Expression Profile and Clinical Significance of MicroRNAs in Papillary Thyroid Carcinoma
Abstract
:1. Introduction
2. Results and Discussion
2.1. Results
2.1.1. Quality Evaluation of the Total miRNAs
2.1.2. Results of miRNA Microarray
| miRNA | A vs. B | A vs. C | B vs. C |
|---|---|---|---|
| hsa-miR-146b-5p | 1.08 | 43.36 | 40.25 |
| hsa-miR-136-5p | 3.11 | 5.46 | 1.75 |
| hsa-miR-199b-5p | 2.05 | 11.48 | 5.61 |
| hsa-miR-30a-3p | 0.49 | 0.21 | 0.42 |
| hsa-miR-122-5p | 1.08 | 0.28 | 0.26 |
| hsa-miR-222-3p | 0.84 | 2.87 | 3.41 |
| hsa-miR-221-3p | 0.71 | 2.44 | 3.42 |
| hsa-miR-10b-5p | 0.97 | 15.60 | 16.01 |
| hsa-miR-199a-3p/hsa-miR-199b-3p | 1.95 | 29.27 | 14.99 |
| hsa-miR-203 | 0.87 | 12.43 | 14.30 |
| hsa-miR-32-5p | 0.89 | 10.87 | 12.22 |
| hsa-miR-183-3p | 1.33 | 0.49 | 0.37 |
| hsa-miR-199a-5p | 2.02 | 3.95 | 1.95 |
| hsa-miR-149-3p | 1.02 | 0.50 | 0.49 |
| hsa-miR-514a-3p | 0.60 | 0.14 | 0.24 |
| hsa-miR-1243 | 0.13 | 0.07 | 0.50 |
| hsa-miR-513b | 0.32 | 0.23 | 0.73 |
2.1.3. Results of the qRT-PCR
2.1.4. Results of Real-Time RT-PCR Verification of the Microarray Analysis
| miRNAs | Tissues | Expression Levels |
|---|---|---|
| miR-30a-3p | T-NG | 3.58 (1.75–4.27) |
| T-PTC | 4.66 (3.54–5.37) * | |
| A-NG | 3.35 (2.27–3.66) ## | |
| A-PTC | 2.05 (0.61–2.96) **,##,Δ | |
| miR-122-5p | T-NG | 10.43 (9.98–12.79) |
| T-PTC | 10.52 (9.20–11.96) | |
| A-NG | 7.36 (6.66–7.98) **,## | |
| A-PTC | 7.55 (6.25–8.52) **,## | |
| miR-136-5p | T-NG | 10.60 (9.09–12.02) |
| T-PTC | 10.69 (9.09–11.34) | |
| A-NG | 7.56 (6.74–8.68) **,## | |
| A-PTC | 7.76 (6.94–8.82) **,## | |
| miR-146b-5p | T-NG | 3.12 (2.23–4.99) |
| T-PTC | −0.34 (−1.58–0.31) ** | |
| A-NG | 2.89 (1.92–4.30) ## | |
| A-PTC | 2.78 (1.52–3.84) ## | |
| miR-199b-5p | T-NG | 7.32 (5.79–7.90) |
| T-PTC | 4.6s7 (3.90–5.87) ** | |
| A-NG | 1.49 (0.68–4.22) **,## | |
| A-PTC | 3.31 (2.21–3.93) **,## |
| miRNAs | Benign Thyroid Nodular Goiter (15 cases) | PTC (36 cases) | p |
|---|---|---|---|
| miR-30a-3p | −0.28 (−1.0–1.11) | −2.89 (−4.15–−1.47) | 0.003 |
| miR-122-5p | −3.66 (−4.41–−1.55) | −3.04 (−5.47–−0.99) | 0.972 |
| miR-136-5p | −2.43 (−4.51–−1.20) | −2.62 (−4.07–−1.42) | 0.973 |
| miR-146b-5p | −1.04 (−2.42–1.31) | 3.72 (1.2-5.04) | 0.001 |
| miR-199b-5p | −4.68 (−7.11–−2.29) | −1.95 (−2.93–−0.04) | 0.007 |
2.1.5. Analysis of the Diagnostic Value of miRNAs for Distinguishing PTC from Benign Thyroid Nodular Goiter Tissues
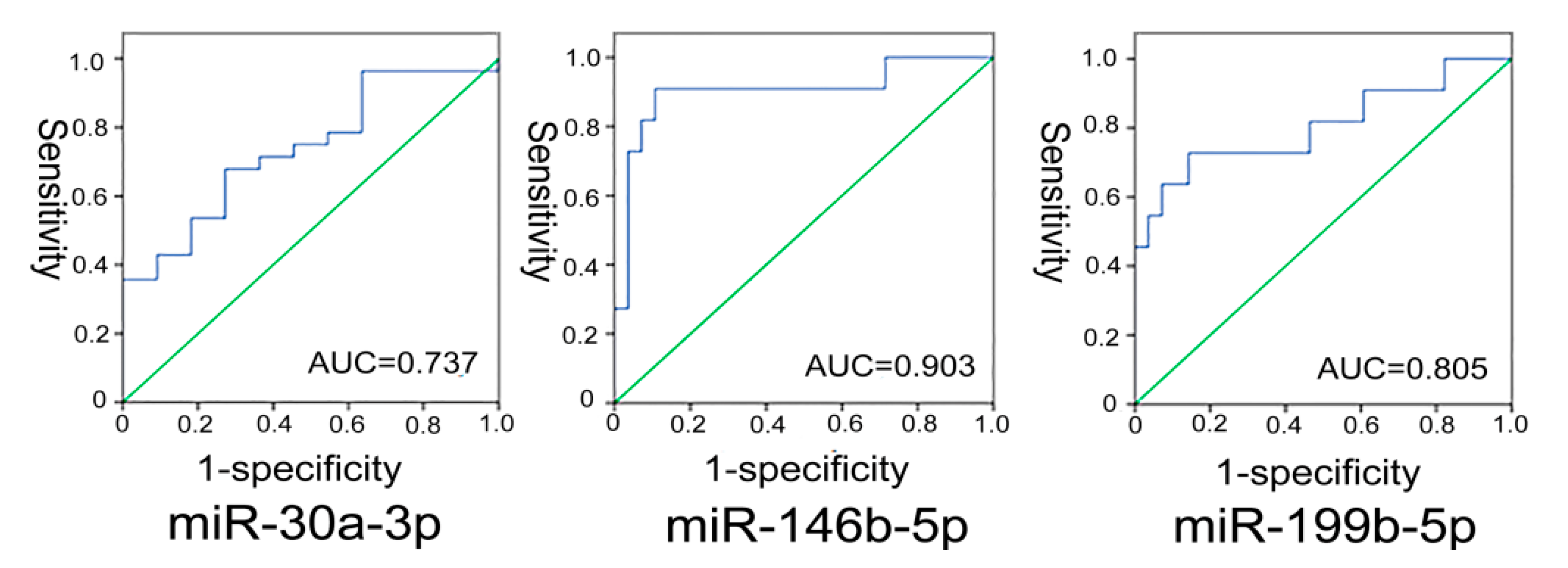
2.1.6. Correlation Analysis between miRNAs and Clinical-Pathological Features
| Clinical Features | Group | n | miR-30a-3p | miR-146b-5p | miR-199b-5p | |||
|---|---|---|---|---|---|---|---|---|
| Expression Level | p | Expression Level | p | Expression Level | p | |||
| Age | ≥45 | 21 | −2.56 (−3.32–−1.27) | 0.547 | 3.44 (0.97–3.69) | 0.705 | −2.54 (−3.39–−1.92) | 0.319 * |
| ˂45 | 15 | −1.54 (−4.39–−0.27) | 1.94 (0.97–4.97) | −1.81 (−3.67–−0.13) | ||||
| Sex | Male | 7 | −1.73 (−3.62–0.60) | 0.670 | 3.62 (1.12–6.12) | 0.279 | −0.56 (−2.84–0.17) | 0.092 * |
| Female | 29 | −2.099 (−3.69–−0.84) | 2.61 (0.16–4.08) | −2.54 (−3.85–−1.33) | ||||
| Tumor size | ≤1 cm | 14 | −1.61 (−3.88–0.33) | 0.482 | 3.57 (1.05–4.97) | 0.352 | −1.63 (−2.97–0.11) | 0.125 * |
| ˃1 cm | 22 | −2.329 (−3.50–−1.26) | 2.51 (−1.4–4.06) | −2.67 (−5.28–−1.73) | ||||
| Number of foci | Single lesion | 20 | −1.97 (−3.17–−1.23) | 0.058 | 3.84 (2.36–5.21) | 0.339 | −1.38 (−3.18–0.17) | 0.650 * |
| Multiple lesions | 16 | −3.60 (−4.72–−2.93) | 3.47 (1.15–4.33) | −1.98 (−2.71–−1.22) | ||||
| Extrathyroidal extension | NO | 27 | −2.93 (−4.56–−1.56) | 0.435 | 3.69 (1.15–5.10) | 0.678 | −2.51 (−2.97–−1.29) | 0.047 * |
| YES | 9 | −1.97 (−3.52–−1.47) | 3.73 (3.05–3.96) | 0.21 (−1.80–0.44) | ||||
| Lymph node metastasis | N0 | 15 | −3.15 (−3.88–−1.56) | 0.679 | 3.63 (1.15–4.06) | 0.303 | −2.54 (−3.39–−1.92) | 0.010 # |
| N1a | 9 | −2.37 (−3.60–−1.97) | 3.80 (3.47–5.45) | –2.61 (−3.65–−1.22) | ||||
| N1b | 12 | −1.65 (−4.56–0.87) | 4.97 (1.27–5.75) | 0.10 (−0.62–0.29) | ||||
2.2. Discussion
2.2.1. miRNA and Diagnosis of PTC
2.2.2. miRNA is Related to the Invasiveness of PTC
3. Experimental Section
3.1. Source of Specimens
3.2. Experiment Procedure
3.2.1. Microarrays Analysis of miRNA
3.2.2. Quantitative Real-Time RT-PCR Detection of miRNAs
3.2.2.1. Design of Forward Primers
| miRNAs | PCR Forward Primer |
|---|---|
| hsa-miR-146b-5p | 5'-TGAGAACTGAATTCCATAGGCT-3' |
| hsa-miR-199b-5p | 5'-CCCAGTGTTTAGACTATCTGTTC-3' |
| hsa-miR-30a-3p | 5'-CTTTCAGTCGGATGTTTGCAGC-3' |
| hsa-miR-122-5p | 5'-TGGAGTGTGACAATGGTGTTTG-3' |
| hsa-miR-136-5p | 5'-ACTCCATTTGTTTTGATGATGGA-3' |
| U6 RNA | 5'-TGCGGGTGCTCGCTTCGGCAGC-3' |
3.2.2.2. Extraction of Total RNA
3.2.2.3. Reverse Transcription
3.2.2.4. Amplification of PCR
3.3. Statistical Analysis
4. Conclusions
Supplementary Materials
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jemal, A.; Siegel, R.; Xu, J.; Ward, E. Cancer statistics. CA Cancer J. Clin. 2010, 60, 277–300. [Google Scholar] [CrossRef]
- Ferlay, J.; Steliarova-Foucher, E.; Lortet-Tieulent, J.; Rosso, S.; Coebergh, J.W.; Comber, H.; Forman, D.; Bray, F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012. Eur. J. Cancer 2013, 49, 1374–1403. [Google Scholar] [CrossRef]
- Cooper, D.S.; Doherty, G.M.; Haugen, B.R.; Kloos, R.T.; Lee, S.L.; Mandel, S.J.; Mazzaferri, E.L.; McIver, B.; Pacini, F.; Schlumberger, M.; et al. Revised American Thyroid Association management guidelines for patients with thyroid nodules and differentiated thyroid cancer: The American Thyroid Association (ATA) Guidelines Taskforce on Thyroid Nodules and Differentiated Thyroid Cancer. Thyroid 2009, 19, 1167–1214. [Google Scholar]
- Loh, K.C.; Greenspan, F.S.; Gee, L.; Miller, T.R.; Yeo, P.P. Pathological tumor-node-metastasis (pTNM) staging for papillary and follicular thyroid carcinomas: A retrospective analysis of 700 patients. J. Clin. Endocrinol. MeTable 1997, 82, 3553–3562. [Google Scholar] [CrossRef]
- Syrenicz, A.; Koziołek, M.; Ciechanowicz, A.; Sieradzka, A.; Bińczak-Kuleta, A.; Parczewski, M. New insights into the diagnosis of nodular goiter. Thyroid Res. 2014, 7. [Google Scholar] [CrossRef]
- Brito, J.P.; Hay, I.D.; Morris, J.C. Low risk papillary thyroid cancer. BMJ. 2014, 348. http://dx.doi.org/10.1136/bmj.g3045..
- Tufano, R.P.; Teixeira, G.V.; Bishop, J.; Carson, K.A.; Xing, M. BRAF mutation in papillary thyroid cancer and its value in tailoring initial treatment: A systematic review and meta-analysis. Medicine (Baltimore) 2012, 91, 274–286. [Google Scholar] [CrossRef]
- Jazdzewski, K.; Liyanarachchi, S.; Swierniak, M.; Pachucki, J.; Ringel, M.D.; Jarzab, B.; de la Chapelle, A. Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc. Natl. Acad. Sci. USA 2009, 106, 1502–1505. [Google Scholar] [CrossRef]
- Lodewijk, L.; Prins, A.M.; Kist, J.W.; Valk, G.D.; Kranenburg, O.; Rinkes, I.H.; Vriens, M.R. The value of miRNA in diagnosing thyroid cancer: A systematic review. Cancer Biomark. 2012, 11, 229–238. [Google Scholar]
- Giulia, C.; Pierfrancesco, F.; Loredana, P.; Salvatore, P.; Gianmauro, N. The Role of microRNA in head and neck cancer: Current knowledge and perspectives. Molecules 2014, 19, 5704–5716. [Google Scholar] [CrossRef]
- Vicentini, C.; Fassan, M.; D’Angelo, E.; Corbo, V.; Silvestris, N.; Nuovo, G.J.; Scarpa, A. Clinical application of microRNA testing in neuroendocrine tumors of the gastrointestinal tract. Molecules 2014, 19, 2458–2468. [Google Scholar] [CrossRef]
- Chun, Z.; Cong, Y.; Haopeng, L.; Guoyu, W.; Xijng, H. Combined elevation of microRNA-196a and microRNA-196b in sera predicts unfavorable prognosis in patients with osteosarcomas. Int. J. Mol. Sci. 2014, 15, 6544–6555. [Google Scholar] [CrossRef]
- Dettmer, M.S.; Perren, A.; Moch, H.; Komminoth, P.; Nikiforov, Y.E.; Nikiforova, M.N. MicroRNA profile of poorly differentiated thyroid carcinomas: New diagnostic and prognostic insights. J. Mol. Endocrinol. 2014, 52, 181–189. [Google Scholar] [CrossRef]
- Wojtas, B.; Ferraz, C.; Stokowy, T.; Hauptmann, S.; Lange, D.; Dralle, H.; Musholt, T.; Jarzab, B.; Paschke, R.; Eszlinger, M. Differential miRNA expression defines migration and reduced apoptosis in follicular thyroid carcinomas. Mol. Cell. Endocrinol. 2014, 388, 1–9. [Google Scholar] [CrossRef]
- Lundstrom, K. Micro-RNA in disease and gene therapy. Curr. Drug Discov. Technol. 2011, 8, 76–86. [Google Scholar] [CrossRef]
- Chou, C.K.; Yang, K.D.; Chou, F.F.; Huang, C.C.; Lan, Y.W.; Lee, Y.F.; Kang, H.Y.; Liu, R.T. Prognostic implications of miR-146b expression and its functional role in papillary thyroid carcinoma. J. Clin. Endocrinol. MeTable 2013, 98, E196–E205. [Google Scholar] [CrossRef]
- Yu, S.; Liu, Y.; Wang, J.; Guo, Z.; Zhang, Q.; Yu, F.; Zhang, Y.; Huang, K.; Li, Y.; Song, E.; et al. Circulating microRNA profiles as potential biomarkers for diagnosis of papillary thyroid carcinoma. J. Clin. Endocrinol. MeTable 2012, 97, 2084–2092. [Google Scholar] [CrossRef]
- He, H.; Jazdzewski, K.; Li, W.; Liyanarachchi, S.; Nagy, R.; Volinia, S.; Calin, G.A.; Liu, C.G.; Franssila, K.; Suster, S.; et al. The role of microRNA genes in papillary thyroid carcinoma. Proc. Natl. Acad. Sci. USA 2005, 102, 19075–19080. [Google Scholar] [CrossRef]
- Edge, S.B.; Compton, C.C. The American Joint Committee on Cancer: The 7th Edition of the AJCC cancer staging manual and the future of TNM. Ann. Surg. Oncol. 2010, 17, 1471–1474. [Google Scholar] [CrossRef]
- Pallante, P.; Visone, R.; Ferracin, M.; Ferraro, A.; Berlingieri, M.T.; Troncone, G.; Chiappetta, G.; Liu, C.G.; Santoro, M.; Negrini, M.; et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr. Relat. Cancer. 2006, 13, 497–508. [Google Scholar] [CrossRef]
- Acibucu, F.; Dökmetaş, H.S.; Tutar, Y.; Elagoz, S.; Kilicli, F. Correlations between the expression levels of Micro-RNA146b, 221, 222 and p27Kip1 protein mRNA and the clinicopathologic parameters in papillary thyroid cancers. Exp. Clin. Endocrinol. Diabetes 2014, 122, 137–143. [Google Scholar] [CrossRef]
- Yang, Z.; Yuan, Z.; Fan, Y.; Deng, X.; Zheng, Q. Integrated analyses of microRNA and mRNA expression profiles in aggressive papillary thyroid carcinoma. Mol. Med. Rep. 2013, 8, 1353–1358. [Google Scholar]
- Lee, J.C.; Zhao, J.T.; Clifton-Bligh, R.J.; Gill, A.; Gundara, J.S.; Ip, J.C.; Glover, A.; Sywak, M.S.; Delbridge, L.W.; Robinson, B.G.; et al. MicroRNA-222 and microRNA-146b are tissue and circulating biomarkers of recurrent papillary thyroid cancer. Cancer 2013, 119, 4358–4365. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, H.; He, L.; Dong, W.; Li, J.; Shan, Z.; Teng, W. Association between the expression of four upregulated miRNAs and extrathyroidal invasion in papillary thyroid carcinoma. Onco Targets Ther. 2013, 6, 281–287. [Google Scholar]
- Lloyd, R.V.; Buehler, D.; Khanafshar, E. Papillary thyroid carcinoma variants. Head Neck Pathol. 2011, 5, 51–56. [Google Scholar] [CrossRef]
- Rossing, M.; Borup, R.; Henao, R.; Winther, O.; Vikesaa, J.; Niazi, O.; Godballe, C.; Krogdahl, A.; Glud, M.; Hjort-Sørensen, C.; et al. Down-regulation of microRNAs controlling tumourigenic factors in follicular thyroid carcinoma. J. Mol. Endocrinol. 2012, 48, 11–23. [Google Scholar] [CrossRef]
- Nikiforova, M.N.; Tseng, G.C.; Steward, D.; Diorio, D.; Nikiforov, Y.E. MicroRNA expression profiling of thyroid tumors: Biological significance and diagnostic utility. J. Clin. Endocrinol. MeTable 2008, 93, 1600–1608. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.; Liu, Z.; Wang, X.M.; Yin, D.T.; Zheng, L.L.; Zhang, D.Y.; Lu, X.B. Differential expression profiling and functional analysis of microRNAs through stage I–III papillary thyroid carcinoma. Int. J. Med. Sci. 2013, 10, 585–592. [Google Scholar] [CrossRef]
- Liu, X.; He, M.; Hou, Y.; Liang, B.; Zhao, L.; Ma, S.; Yu, Y.; Liu, X. Expression profiles of microRNAs and their target genes in papillary thyroid carcinoma. Oncol. Rep. 2013, 29, 1415–1420. [Google Scholar]
- Yip, L.; Kelly, L.; Shuai, Y.; Armstrong, M.J.; Nikiforov, Y.E.; Carty, S.E.; Nikiforova, M.N. MicroRNA signature distinguishes the degree of aggressiveness of papillary thyroid carcinoma. Ann. Surg. Oncol. 2011, 18, 2035–2041. [Google Scholar]
- Chou, C.K; Chen, R.F.; Chou, F.F.; Chang, H.W.; Chen, Y.J.; Lee, Y.F.; Yang, K.D.; Cheng, J.T.; Huang, C.C.; Liu, R.T. miR-146b is highly expressed in adult papillary thyroid carcinomas with high risk features including extrathyroidal invasion and the BRAF(V600E) mutation. Thyroid 2010, 20, 489–494. [Google Scholar] [CrossRef]
- Natali, P.G.; Berlingieri, M.T.; Nicotra, M.R.; Fusco, A.; Santoro, E.; Bigotti, A.; Vecchio, G. Transformation of thyroid epithelium is associated with loss of c-kit receptor. Cancer Res. 1995, 55, 1787–1791. [Google Scholar]
- Hurst, D.R.; Edmonds, M.D.; Scott, G.K.; Benz, C.C.; Vaidya, K.S.; Welch, D.R. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 2009, 69, 1279–1283. [Google Scholar] [CrossRef]
- Katakowski, M.; Zheng, X.; Jiang, F.; Rogers, T.; Szalad, A.; Chopp, M. MiR-146b-5p suppresses EGFR expression and reduces in vitro migration and invasion of glioma. Cancer Investig. 2010, 28, 1024–1030. [Google Scholar] [CrossRef]
- Braun, J.; Hoang-Vu, C.; Dralle, H.; Hüttelmaier, S. Downregulation of microRNAs directs the EMT and invasive potential of anaplastic thyroid carcinomas. Oncogene 2010, 29, 4237–4244. [Google Scholar] [CrossRef]
- Wang, C.; Song, B.; Song, W.; Liu, J.; Sun, A.; Wu, D.; Yu, H.; Lian, J.; Chen, L.; Han, J. Underexpressed microRNA-199b-5p targets hypoxia-inducible factor-1α in hepatocellular carcinoma and predicts prognosis of hepatocellular carcinoma patients. J. Gastroenterol. Hepatol. 2011, 26, 1630–1637. [Google Scholar] [CrossRef]
- Flamant, S.; Ritchie, W.; Guilhot, J.; Holst, J.; Bonnet, M.L.; Chomel, J.C.; Guilhot, F.; Turhan, A.G.; Rasko, J.E. Micro-RNA response to imatinib mesylate in patients with chronic myeloid leukemia. Haematologica 2010, 95, 1325–1333. [Google Scholar] [CrossRef]
- Nymark, P.; Guled, M.; Borze, I.; Faisal, A.; Lahti, L.; Salmenkivi, K.; Kettunen, E.; Anttila, S.; Knuutila, S. Integrative analysis of microRNA, mRNA and aCGH data reveals asbestos- and histology-related changes in lung cancer. Gene. Chromosome. Canc. 2011, 50, 585–597. [Google Scholar] [CrossRef]
- The Diana Lab Homepage. Available online: http://diana.cslab.ece.ntua.gr/ (accessed on 4 August 2014).
- miRBase: The microRNA Database. Available online: http://www.mirbase.org/ (accessed on 4 August 2014).
- Sample Availability: Not available.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Peng, Y.; Li, C.; Luo, D.-C.; Ding, J.-W.; Zhang, W.; Pan, G. Expression Profile and Clinical Significance of MicroRNAs in Papillary Thyroid Carcinoma. Molecules 2014, 19, 11586-11599. https://doi.org/10.3390/molecules190811586
Peng Y, Li C, Luo D-C, Ding J-W, Zhang W, Pan G. Expression Profile and Clinical Significance of MicroRNAs in Papillary Thyroid Carcinoma. Molecules. 2014; 19(8):11586-11599. https://doi.org/10.3390/molecules190811586
Chicago/Turabian StylePeng, You, Chen Li, Ding-Cun Luo, Jin-Wang Ding, Wo Zhang, and Gang Pan. 2014. "Expression Profile and Clinical Significance of MicroRNAs in Papillary Thyroid Carcinoma" Molecules 19, no. 8: 11586-11599. https://doi.org/10.3390/molecules190811586




