Addition of Bases to the 5'-end of Human Telomeric DNA: Influences on Thermal Stability and Energetics of Unfolding
Abstract
:1. Introduction
| Name | Sequence | a ΔHtot (kJ/mol) | b ΔΔHtot | c Tm(tot) (°C) | d ΔH(1) (kJ/mol) | e Tm(1) (°C) | d ΔH(2) (kJ/mol) | e Tm(2) (°C) |
|---|---|---|---|---|---|---|---|---|
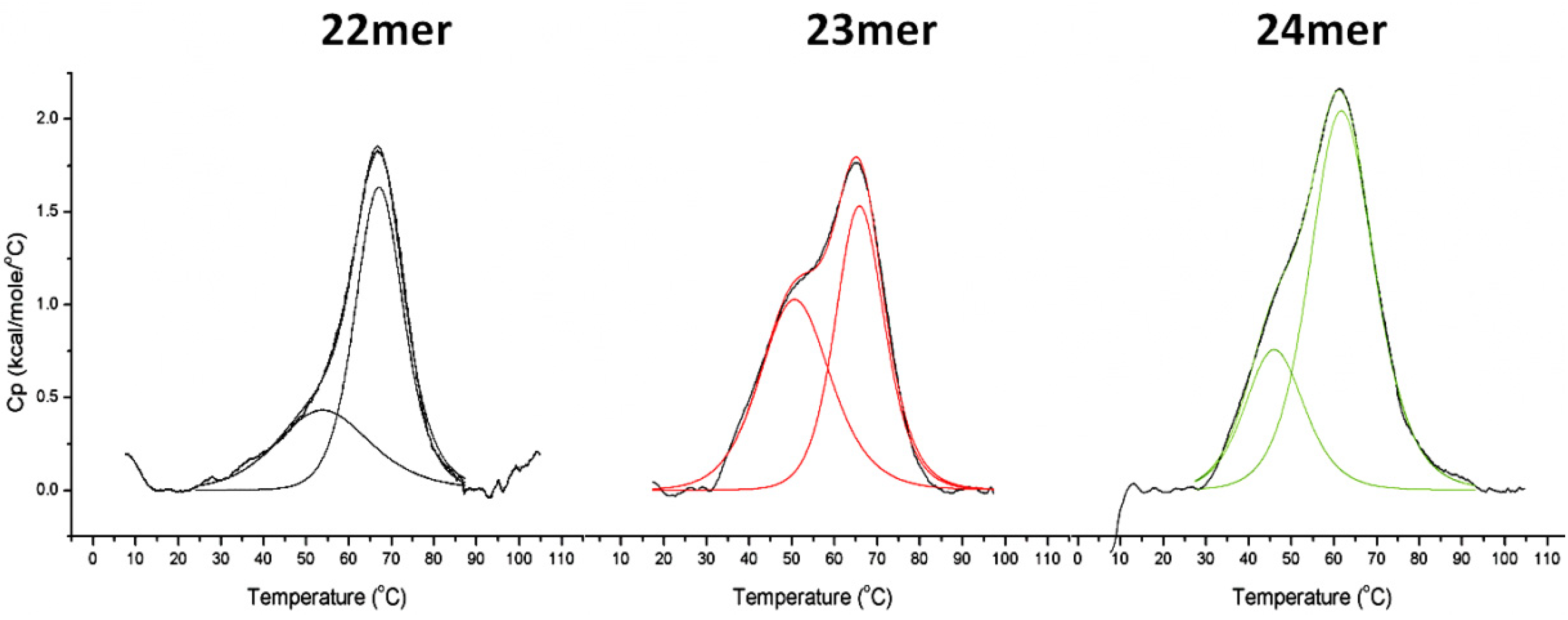
| 22mer | 5'-AGGG(TTAGGG)3-3' | 154.5 | NA | 67.2 | 57.4 | 53.5 | 98.9 | 67.2 |
| 23mer | 5'-TAGGG(TTAGGG)3-3' | 189.2 | 34.7 | 62.4 | 72.3 | 52.3 | 98.5 | 66.5 |
| 24mer | 5'-TTAGGG(TTAGGG)3-3' | 229.7 | 75.2 | 61.3 | 60.0 | 46.2 | 174.3 | 62.4 |
2. Results and Discussion
2.1. CD Analysis of G-Quadruplex Structures

2.2. DSC Analysis of G-Quadruplex Stability
2.3. Molecular Dynamics Simulation on the Parent h-Tel22 and the 23-mer 24-mer G-quadruplexes

2.4. DSC with Osmotic Perturbation


| Name | Sequence | R2 | Δnw | Error |
|---|---|---|---|---|
| 22-mer | 5'-AGGG(TTAGGG)3-3' | 0.9803 | 61.1 | ±4.3 |
| 23-mer | 5'-TAGGG(TTAGGG)3-3' | 0.9855 | 73.2 | ±4.4 |
| 24-mer | 5'-TTAGGG(TTAGGG)3-3' | 0.9916 | 59.2 | ±2.7 |
2.5. Pressure Perturbation Calorimetry (PPC)



3. Experimental
3.1. Circular Dichroism Spectropolarimetry
3.2. Differential Scanning Calorimetry
3.3. Molecular Dynamics Simulations
3.4. Differential Scanning Calorimetry with Osmolyte Perturbation
3.5. Pressure Pertrubration Calorimetry
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Petraccone, L.; Spink, C.; Trent, J.O.; Garbett, N.C.; Mekmaysy, C.S.; Giancola, C.; Chaires, J.B. Structure and stability of higher-order human telomeric quadruplexes. J. Am. Chem. Soc. 2011, 133, 20951–20961. [Google Scholar]
- Parkinson, G.N.; Lee, M.P.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef]
- Luu, K.N.; Phan, A.T.; Kuryavyi, V.; Lacroix, L.; Patel, D.J. Structure of the human telomere in K+ solution: An intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006, 128, 9963–9970. [Google Scholar] [CrossRef]
- Lim, K.W.; Amrane, S.; Bouaziz, S.; Xu, W.; Mu, Y.; Patel, D.J.; Luu, K.N.; Phan, A.T. Structure of the human telomere in K+ solution: A stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 2009, 131, 4301–4309. [Google Scholar]
- Gray, R.D.; Chaires, J.B. Kinetics and mechanism of K+- and Na+-induced folding of models of human telomeric DNA into G-quadruplex structures. Nucl. Acid. Res. 2008, 36, 4191–4203. [Google Scholar] [CrossRef]
- Gray, R.D.; Li, J.; Chaires, J.B. Energetics and kinetics of a conformational switch in G-quadruplex DNA. J. Phys. Chem. B 2009, 113, 2676–2683. [Google Scholar] [CrossRef]
- Wang, Y.; Patel, D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure 1993, 1, 263–282. [Google Scholar] [CrossRef]
- Wang, Y.; Patel, D.J. Solution structure of the Oxytricha telomeric repeat d[G4(T4G4)3] G-tetraplex. J. Mol. Biol. 1995, 251, 76–94. [Google Scholar] [CrossRef]
- Gray, D.M.; Wen, J.D.; Gray, C.W.; Repges, R.; Repges, C.; Raabe, G.; Fleischhauer, J. Measured and calculated CD spectra of G-quartets stacked with the same or opposite polarities. Chirality 2008, 20, 431–440. [Google Scholar] [CrossRef]
- Gottarelli, G.; Lena, S.; Masiero, S.; Pieraccini, S.; Spada, G.P. The use of circular dichroism spectroscopy for studying the chiral molecular self-assembly: An overview. Chirality 2008, 20, 471–485. [Google Scholar] [CrossRef]
- Karsisiotis, A.I.; Hessari, N.M.; Novellino, E.; Spada, G.P.; Randazzo, A.; Webba da Silva, M. Topological characterization of nucleic acid G-quadruplexes by UV absorption and circular dichroism. Angew. Chem. 2011, 50, 10645–10648. [Google Scholar] [CrossRef]
- Parsegian, V.A.; Rand, R.P.; Rau, D.C. Osmotic stress, crowding, preferential hydration, and binding: A comparison of perspectives. Proc. Natl. Acad. Sci. USA 2000, 97, 3987–3992. [Google Scholar] [CrossRef]
- Spink, C.H.; Garbett, N.; Chaires, J.B. Enthalpies of DNA melting in the presence of osmolytes. Biophys. Chem. 2007, 126, 176–185. [Google Scholar] [CrossRef]
- Qu, X.; Chaires, J.B. Hydration changes for DNA intercalation reactions. J. Am. Chem. Soc. 2001, 123, 1–7. [Google Scholar] [CrossRef]
- Kiser, J.R.; Monk, R.W.; Smalls, R.L.; Petty, J.T. Hydration changes in the association of Hoechst 33258 with DNA. Biochemistry 2005, 44, 16988–16997. [Google Scholar] [CrossRef]
- Nakano, S.; Yamaguchi, D.; Tateishi-Karimata, H.; Miyoshi, D.; Sugimoto, N. Hydration changes upon DNA folding studied by osmotic stress experiments. Biophys. J. 2012, 102, 2808–2817. [Google Scholar] [CrossRef]
- Cantor, C.R.; Warshaw, M.M.; Shapiro, H. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers 1970, 9, 1059–1077. [Google Scholar] [CrossRef]
- Olsen, C.M.; Gmeiner, W.H.; Marky, L.A. Unfolding of G-quadruplexes: energetic, and ion and water contributions of G-quartet stacking. J. Phys. Chem. B 2006, 110, 6962–6969. [Google Scholar] [CrossRef]
- Miyoshi, D.; Karimata, H.; Sugimoto, N. Hydration regulates thermodynamics of G-quadruplex formation under molecular crowding conditions. J. Am. Chem. Soc. 2006, 128, 7957–7963. [Google Scholar] [CrossRef]
- Miller, M.C.; Buscaglia, R.; Chaires, J.B.; Lane, A.N.; Trent, J.O. Hydration is a major determinant of the G-quadruplex stability and conformation of the human telomere 3' sequence of d(AG3(TTAG3)3). J. Am. Chem. Soc. 2010, 132, 17105–17107. [Google Scholar]
- Spink, C.H.; Chaires, J.B. Effects of hydration, ion release, and excluded volume on the melting of triplex and duplex DNA. Biochemistry 1999, 38, 496–508. [Google Scholar] [CrossRef]
- Lin, L.N.; Brandts, J.F.; Brandts, J.M.; Plotnikov, V. Determination of the volumetric properties of proteins and other solutes using pressure perturbation calorimetry. Anal. Biochem. 2002, 302, 144–160. [Google Scholar] [CrossRef]
- Fan, H.Y.; Shek, Y.L.; Amiri, A.; Dubins, D.N.; Heerklotz, H.; Macgregor, R.B., Jr.; Chalikian, T.V. Volumetric characterization of sodium-induced G-quadruplex formation. J. Am. Chem. Soc. 2011, 133, 4518–4526. [Google Scholar]
- Chalikian, T.V.; Breslauer, K.J. Volumetric properties of nucleic acids. Biopolymers 1998, 48, 264–280. [Google Scholar]
- Schweiker, K.L.; Makhatadze, G.I. Use of pressure perturbation calorimetry to characterize the volumetric properties of proteins. Meth. Enzymol. 2009, 466, 527–547. [Google Scholar] [CrossRef]
- Wu, J.Q.; Macgregor, R.B., Jr. Pressure dependence of the melting temperature of dA·dT polymers. Biochemistry 1993, 32, 12531–12537. [Google Scholar] [CrossRef]
- Case, D.A.; Darden, T.A.; Cheatham, T.E.; Simmerling, C.L.; Wang, J.; Duke, R.E.; Luo, R.; Walker, R.C.; Zhang, W.; Merz, K.M.; et al. AMBER 12; University of California: San Francisco, CA, USA, 2012. [Google Scholar]
- Freier, S.M.; Alkema, D.; Sinclair, A.; Neilson, T.; Turner, D.H. Contributions of dangling end stacking and terminal base-pair formation to the stabilities of XGGCCp, XCCGGp, XGGCCYp, and XCCGGYp helixes. Biochemistry 1985, 24, 4533–4539. [Google Scholar] [CrossRef]
- Freier, S.M.; Albergo, D.D.; Turner, D.H. Solvent effects on the dynamics of (dG-dC)3. Biopolymers 1983, 22, 1107–1131. [Google Scholar] [CrossRef]
- Sample Availability: Not available.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hayden, K.L.; Graves, D.E. Addition of Bases to the 5'-end of Human Telomeric DNA: Influences on Thermal Stability and Energetics of Unfolding. Molecules 2014, 19, 2286-2298. https://doi.org/10.3390/molecules19022286
Hayden KL, Graves DE. Addition of Bases to the 5'-end of Human Telomeric DNA: Influences on Thermal Stability and Energetics of Unfolding. Molecules. 2014; 19(2):2286-2298. https://doi.org/10.3390/molecules19022286
Chicago/Turabian StyleHayden, Katherine L., and David E. Graves. 2014. "Addition of Bases to the 5'-end of Human Telomeric DNA: Influences on Thermal Stability and Energetics of Unfolding" Molecules 19, no. 2: 2286-2298. https://doi.org/10.3390/molecules19022286




