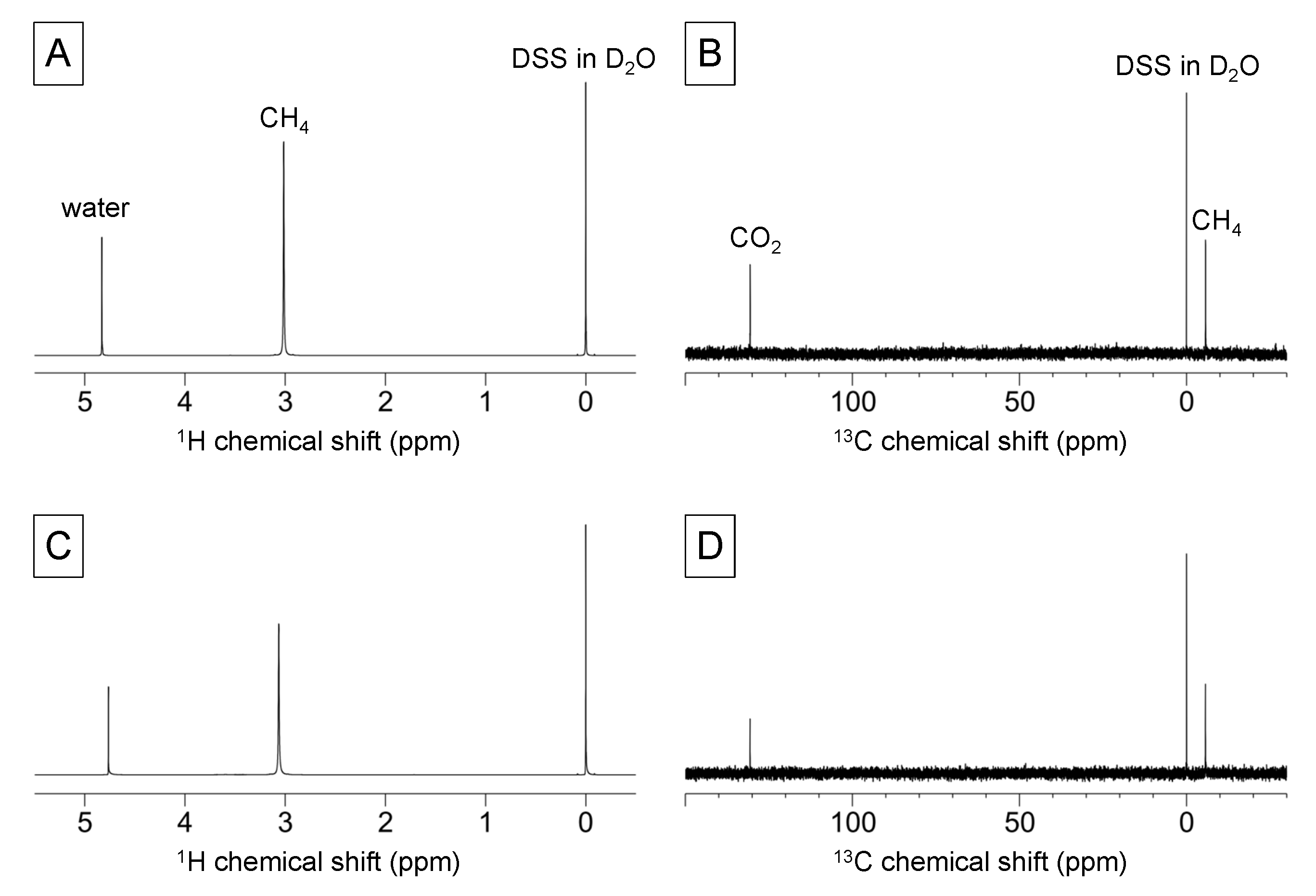
2.4. Gas-State NMR Spectroscopy
2.5. Triple-Phase Monitoring of Anaerobic Digestion Using NMR Spectroscopy
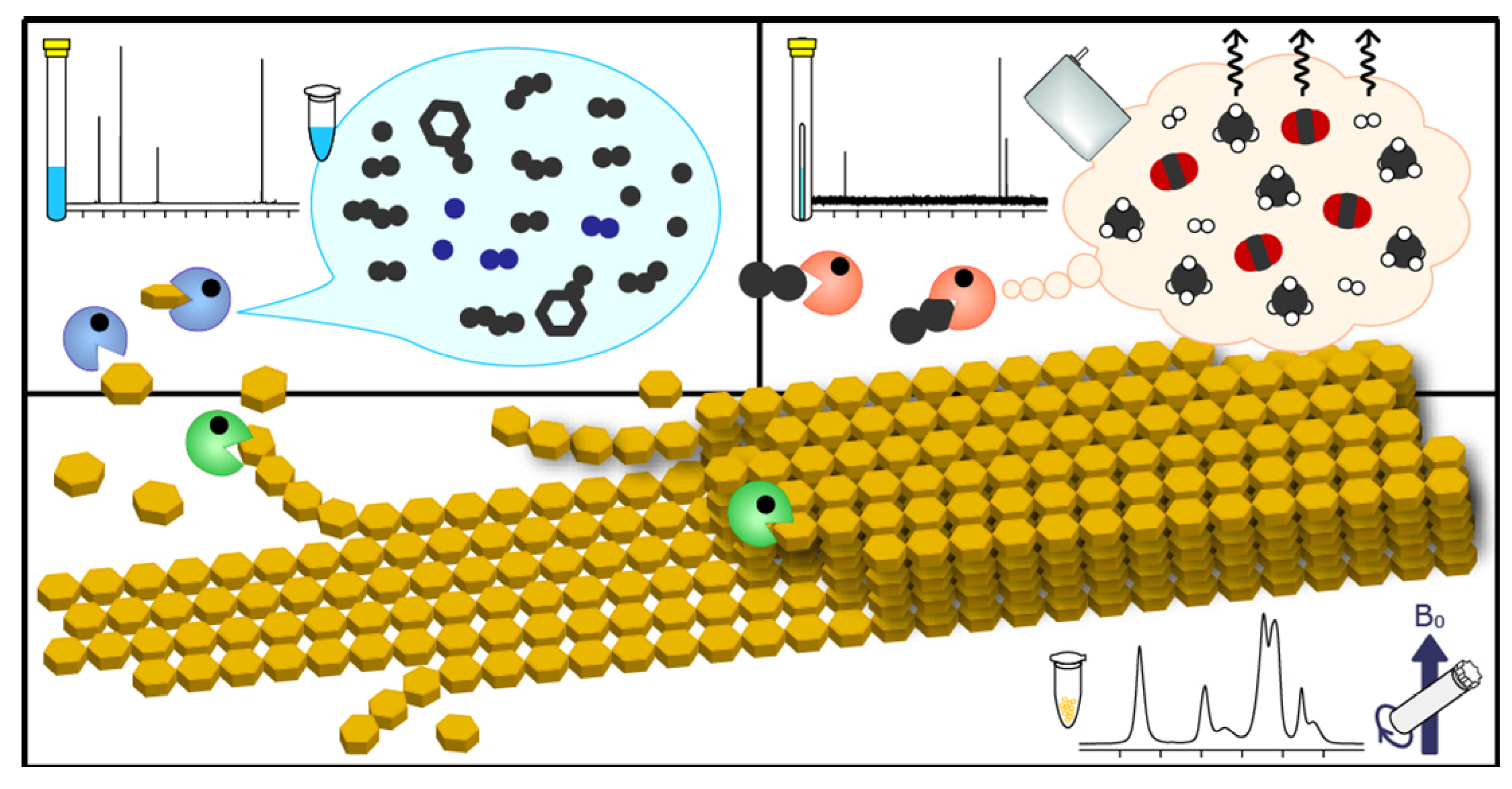
3. Experimental
3.1. General
3.2. Materials
3.3. Solid-State NMR Spectroscopy
3.4. Solution-State NMR Spectroscopy
3.5. Gas-State NMR Spectroscopy
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Conflicts of Interest
References
- Schink, B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol. Mol. Biol. Rev. 1997, 61, 262–280. [Google Scholar]
- Wahlen, M. The Global Methane Cycle. Annu. Rev. Earth Planet. Sci. 1993, 21, 407–426. [Google Scholar] [CrossRef]
- Chynoweth, D.P.; Owens, J.M.; Legrand, R. Renewable methane from anaerobic digestion of biomass. Renewable Energy 2001, 22, 1–8. [Google Scholar] [CrossRef]
- Klemm, D.; Heublein, B.; Fink, H.P.; Bohn, A. Cellulose: Fascinating biopolymer and sustainable raw material. Angew. Chem. Int. Ed. Engl. 2005, 44, 3358–3393. [Google Scholar] [CrossRef]
- Rubin, E.M. Genomics of cellulosic biofuels. Nature 2008, 454, 841–845. [Google Scholar] [CrossRef]
- Kikuchi, J.; Asakura, T. Use of 13C conformation-dependent chemical shifts to elucidate the local structure of a large protein with homologous domains in solution and solid state. J. Biochem. Biophys. Methods 1999, 38, 203–208. [Google Scholar] [CrossRef]
- Mao, J.D.; Holtman, K.M.; Franqui-Villanueva, D. Chemical structures of corn stover and its residue after dilute acid prehydrolysis and enzymatic hydrolysis: Insight into factors limiting enzymatic hydrolysis. J. Agric. Food Chem. 2010, 58, 11680–11687. [Google Scholar] [CrossRef]
- Mao, J.D.; Schmidt-Rohr, K. Accurate quantification of aromaticity and nonprotonated aromatic carbon fraction in natural organic matter by 13C solid-state nuclear magnetic resonance. Environ. Sci. Technol. 2004, 38, 2680–2684. [Google Scholar] [CrossRef]
- Mori, T.; Chikayama, E.; Tsuboi, Y.; Ishida, N.; Shisa, N.; Noritake, Y.; Moriya, S.; Kikuchi, J. Exploring the conformational space of amorphous cellulose using NMR chemical shifts. Carbohydr. Polym. 2012, 90, 1197–1203. [Google Scholar] [CrossRef]
- Ogura, T.; Date, Y.; Kikuchi, J. Differences in cellulosic supramolecular structure of compositionally similar rice straw affect biomass metabolism by paddy soil microbiota. PLoS One 2013, 8, e66919. [Google Scholar] [CrossRef]
- Okushita, K.; Chikayama, E.; Kikuchi, J. Solubilization mechanism and characterization of the structural change of bacterial cellulose in regenerated states through ionic liquid treatment. Biomacromolecules 2012, 13, 1323–1330. [Google Scholar] [CrossRef]
- Okushita, K.; Komatsu, T.; Chikayama, E.; Kikuchi, J. Statistical approach for solid-state NMR spectra of cellulose derived from a series of variable parameters. Polym. J. 2012, 44, 895–900. [Google Scholar] [CrossRef]
- Everroad, R.C.; Yoshida, S.; Tsuboi, Y.; Date, Y.; Kikuchi, J.; Moriya, S. Concentration of metabolites from low-density planktonic communities for environmental metabolomics using nuclear magnetic resonance spectroscopy. J. Vis. Exp. 2012, 62, e3163. [Google Scholar]
- Nakanishi, Y.; Fukuda, S.; Chikayama, E.; Kimura, Y.; Ohno, H.; Kikuchi, J. Dynamic omics approach identifies nutrition-mediated microbial interactions. J. Proteome Res. 2010, 10, 824–836. [Google Scholar]
- Ellis, J.K.; Athersuch, T.J.; Thomas, L.D.K.; Teichert, F.; Perez-Trujillo, M.; Svendsen, C.; Spurgeon, D.J.; Singh, R.; Jarup, L.; Bundy, J.G.; et al. Metabolic profiling detects early effects of environmental and lifestyle exposure to cadmium in a human population. BMC Med. 2012, 10, 61. [Google Scholar] [CrossRef] [Green Version]
- Sekiyama, Y.; Chikayama, E.; Kikuchi, J. Evaluation of a semipolar solvent system as a step toward heteronuclear multidimensional NMR-based metabolomics for 13C-labeled bacteria, plants, and animals. Anal. Chem. 2011, 82, 1643–1652. [Google Scholar] [CrossRef]
- Viant, M.R.; Bearden, D.W.; Bundy, J.G.; Burton, I.W.; Collette, T.W.; Ekman, D.R.; Ezernieks, V.; Karakach, T.K.; Lin, C.Y.; Rochfort, S.; et al. International NMR-based environmental metabolomics intercomparison exercise. Environ. Sci. Technol. 2009, 43, 219–225. [Google Scholar] [CrossRef]
- Date, Y.; Nakanishi, Y.; Fukuda, S.; Kato, T.; Tsuneda, S.; Ohno, H.; Kikuchi, J. New monitoring approach for metabolic dynamics in microbial ecosystems using stable-isotope-labeling technologies. J. Biosci. Bioeng. 2010, 110, 87–93. [Google Scholar] [CrossRef]
- Ebrahimi, H.P.; Shaghaghi, H.; Tafazzoli, M. Gauge invariant atomic orbital-density functional theory prediction of accurate gas phase H-1 and C-13 NMR chemical shifts. Concepts Magn. Reson. Part A 2011, 38A, 269–279. [Google Scholar] [CrossRef]
- LeMaster, C.B. Nuclear magnetic resonance spectroscopy of molecules in the gas phase. Prog. Nucl. Magn. Reson. Spectrosc. 1997, 31, 119–154. [Google Scholar]
- Makulski, W. An 2H(D) isotope shift in the 1H NMR spectra of water in gaseous environment of fluoromethanes. J. Mol. Struct. 2007, 839, 90–93. [Google Scholar] [CrossRef]
- Tafazzoli, M.; Shaghaghi, H.; Jalali-Heravi, M. Calculations of gas phase 1H NMR chemical shifts of alcohols: An approach to optimize basis functions using factorial design. Concepts Magn. Reson. Part A 2008, 32A, 157–167. [Google Scholar] [CrossRef]
- Date, Y.; Iikura, T.; Yamazawa, A.; Moriya, S.; Kikuchi, J. Metabolic sequences of anaerobic fermentation on glucose-based feeding substrates based on correlation analyses of microbial and metabolite profiling. J. Proteome Res. 2012, 11, 5602–5610. [Google Scholar]
- Kikuchi, J.; Williamson, M.P.; Shimada, K.; Asakura, T. Structure and dynamics of photosynthetic membrane-bound proteins in Rhodobacter Sphaeroides, studied with solid-state NMR spectroscopy. Photosynth. Res. 2000, 63, 259–267. [Google Scholar] [CrossRef]
- Larsson, P.T.; Westermark, U.; Iversen, T. Determination of the cellulose I alpha allomorph content in a tunicate cellulose by CP/MAS C-13-NMR spectroscopy. Carbohydr. Res. 1995, 278, 339–343. [Google Scholar] [CrossRef]
- Larsson, P.T.; Wickholm, K.; Iversen, T. A CP/MAS C-13 NMR investigation of molecular ordering in celluloses. Carbohydr. Res. 1997, 302, 19–25. [Google Scholar] [CrossRef]
- Wickholm, K.; Larsson, P.T.; Iversen, T. Assignment of non-crystalline forms in cellulose I by CP/MAS C-13 NMR spectroscopy. Carbohydr. Res. 1998, 312, 123–129. [Google Scholar] [CrossRef]
- Ek, R.; Wormald, P.; Ostelius, J.; Iversen, T.; Nystrom, C. Crystallinity Index of Microcrystalline Cellulose Particles Compressed into Tablets. Int. J. Pharm. 1995, 125, 257–264. [Google Scholar] [CrossRef]
- Morrow, J.S.; Keim, P.; Visscher, R.B.; Marshall, R.C.; Gurd, F.R. Interaction of 13CO2 and bicarbonate with human hemoglobin preparations. Proc. Natl. Acad. Sci. USA 1973, 70, 1414–1418. [Google Scholar] [CrossRef]
- O'Leary M, H.; Jaworski, R.J.; Hartman, F.C. C nuclear magnetic resonance study of the CO2 activation of ribulosebisphosphate carboxylase from rhodospirillum rubrum. Proc. Natl. Acad. Sci. USA 1979, 76, 673–675. [Google Scholar] [CrossRef]
- Wojdyr, M. Fityk: A general-purpose peak fitting program. J. Appl. Cryst. 2010, 43, 1126–1128. [Google Scholar] [CrossRef]
- Kono, H.; Erata, T.; Takai, M. Complete assignment of the CP/MAS C-13 NMR spectrum of cellulose IIII. Macromolecules 2003, 36, 3589–3592. [Google Scholar] [CrossRef]
- Kono, H.; Yunoki, S.; Shikano, T.; Fujiwara, M.; Erata, T.; Takai, M. CP/MAS C-13 NMR study of cellulose and cellulose derivatives. 1. Complete assignment of the CP/MAS C-13 NMR spectrum of the native cellulose. J. Am. Chem. Soc. 2002, 124, 7506–7511. [Google Scholar]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on unix pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar]
- Akiyama, K.; Chikayama, E.; Yuasa, H.; Shimada, Y.; Tohge, T.; Shinozaki, K.; Hirai, M.Y.; Sakurai, T.; Kikuchi, J.; Saito, K. PRIMe: A web site that assembles tools for metabolomics and transcriptomics. In Silico Biol. 2008, 8, 339–345. [Google Scholar]
- Chikayama, E.; Sekiyama, Y.; Okamoto, Y.; Nakanishi, Y.; Tsuboi, Y.; Akiyama, K.; Saito, K.; Shinozaki, K.; Kikuchi, J. Statistical indices for simultaneous large-scale metabolite detections for a single NMR spectrum. Anal. Chem. 2010, 82, 1653–1658. [Google Scholar] [CrossRef]
- Chikayama, E.; Suto, M.; Nishihara, T.; Shinozaki, K.; Kikuchi, J. Systematic NMR analysis of stable isotope labeled metabolite mixtures in plant and animal systems: coarse grained views of metabolic pathways. PLoS One 2008, 3, e3805. [Google Scholar]
- Date, Y.; Sakata, K.; Kikuchi, J. Chemical profiling of complex biochemical mixtures from various seaweeds. Polym. J. 2012, 44, 888–894. [Google Scholar] [CrossRef]
- Sample Availability: Not available.
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yamazawa, A.; Iikura, T.; Shino, A.; Date, Y.; Kikuchi, J. Solid-, Solution-, and Gas-state NMR Monitoring of 13C-Cellulose Degradation in an Anaerobic Microbial Ecosystem. Molecules 2013, 18, 9021-9033. https://doi.org/10.3390/molecules18089021
Yamazawa A, Iikura T, Shino A, Date Y, Kikuchi J. Solid-, Solution-, and Gas-state NMR Monitoring of 13C-Cellulose Degradation in an Anaerobic Microbial Ecosystem. Molecules. 2013; 18(8):9021-9033. https://doi.org/10.3390/molecules18089021
Chicago/Turabian StyleYamazawa, Akira, Tomohiro Iikura, Amiu Shino, Yasuhiro Date, and Jun Kikuchi. 2013. "Solid-, Solution-, and Gas-state NMR Monitoring of 13C-Cellulose Degradation in an Anaerobic Microbial Ecosystem" Molecules 18, no. 8: 9021-9033. https://doi.org/10.3390/molecules18089021




