Antiviral, Antifungal and Antibacterial Activities of a BODIPY-Based Photosensitizer
Abstract
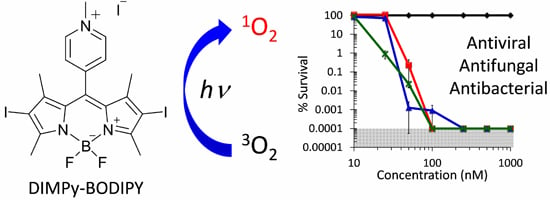
:1. Introduction

2. Results and Discussion
2.2. Antibacterial Photodynamic Inactivation Studies


2.3. Antifungal Photodynamic Inactivation Studies

3. Experimental Section
3.1. Materials
3.2. Cell Culture
3.3. Viral Propagation
3.4. Photodynamic Inactivation Assay
3.4.1. Bacteria and Yeast
3.4.2. Vesicular Stomatitis Virus
3.4.3. Dengue-1 Virus
3.4.4. Human Adenovirus-5
3.5. Red Blood Cell Hemolysis Assay
3.6. Singlet Oxygen Quantum Yield Determination
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Vicente, M.; Hodgson, J.; Massidda, O.; Tonjum, T.; Henriques-Normark, B.; Ron, E.Z. The fallacies of hope: Will we discover new antibiotics to combat pathogenic bacteria in time? FEMS Microbiol. Rev. 2006, 30, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Kraus, C.N. Low hanging fruit in infectious disease drug development. Curr. Opin. Microbiol. 2008, 11, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Vera, D.M.; Haynes, M.H.; Ball, A.R.; Dai, T.; Astrakas, C.; Kelso, M.J.; Hamblin, M.R.; Tegos, G.P. Strategies to potentiate antimicrobial photoinactivation by overcoming resistant phenotypes. Photochem. Photobiol. 2012, 88, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Dai, T.; Fuchs, B.B.; Coleman, J.J.; Prates, R.A.; Astrakas, C.; St Denis, T.G.; Ribeiro, M.S.; Mylonakis, E.; Hamblin, M.R.; Tegos, G.P. Concepts and principles of photodynamic therapy as an alternative antifungal discovery platform. Front. Microbiol. 2012, 3, 1–16. [Google Scholar] [CrossRef] [PubMed]
- St Denis, T.G.; Dai, T.; Izikson, L.; Astrakas, C.; Anderson, R.R.; Hamblin, M.R.; Tegos, G.P. All you need is light: Antimicrobial photoinactivation as an evolving and emerging discovery strategy against infectious disease. Virulence 2011, 2, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Dai, T.; Huang, Y.Y.; Hamblin, M.R. Photodynamic therapy for localized infections—State of the art. Photodiagn. Photodyn. Ther. 2009, 6, 170–188. [Google Scholar] [CrossRef] [PubMed]
- Wainwright, M. ‘Safe’ photoantimicrobials for skin and soft-tissue infections. Int. J. Antimicrob. Agents 2010, 36, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Merkel, P.B.; Kearns, D.R. Radiationless decay of singlet molecular oxygen in solution. Experimental and theoretical study of electronic-to-vibrational energy transfer. J. Am. Chem. Soc. 1972, 94, 7244–7253. [Google Scholar] [CrossRef]
- Jori, G.; Fabris, C.; Soncin, M.; Ferro, S.; Coppellotti, O.; Dei, D.; Fantetti, L.; Chiti, G.; Roncucci, G. Photodynamic therapy in the treatment of microbial infections: Basic principles and perspective applications. Lasers Surg. Med. 2006, 38, 468–481. [Google Scholar] [CrossRef] [PubMed]
- Tavares, A.; Carvalho, C.M.; Faustino, M.A.; Neves, M.G.; Tome, J.P.; Tome, A.C.; Cavaleiro, J.A.; Cunha, A.; Gomes, N.C.; Alves, E.; et al. Antimicrobial photodynamic therapy: Study of bacterial recovery viability and potential development of resistance after treatment. Mar. Drugs 2010, 8, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Maisch, T. A new strategy to destroy antibiotic resistant microorganisms: Antimicrobial photodynamic treatment. Mini Rev. Med. Chem. 2009, 9, 974–983. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, B.L.; Feese, E.; Sadeghifar, H.; Argyropoulos, D.S.; Ghiladi, R.A. Porphyrin-cellulose nanocrystals: A photobactericidal material that exhibits broad spectrum antimicrobial activity. Photochem. Photobiol. 2012, 88, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Feese, E.; Ghiladi, R.A. Highly efficient in vitro photodynamic inactivation of mycobacterium smegmatis. J. Antimicrob. Chemother. 2009, 64, 782–785. [Google Scholar] [CrossRef] [PubMed]
- Bertoloni, G.; Rossi, F.; Valduga, G.; Jori, G.; van Lier, J. Photosensitizing activity of water- and lipid-soluble phthalocyanines on escherichia coli. FEMS Microbiol. Lett. 1990, 59, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Wainwright, M.; Phoenix, D.A.; Marland, J.; Wareing, D.R.A.; Bolton, F.J. A study of photobactericidal activity in the phenothiazinium series. FEMS Immunol. Med. Microbiol. 1997, 19, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.Y.; Balasubramanian, T.; Yang, E.; Luo, D.; Diers, J.R.; Bocian, D.F.; Lindsey, J.S.; Holten, D.; Hamblin, M.R. Stable synthetic bacteriochlorins for photodynamic therapy: Role of dicyano peripheral groups, central metal substitution (2h, Zn, Pd), and cremophor el delivery. Chem. Med. Chem. 2012, 7, 2155–2167. [Google Scholar] [CrossRef] [PubMed]
- Wainwright, M.; Antczak, J.; Baca, M.; Loughran, C.; Meegan, K. Phenothiazinium photoantimicrobials with basic side chains. J. Photochem. Photobiol. B 2015, in press. [Google Scholar] [CrossRef] [PubMed]
- Wainwright, M. In defence of ‘dye therapy’. Int. J. Antimicrob. Agents 2014, 44, 26–29. [Google Scholar] [CrossRef] [PubMed]
- De Menezes, H.D.; Rodrigues, G.B.; Teixeira Sde, P.; Massola, N.S., Jr.; Bachmann, L.; Wainwright, M.; Braga, G.U. In vitro photodynamic inactivation of plant-pathogenic fungi colletotrichum acutatum and colletotrichum gloeosporioides with novel phenothiazinium photosensitizers. Appl. Environ. Microbiol. 2014, 80, 1623–1632. [Google Scholar] [CrossRef] [PubMed]
- Bacellar, I.O.; Pavani, C.; Sales, E.M.; Itri, R.; Wainwright, M.; Baptista, M.S. Membrane damage efficiency of phenothiazinium photosensitizers. Photochem. Photobiol. 2014, 90, 801–813. [Google Scholar] [CrossRef] [PubMed]
- Ziessel, R.; Ulrich, G.; Harriman, A. The chemistry of bodipy: A new el dorado for fluorescence tools. New J. Chem. 2007, 31, 496–501. [Google Scholar] [CrossRef]
- Loudet, A.; Burgess, K. Bodipy dyes and their derivatives: Syntheses and spectroscopic properties. Chem. Rev. 2007, 107, 4891–4932. [Google Scholar] [CrossRef] [PubMed]
- Hendricks, J.A.; Keliher, E.J.; Wan, D.; Hilderbrand, S.A.; Weissleder, R.; Mazitschek, R. Synthesis of [18f]bodipy: Bifunctional reporter for hybrid optical/positron emission tomography imaging. Angew. Chem. 2012, 51, 4603–4606. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, M.J.; Agarrabeitia, A.R.; Duran-Sampedro, G.; Bañuelos Prieto, J.; Lopez, T.A.; Massad, W.A.; Montejano, H.A.; García, N.A.; Lopez Arbeloa, I. Synthesis and functionalization of new polyhalogenated bodipy dyes. Study of their photophysical properties and singlet oxygen generation. Tetrahedron 2012, 68, 1153–1162. [Google Scholar] [CrossRef]
- Banfi, S.; Nasini, G.; Zaza, S.; Caruso, E. Synthesis and photo-physical properties of a series of bodipy dyes. Tetrahedron 2013, 69, 4845–4856. [Google Scholar] [CrossRef] [Green Version]
- Caruso, E.; Banfi, S.; Barbieri, P.; Leva, B.; Orlandi, V.T. Synthesis and antibacterial activity of novel cationic bodipy photosensitizers. J. Photochem. Photobiol. B 2012, 114, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Frimannsson, D.O.; Grossi, M.; Murtagh, J.; Paradisi, F.; O’Shea, D.F. Light induced antimicrobial properties of a brominated boron difluoride (bf(2)) chelated tetraarylazadipyrromethene photosensitizer. J. Med. Chem. 2010, 53, 7337–7343. [Google Scholar] [CrossRef] [PubMed]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No eskape! An update from the infectious diseases society of america. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hidron, A.I.; Edwards, J.R.; Patel, J.; Horan, T.C.; Sievert, D.M.; Pollock, D.A.; Fridkin, S.K. Nhsn annual update: Antimicrobial-resistant pathogens associated with healthcare-associated infections: Annual summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2006–2007. Infect. Control Hosp. Epidemiol. 2008, 29, 996–1011. [Google Scholar] [CrossRef] [PubMed]
- Bartelmess, J.; Weare, W.W. Preparation and characterization of multi-cationic bodipys and their synthetically versatile precursors. Dyes Pigment. 2013, 97, 1–8. [Google Scholar] [CrossRef]
- Nicolau, D.P. Current challenges in the management of the infected patient. Curr. Opin. Infect. Dis. 2011, 24, S1–S10. [Google Scholar] [CrossRef] [PubMed]
- Horowitz, B.; Williams, B.; Rywkin, S.; Prince, A.M.; Pascual, D.; Geacintov, N.; Valinsky, J. Inactivation of viruses in blood with aluminum phthalocyanine derivatives. Transfusion 1991, 31, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Ben-Hur, E.; Hoeben, R.C.; van Ormondt, H.; Dubbelman, T.M.; van Steveninck, J. Photodynamic inactivation of retroviruses by phthalocyanines: The effects of sulphonation, metal ligand and fluoride. J. Photochem. Photobiol. B 1992, 13, 145–152. [Google Scholar] [CrossRef]
- Smetana, Z.; Mendelson, E.; Manor, J.; van Lier, J.E.; Ben-Hur, E.; Salzberg, S.; Malik, Z. Photodynamic inactivation of herpes viruses with phthalocyanine derivatives. J. Photochem. Photobiol. B 1994, 22, 37–43. [Google Scholar] [CrossRef]
- Gaspard, S.; Tempete, C.; Werner, G.H. Studies on photoinactivation by various phthalocyanines of a free or replicating non-enveloped virus. J. Photochem. Photobiol. B 1995, 31, 159–162. [Google Scholar] [CrossRef]
- Macdonald, I.J.; Dougherty, T.J. Basic principles of photodynamic therapy. J. Porphyr. Phthalocyanines 2001, 5, 105–129. [Google Scholar] [CrossRef]
- Trias, J.; Benz, R. Permeability of the cell wall of mycobacterium smegmatis. Mol. Microbiol. 1994, 14, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Padilla, E.; Llobet, E.; Domenech-Sanchez, A.; Martinez-Martinez, L.; Bengoechea, J.A.; Alberti, S. Klebsiella pneumoniae acrab efflux pump contributes to antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 2010, 54, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Park, B.J.; Wannemuehler, K.A.; Marston, B.J.; Govender, N.; Pappas, P.G.; Chiller, T.M. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/aids. AIDS 2009, 23, 525–530. [Google Scholar] [CrossRef] [PubMed]
- McFadden, D.; Zaragoza, O.; Casadevall, A. The capsular dynamics of cryptococcus neoformans. Trends Microbiol. 2006, 14, 497–505. [Google Scholar] [CrossRef] [PubMed]
- McFadden, D.C.; de Jesus, M.; Casadevall, A. The physical properties of the capsular polysaccharides from cryptococcus neoformans suggest features for capsule construction. J. Biol. Chem. 2006, 281, 1868–1875. [Google Scholar] [CrossRef] [PubMed]
- Zaragoza, O.; Rodrigues, M.L.; de Jesus, M.; Frases, S.; Dadachova, E.; Casadevall, A. The capsule of the fungal pathogen cryptococcus neoformans. Adv. Appl. Microbiol. 2009, 68, 133–216. [Google Scholar] [PubMed]
- Frases, S.; Pontes, B.; Nimrichter, L.; Rodrigues, M.L.; Viana, N.B.; Casadevall, A. The elastic properties of the cryptococcus neoformans capsule. Biophys. J. 2009, 97, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Bartelmess, J.; Weare, W.W.; Latortue, N.; Duong, C.; Jones, D.S. Meso-pyridyl bodipys with tunable chemical, optical and electrochemical properties. New J. Chem. 2013, 37, 2663–2668. [Google Scholar] [CrossRef]
- Bacon, J.; Hatch, K.A. Continuous culture of mycobacteria. In Methods in Molecular Biology, 2nd ed.; Parish, T., Brown, A.C., Eds.; Humana Press: New York, NY, USA, 2009; pp. 153–171. [Google Scholar]
- Budhathoki-Uprety, J.; Peng, L.; Melander, C.; Novak, B.M. Synthesis of guanidinium functionalized polycarbodiimides and their antibacterial activities. ACS Macro Lett. 2012, 1, 370–374. [Google Scholar] [CrossRef]
- Wilkinson, F.; Helman, W.P.; Ross, A.B. Quantum yields of photosensitized formation of the lowest electronically excited singlet state of molecular oxygen in solution. J. Phys. Chem. Ref. Data 1993, 22, 113–262. [Google Scholar] [CrossRef]
- Rachford, A.A.; Goeb, S.; Castellano, F.N. Accessing the triplet excited state in perylenediimides. J. Am. Chem. Soc. 2008, 130, 2766–2767. [Google Scholar] [CrossRef] [PubMed]
- Feese, E.; Sadeghifar, H.; Gracz, H.S.; Argyropoulos, D.S.; Ghiladi, R.A. Photobactericidal porphyrin-cellulose nanocrystals: Synthesis, characterization, and antimicrobial properties. Biomacromolecules 2011, 12, 3528–3539. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compound DIMPy-BODIPY are available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carpenter, B.L.; Situ, X.; Scholle, F.; Bartelmess, J.; Weare, W.W.; Ghiladi, R.A. Antiviral, Antifungal and Antibacterial Activities of a BODIPY-Based Photosensitizer. Molecules 2015, 20, 10604-10621. https://doi.org/10.3390/molecules200610604
Carpenter BL, Situ X, Scholle F, Bartelmess J, Weare WW, Ghiladi RA. Antiviral, Antifungal and Antibacterial Activities of a BODIPY-Based Photosensitizer. Molecules. 2015; 20(6):10604-10621. https://doi.org/10.3390/molecules200610604
Chicago/Turabian StyleCarpenter, Bradley L., Xingci Situ, Frank Scholle, Juergen Bartelmess, Walter W. Weare, and Reza A. Ghiladi. 2015. "Antiviral, Antifungal and Antibacterial Activities of a BODIPY-Based Photosensitizer" Molecules 20, no. 6: 10604-10621. https://doi.org/10.3390/molecules200610604