A Novel Phage-Library-Selected Peptide Inhibits Human TNF-α Binding to Its Receptors
Abstract
:1. Introduction
2. Results and Discussion
2.1. Phage Display and Synthesis

2.2. Binding Experiments by SPR

2.3. Human TNF-α Binding to Adalimumab was Inhibited by Tetra-Branched Anti-TNF-α Peptide

2.4. Branched Anti-TNF-α Peptide Inhibited Binding of TNF-α to TNF Receptor2

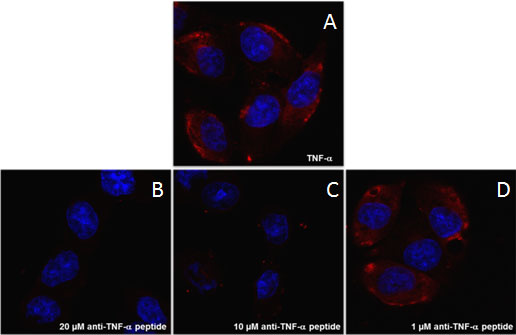
2.5. Branched Anti-TNF-α Peptide Inhibited Binding of TNF-α to Cells Expressing TNF-α Receptors

2.6. Branched Anti-TNF-α Peptide Dissociated Homotrimeric Form of hTNF-α
3. Experimental Section
3.1. Biotinylation of Human TNF-α

3.2. Phage Display
3.3. Peptide Synthesis
3.4. Surface Plasmon Resonance (SPR).
3.5. Cell Cultures
3.6. Confocal Microscopy
3.7. Mass Spectrometry Analysis of TNF-α Dissociation
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Baud, V.; Karin, M. Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 2001, 11, 372–377. [Google Scholar] [CrossRef]
- Beutler, B.A. The role of tumor necrosis factor in health and disease. J. Rheumatol. 1999, 26, 16–21. [Google Scholar]
- Tracey, K.J.; Cerami, A. Tumor necrosis factor, other cytokines and disease. Annu. Rev. Cell Biol. 1993, 9, 317–343. [Google Scholar] [CrossRef]
- Kriegler, M.; Perez, C.; de Fay, K.; Albert, I.; Lu, S.D. A novel form of TNF/cachectin is a cell surface cytotoxic transmembrane protein: Ramifications for the complex physiology of TNF. Cell 1998, 53, 45–53. [Google Scholar]
- Luettiq, B.; Decker, T.; Lohmann-Matthes, M.L. Evidence for the existence of two forms of membrane tumor necrosis factor: An integral protein and a molecule attached to its receptor. J. Immunol. 1989, 143, 4034–4038. [Google Scholar]
- Tracey, D.; Klareskog, L.; Sasso, E.H.; Salfeld, J.G.; Tak, P.P. Tumor necrosis factor antagonist mechanisms of action: A comprehensive review. Pharmacol. Therapeut. 2008, 117, 244–279. [Google Scholar] [CrossRef]
- Blüml, S.; Scheinecker, C.; Smolen, J.S.; Redlich, K. Targeting TNF receptors in rheumatoid arthritis. Int. Immunol. 2012, 24, 275–281. [Google Scholar]
- Bazzoni, F.; Beutler, B. The tumor necrosis factor ligand and receptor families. N. Engl. J. Med. 1996, 334, 1717–1725. [Google Scholar] [CrossRef]
- Moss, M.L.; Jin, S.L.; Milla, M.E.; Bickett, D.M.; Burkhart, W.; Carter, H.L.; Chen, W.J.; Clay, W.C.; Didsbury, J.R.; Hassler, D.; et al. Cloning of a disintegrin metalloproteinase that processes precursor tumour necrosis factor-alpha. Nature 1997, 385, 733–736. [Google Scholar] [CrossRef]
- Black, R.A.; Rauch, C.T.; Kozlosky, C.J.; Peschon, J.J.; Slack, J.L.; Wolfson, M.F.; Castner, B.J.; Stocking, K.L.; Reddy, P.; Srinivasan, S.; et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature 1997, 385, 729–733. [Google Scholar] [CrossRef]
- Vandenabeele, P.; Declercq, W.; Beyaert, R.; Fiers, W. Two tumour necrosis factor receptors: Structure and function. Trends Cell. Biol. 1995, 5, 392–399. [Google Scholar] [CrossRef]
- Tang, P.; Hung, M.C.; Klostergaard, J. Human pro-tumor necrosis factor is a homotrimer. Biochemistry 1996, 35, 8216–8225. [Google Scholar] [CrossRef]
- Kollias, G.; Douni, E.; Kassiotis, G.; Kontoyiannis, D. On the role of tumor necrosis factor and receptors in models of multiorgan failure, rheumatoid arthritis, multiple sclerosis and inflammatory bowel disease. Immunol. Rev. 1999, 169, 175–194. [Google Scholar] [CrossRef]
- Benucci, M.; Saviola, G.; Manfredi, M.; Sarzi-Puttini, P.; Atzeni, F. Tumor necrosis factors blocking agents: Analogies and differences. Acta Biomed. 2012, 83, 72–80. [Google Scholar]
- Tam, J.P. Synthetic peptide vaccine design: Synthesis and properties of a high-density multiple antigenic peptide system. Proc. Natl. Acad. Sci. USA 1998, 85, 5409–5413. [Google Scholar] [CrossRef]
- Bracci, L.; Falciani, C.; Lelli, B.; Lozzi, L.; Runci, Y.; Pini, A.; de Montis, M.G.; Tagliamonte, A.; Neri, P. Synthetic peptides in the form of dendrimers become resistant to protease activity. J. Biol. Chem. 2003, 278, 46590–46595. [Google Scholar] [CrossRef]
- Falciani, C.; Lozzi, L.; Pini, A.; Corti, F.; Fabbrini, M.; Bernini, A.; Lelli, B.; Niccolai, N.; Bracci, L. Molecular basis of branched peptides resistance to enzyme proteolysis. Chem. Biol. Drug Des. 2007, 69, 216–221. [Google Scholar] [CrossRef]
- Pini, A.; Falciani, C.; Bracci, L. Branched peptides as therapeutics. Curr. Protein Pept. Sci. 2008, 9, 468–477. [Google Scholar] [CrossRef]
- Pini, A.; Giuliani, A.; Falciani, C.; Fabbrini, M.; Pileri, S.; Lelli, B.; Bracci, L. Characterization of the branched antimicrobial peptide M6 by analyzing its mechanism of action and in vivo toxicity. J. Pept. Sci. 2007, 13, 393–399. [Google Scholar] [CrossRef]
- Lozzi, L.; Lelli, B.; Runci, Y.; Scali, S.; Bernini, A.; Falciani, C.; Pini, A.; Niccolai, N.; Neri, P.; Bracci, L. Rational design and molecular diversity for the construction of anti-alpha-bungarotoxin antidotes with high affinity and in vivo efficiency. Chem. Biol. 2003, 10, 411–417. [Google Scholar] [CrossRef]
- Pini, A.; Runci, Y.; Falciani, C.; Lelli, B.; Brunetti, J.; Pileri, S.; Fabbrini, M.; Lozzi, L.; Ricci, C.; Bernini, A.; et al. Stable peptide inhibitors prevent binding of lethal and oedema factors to protective antigen and neutralize anthrax toxin in vivo. Biochem. J. 2006, 395, 157–163. [Google Scholar] [CrossRef]
- Pini, A.; Falciani, C.; Mantengoli, E.; Bindi, S.; Brunetti, J.; Iozzi, S.; Rossolini, G.M.; Bracci, L. A novel tetrabranched antimicrobial peptide that neutralizes bacterial lipopolysaccharide and prevents septic shock in vivo. Faseb. J. 2010, 24, 1015–1022. [Google Scholar] [CrossRef]
- Falciani, C.; Fabbrini, M.; Pini, A.; Lozzi, L.; Lelli, B.; Pileri, S.; Brunetti, J.; Bindi, S.; Scali, S.; Bracci, L. Synthesis and biological activity of stable branched neurotensin peptides for tumor targeting. Mol. Cancer Ther. 2007, 6, 2441–2448. [Google Scholar]
- Falciani, C.; Lelli, B.; Brunetti, J.; Pileri, S.; Cappelli, A.; Pini, A.; Pagliuca, C.; Ravenni, N.; Bencini, L.; Menichetti, S.; et al. Modular branched neurotensin peptides for tumor target tracing and receptor-mediated therapy: A proof-of-concept. Curr. Cancer Drug Targets. 2010, 10, 695–704. [Google Scholar] [CrossRef]
- Falciani, C.; Brunetti, J.; Lelli, B.; Ravenni, N.; Lozzi, L.; Depau, L.; Scali, S.; Bernini, A.; Pini, A.; Bracci, L. Cancer selectivity of tetrabranched neurotensin peptides is generated by simultaneous binding to sulfated glycosaminoglycans and protein receptors. J. Med. Chem. 2013, 56, 5009–5018. [Google Scholar] [CrossRef]
- Liu, Y.; Cheung, L.H.; Marks, J.W.; Rosenblum, M.G. Recombinant single-chain antibody fusion construct targeting human melanoma cells and containing tumor necrosis factor. Int. J. Cancer 2004, 108, 549–557. [Google Scholar] [CrossRef]
- Mitoma, H.; Horiuchi, T.; Tsukamoto, H.; Tamimoto, Y.; Kimoto, Y.; Uchino, A.; To, K.; Harashima, S.; Hatta, N.; Harada, M. Mechanisms for cytotoxic effects of anti-tumor necrosis factor agents on transmembrane tumor necrosis factor alpha-expressing cells: Comparison among infliximab, etanercept, and adalimumab. Arthritis Rheum. 2008, 58, 1248–1257. [Google Scholar] [CrossRef]
- Chappuis, S.; Ribi, C.; Spertini, F. Management of the adverse events induced by TNF-α blocking agents. Rev. Med. Suisse. 2013, 9, 12–16. [Google Scholar]
- Sand, F.L.; Thomsen, S.F. TNF-alpha inhibitors for chronic urticaria: Experience in 20 patients. J. Allergy 2013. [Google Scholar] [CrossRef]
- Ozkan, H.; Cetinkaya, A.S.; Yildiz, T.; Bozat, T. A rare side effect due to TNF-α blocking agent: Acute pleuropericarditis with adalimumab. Case Rep. Rheumatol 2013, 2013, 985914:1–985914:2. [Google Scholar]
- Efe, C. Drug induced autoimmune hepatitis and TNF-α blocking agents: Is there a real relationship? Autoimmun. Rev 2012, 12, 337–339. [Google Scholar] [CrossRef]
- Sclavons, C.; Burtea, C.; Boutry, S.; Laurent, S.; Vander Elst, L.; Muller, R.N. Phage Display screening for tumor necrosis factor-𝛼-binding peptides: Detection of inflammation in a mouse model of hepatitis. Int. J. Pept. 2013. [Google Scholar] [CrossRef]
- Bracci, L.; Pini, A.; Lozzi, L.; Lelli, B.; Battestin, P.; Spreafico, A.; Bernini, A.; Niccolai, N.; Neri, P. Mimicking the nicotinic receptor binding site by a single chain Fv selected by competitive panning from a synthetic phage library. J. Neurochem. 2001, 78, 24–31. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the anti-TNF-α compounds are available from the authors with the signature of an appropriate Material Transfer Agreement.
© 2014 by the authors. licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Brunetti, J.; Lelli, B.; Scali, S.; Falciani, C.; Bracci, L.; Pini, A. A Novel Phage-Library-Selected Peptide Inhibits Human TNF-α Binding to Its Receptors. Molecules 2014, 19, 7255-7268. https://doi.org/10.3390/molecules19067255
Brunetti J, Lelli B, Scali S, Falciani C, Bracci L, Pini A. A Novel Phage-Library-Selected Peptide Inhibits Human TNF-α Binding to Its Receptors. Molecules. 2014; 19(6):7255-7268. https://doi.org/10.3390/molecules19067255
Chicago/Turabian StyleBrunetti, Jlenia, Barbara Lelli, Silvia Scali, Chiara Falciani, Luisa Bracci, and Alessandro Pini. 2014. "A Novel Phage-Library-Selected Peptide Inhibits Human TNF-α Binding to Its Receptors" Molecules 19, no. 6: 7255-7268. https://doi.org/10.3390/molecules19067255