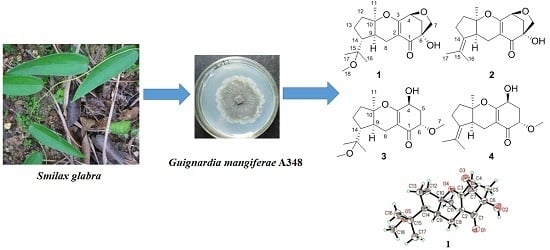
Guignardones P–S, New Meroterpenoids from the Endophytic Fungus Guignardia mangiferae A348 Derived from the Medicinal Plant Smilax glabra
Abstract
:1. Introduction

2. Results and Discussion
2.1. Identification of New Compounds
| Position | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| H-4 | 4.55, d (5.4) | 4.58, d (5.4) | 4.27, m | 4.31, t (5.4) |
| H-5 | 2.45, dd (10.7, 5.5) | 2.44, dd (10.7, 5.5) | 2.41, m | 2.43, m |
| 2.02, d (10.7) | 2.04, d (10.7) | 2.24, m | 2.19, m | |
| H-6 | 3.72, dd (7.5, 3.8) | 3.73 dd (7.5, 3.8) | ||
| H-7 | 3.79, d (7.9) | 3.80, d (7.9) | 3.49, s | 3.49, s |
| 3.48, d(7.9) | 3.51, d (7.9) | |||
| H-8 | 2.66, dd (17.0, 1.2) | 2.63, dd (17.2, 7.2) | 2.66, d (17.2) | 2.64, dd (17.2, 7.2) |
| 2.20, dd (17.0, 6.1) | 1.88, m | 2.20, m | 1.85, m | |
| H-9 | 2.04, m | 2.39, t (8.3) | 2.04, m | 2.47, m |
| H-11 | 1.29, s | 1.35, s | 1.33, s | 1.39, s |
| H-12 | 1.98, m | 1.85, m | 2.01, m | 1.89, m |
| 1.62, m | 1.81, m | 1.60, m | 1.80, m | |
| H-13 | 1.72, m | 2.34, m | 1.72, m | 2.32, m |
| 1.52, m | 2.21, m | 1.60, m | 2.24, m | |
| H-14 | 1.71, m | 1.74, m | ||
| H-16 | 1.09, s | 1.69, s | 1.12, s | 1.71, s |
| H-17 | 1.08, s | 1.57, s | 1.12, s | 1.59, s |
| H-18 | 3.08, s | 3.12, s | ||
| OH | 4.26, brs | 4.24, brs | 3.24, d (7.2) |
 ), HMBC (
), HMBC (  ), and NOE (
), and NOE (  ) correlations of compounds 1–4.
) correlations of compounds 1–4.

| Position | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| 1 | 198.6 | 198.1 | 194.9 | 194.5 |
| 2 | 102.8 | 104.5 | 105.7 | 107.2 |
| 3 | 173.7 | 172.2 | 168.3 | 168.0 |
| 4 | 78.4 | 78.2 | 65.8 | 66.0 |
| 5 | 43.9 | 44.0 | 34.6 | 34.7 |
| 6 | 81.6 | 81.6 | 79.1 | 78.9 |
| 7 | 70.5 | 70.4 | 58.4 | 58.3 |
| 8 | 17.6 | 19.6 | 18.3 | 19.8 |
| 9 | 41.1 | 41.4 | 41.1 | 41.9 |
| 10 | 90.7 | 86.5 | 89.0 | 85.7 |
| 11 | 22.9 | 25.4 | 22.3 | 25.3 |
| 12 | 38.0 | 34.7 | 38.2 | 36.4 |
| 13 | 24.4 | 25.2 | 24.4 | 25.3 |
| 14 | 48.3 | 133.8 | 49.2 | 134.0 |
| 15 | 76.7 | 125.1 | 76.8 | 125.2 |
| 16 | 21.9 | 20.3 | 22.0 | 20.5 |
| 17 | 23.3 | 20.9 | 23.0 | 21.0 |
| 18 | 48.9 | 49.0 |
2.2. Cytotoxicity Assay
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Extraction and Isolation
3.4. Spectroscopic Data
3.5. X-ray Crystallographic Data
3.6. Cytotoxicity Assay
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Aly, A.H.; Debbab, A.; Proksch, P. Fungal endophytes: Unique plant inhabitants with great promises. Appl. Microbiol. Biot. 2011, 90, 1829–1845. [Google Scholar] [CrossRef] [PubMed]
- Aly, A.H.; Debbab, A.; Kjer, J.; Proksch, P. Fungal endophytes from higher plants: A prolific source of phytochemicals and other bioactive natural products. Fungal Divers. 2010, 41, 1–16. [Google Scholar] [CrossRef]
- Amrani, M.E.; Lai, D.; Debbab, A.; Aly, A.H.; Siems, K.; Seidel, C.; Schnekenburger, M.; Gaigneaux, A.; Diederich, M.; Feger, D.; et al. Protein kinase and HDAC inhibitors from the endophytic fungus Epicoccum nigrum. J. Nat. Prod. 2014, 77, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Chu, K.T.; Ng, T.B. Smilaxin, a novel protein with immunostimulatory, antiproliferative, and HIV-1-reverse transcriptase inhibitory activities from fresh Smilax glabra rhizomes. Biochem. Biophys. Res. Commun. 2006, 340, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Li, J.X.; Xu, Q. Phenylpropanoid glycosides from Smilax glabra. Phytochemistry 2000, 53, 1051–1055. [Google Scholar] [CrossRef]
- Xia, D.; Yu, X.; Liao, S.; Shao, Q.; Mou, H.; Ma, W. Protective effect of Smilax glabra extract against lead-induced oxidative stress in rats. J. Ethnopharmacol. 2010, 130, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Shang, M.Y.; Liu, G.X.; Xu, F.; Wang, X.; Shou, C.C.; Cai, S.Q. Chemical constituents from the rhizomes of Smilax glabra and their antimicrobial activity. Molecules 2013, 18, 5265–5287. [Google Scholar] [CrossRef] [PubMed]
- Tan, R.X.; Zou, W.X. Endophytes: A rich source of functional metabolites. Nat. Prod. Rep. 2001, 18, 448–459. [Google Scholar] [CrossRef] [PubMed]
- Ai, W.; Wei, X.; Lin, X.; Sheng, L.; Wang, Z.; Tu, Z.; Yang, X.; Zhou, X.; Li, J.; Liu, Y. Guignardins A–F, spirodioxynaphthalenes from the endophytic fungus Guignardia sp. KcF8 as a new class of PTP1B and SIRT1 inhibitors. Tetrahedron 2014, 70, 5806–5814. [Google Scholar] [CrossRef]
- Xia, X.K.; liu, F.; She, Z.G.; Yang, L.G.; Li, M.F.; Vrijmoed, L.L.P.; Lin, Y.C. 1H- and 13C-NMR assignments for 6-demethylvermistatin and two penicillide derivatives from the mangrove fungus Guignardia sp. (No. 4382) from the South China Sea. Magn. Reson. Chem. 2008, 46, 693–696. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.K.; Huang, H.R.; She, Z.G.; Cai, J.W.; Lan, L.; Zhang, J.Y.; Fu, L.W.; Vrijmoed, L.L.P.; Lin, Y.C. Structural and biological properties of vermistatin and two new vermistatin derivatives isolated from the marine-mangrove endophytic fungus Guignardia sp. No. 4382. Helv. Chim. Acta 2007, 90, 1925–1931. [Google Scholar] [CrossRef]
- Li, T.X.; Yang, M.H.; Wang, X.B.; Wang, Y.; Kong, L.Y. Synergistic antifungal meroterpenes and dioxolanone derivatives from the endophytic fungus Guignardia sp. J. Nat. Prod. 2015, 78, 2511–2520. [Google Scholar] [CrossRef] [PubMed]
- Liang, F.L.; Li, D.L.; Tao, M.H.; Zhang, D.Z.; Zhang, W.M. Chemical constituents of guignardia mangiferae, an endophyte from Smilax glabra. Guangdong Yaoxueyuan Xuebao 2011, 27, 256–259. [Google Scholar]
- Yuan, W.H.; Liu, M.; Jiang, N.; Guo, Z.K.; Ma, J.; Zhang, J.; Song, Y.C.; Tan, R.X. Guignardones A–C: Three meroterpenes from Guignardia mangiferae. Eur. J. Org. Chem. 2010, 33, 6348–6353. [Google Scholar] [CrossRef]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New colorimetric cytotoxicity assay for anticancer-drug sceenimg. J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.Q.; Lin, X.P.; Wang, J.F.; Zhou, X.F.; Liu, J.; Yang, B.; Yang, X.W.; Liao, S.R.; Wang, L.S.; Liu, Y.H. New meroterpenoids from the endophytic fungus aspergillus flavipes AIL8 derived from the mangrove plant Acanthus ilicifolius. Mar. Drugs 2015, 13, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Debbab, A.; Aly, A.H.; Proksch, P. Mangrove derived fungal endophytes—A chemical and biological perception. Fungal Divers. 2013, 61, 1–27. [Google Scholar] [CrossRef]
- Guimaraes, D.O.; Lopes, N.P.; Pupo, M.T. Meroterpenes isolated from the endophytic fungus Guignardia mangiferae. Phytochem. Lett. 2012, 5, 519–523. [Google Scholar] [CrossRef]
- Sommart, U.; Rukachaisirikul, V.; Trisuwan, K.; Tadpetch, K.; Phongpaichit, S.; Preedanon, S.; Sakayaroj, J. Tricycloalternarene derivatives from the endophytic fungus Guignardia bidwellii PSU-G11. Phytochem. Lett. 2012, 5, 139–143. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds 1–7 are available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Z.-H.; Liang, F.-L.; Wu, W.; Chen, Y.-C.; Pan, Q.-L.; Li, H.-H.; Ye, W.; Liu, H.-X.; Li, S.-N.; Tan, G.-H.; et al. Guignardones P–S, New Meroterpenoids from the Endophytic Fungus Guignardia mangiferae A348 Derived from the Medicinal Plant Smilax glabra. Molecules 2015, 20, 22900-22907. https://doi.org/10.3390/molecules201219890
Sun Z-H, Liang F-L, Wu W, Chen Y-C, Pan Q-L, Li H-H, Ye W, Liu H-X, Li S-N, Tan G-H, et al. Guignardones P–S, New Meroterpenoids from the Endophytic Fungus Guignardia mangiferae A348 Derived from the Medicinal Plant Smilax glabra. Molecules. 2015; 20(12):22900-22907. https://doi.org/10.3390/molecules201219890
Chicago/Turabian StyleSun, Zhang-Hua, Fa-Liang Liang, Wen Wu, Yu-Chan Chen, Qing-Ling Pan, Hao-Hua Li, Wei Ye, Hong-Xin Liu, Sai-Ni Li, Guo-Hui Tan, and et al. 2015. "Guignardones P–S, New Meroterpenoids from the Endophytic Fungus Guignardia mangiferae A348 Derived from the Medicinal Plant Smilax glabra" Molecules 20, no. 12: 22900-22907. https://doi.org/10.3390/molecules201219890