Synthesis, Antiphospholipase A2, Antiprotease, Antibacterial Evaluation and Molecular Docking Analysis of Certain Novel Hydrazones
Abstract
:1. Introduction
2. Results and Discussion
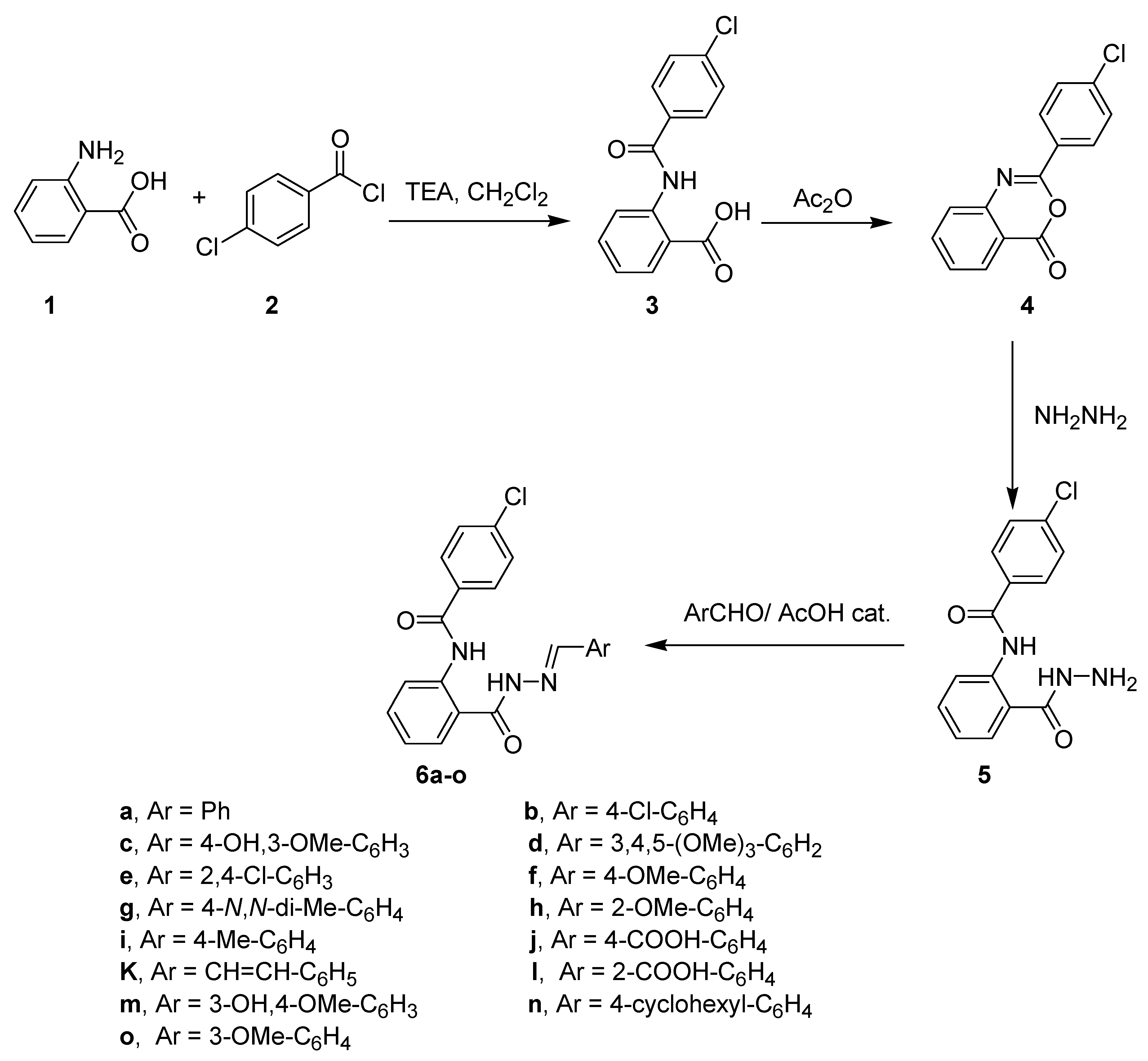
2.1. Chemistry
2.2. Biological Evaluation
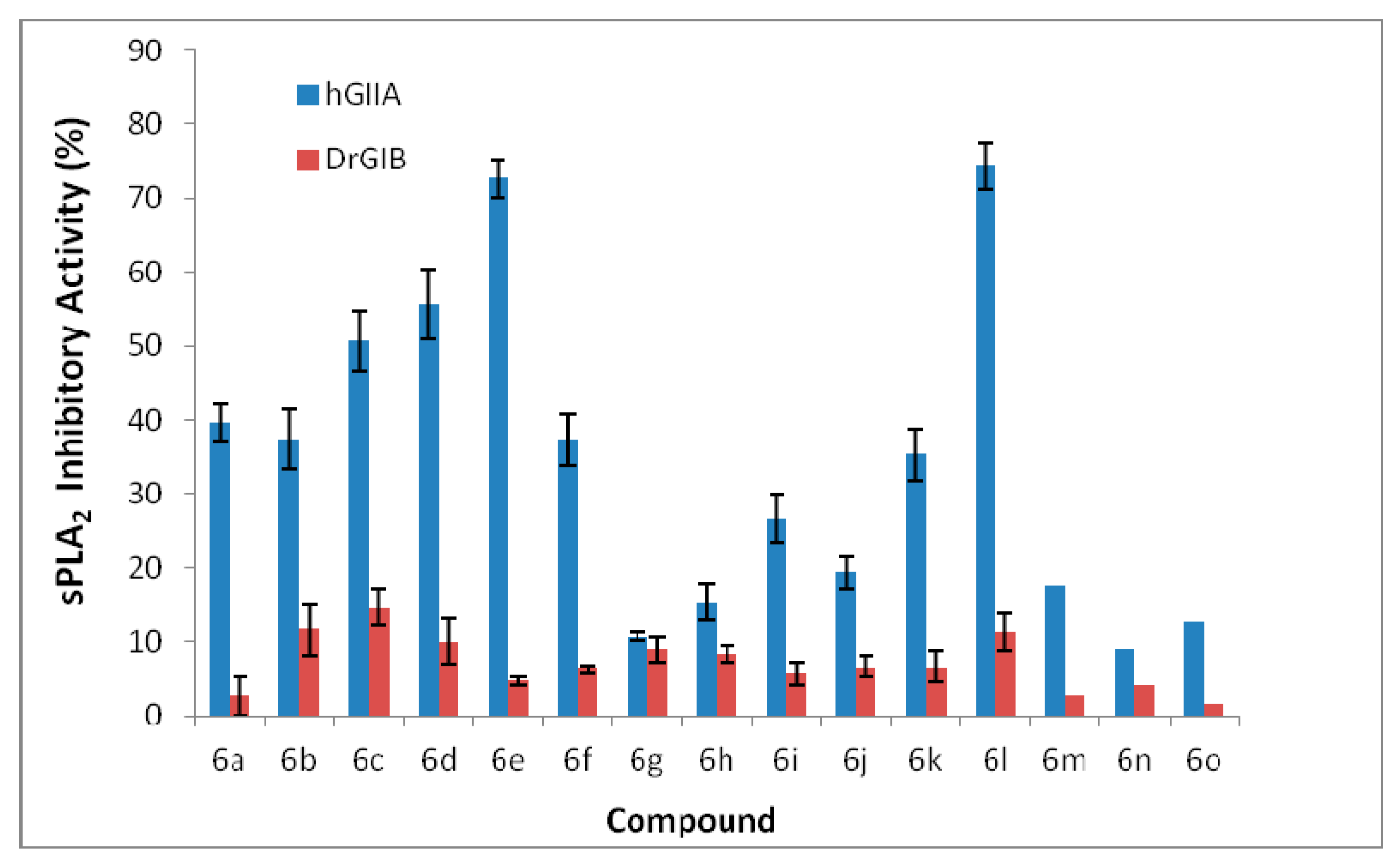
2.2.1. Phospholipases Inhibitory Activity
2.2.2. Proteases Inhibitory Activity
2.2.3. Antibacterial Screening
2.3. Molecular Docking Analysis
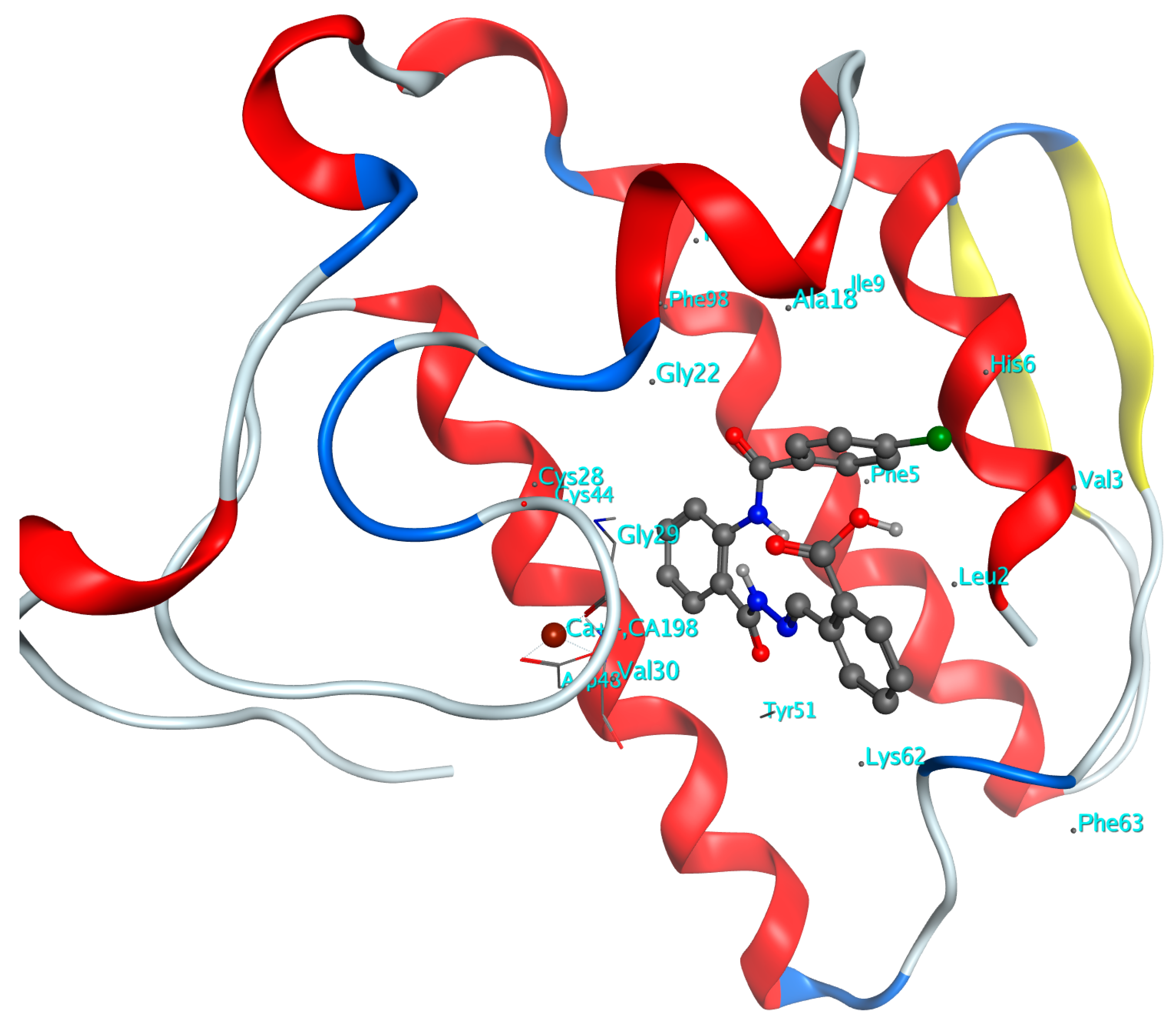
2.3.1. Docking Simulations for Compounds 6a–o in Active site of GIIAsPLA2
2.3.2. Docking Simulations for Compounds 6a–o in Active Site of Proteinase K
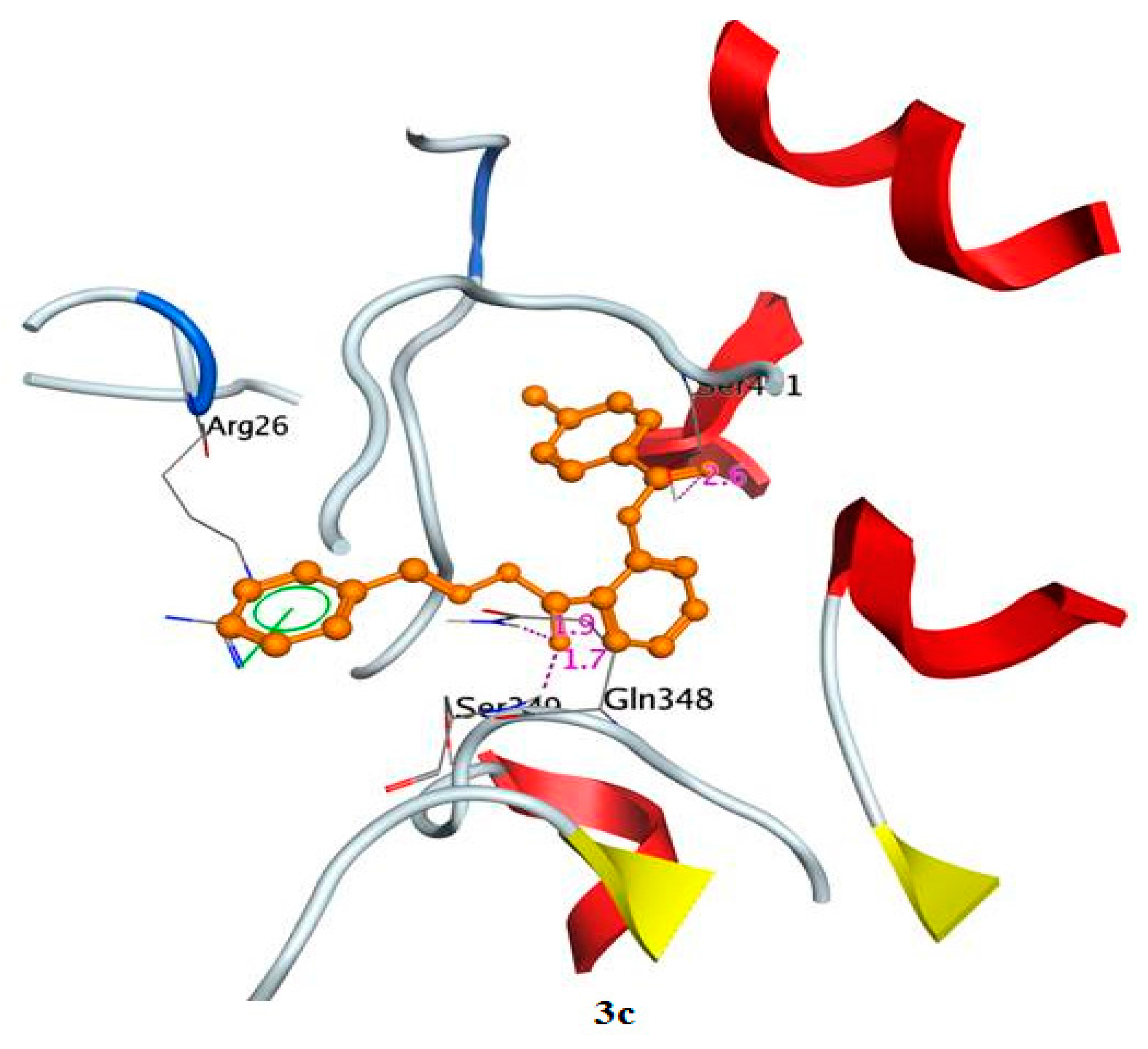
2.3.3. Docking Simulations for Compounds 6a–e in Active Site of Glutamine-Fructose-6-Phosphate Transaminase
3. Experimental Section
3.1. Chemistry
3.1.1. General
3.1.2. Synthetic Procedures
4-Chloro-N-(2-(hydrazinecarbonyl)phenyl)benzamide 5
Synthesis of Compounds 6a–o
3.2. Biological Evalustion
3.2.1. Inhibition of sPLA2 Activity
3.2.2. Protease Inhibitor Assay
3.2.3. Antibacterial Activity
Culture of Microbial Strains Preparation
Antibacterial Assay
3.3. Molecular Docking
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Miklos, F.; Fulop, F. A Simple Green Protocol for the Condensation of Anthranilic Hydrazide with Cyclohexanone and N-Benzylpiperidinone in Water. J. Heterocycl. Chem. 2016, 53, 32–37. [Google Scholar] [CrossRef]
- Jamil, W.; Perveen, S.; Shah, S.A.A.; Taha, M.; Ismail, N.H.; Perveen, S.; Ambreen, N.; Khan, K.M.; Choudhary, M.I. Phenoxyacetohydrazide Schiff Bases: β-Glucuronidase Inhibitors. Molecules 2014, 19, 8788–8802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Guo, D.G.; Wang, Y.Y.; Zheng, C.Z. 4-Hydroxy-3-methoxy-benzaldehyde series aroyl hydrazones: Synthesis, thermostability and antimicrobial activities. RSC Adv. 2014, 4, 58895–58901. [Google Scholar] [CrossRef]
- Ienascu, I.M.C.; Lupea, A.X.; Popescu, I.M.; Padure, M.A.; Zamfir, A.D. The synthesis and characterization of some novel 5-chloro-2-(substituted alkoxy)-N-phenylbenzamide derivatives. J. Serbian Chem. Soc. 2009, 74, 847–855. [Google Scholar] [CrossRef]
- Jin, L.; Chen, J.; Song, B.; Chen, Z.; Yang, S.; Li, Q.; Hu, D.; Xu, R. Synthesis, structure, and bioactivity of N′-substituted benzylidene-3,4,5-trimethoxybenzohydrazide and 3-acetyl-2-substituted phenyl-5-(3,4,5-trimethoxyphenyl)-2,3-dihydro-1,3,4-oxadiazole derivatives. Bioorg. Med. Chem. Lett. 2006, 16, 5036–5040. [Google Scholar] [CrossRef] [PubMed]
- Perdicchia, D.; Licandro, E.; Maiorana, S.; Baldoli, C.; Giannini, C. A new “one-pot” synthesis of hydrazides by reduction of hydrazones. Tetrahedron 2003, 59, 7733–7742. [Google Scholar] [CrossRef]
- Khurana, J.M.; Kandpal, B.M.; Sharma, P.; Gupta, M. A novel method of reduction of >C=N-group in hydrazones, phenylhydrazones, azines, and tosylhydrazones by Mg–methanol. Monatshefte Chem. 2015, 146, 187–190. [Google Scholar] [CrossRef]
- Desai, S.R.; Laddi, U.V.; Benur, R.B.; Bennur, S.C. Synthesis and Antimicrobial Activities of Some New Azetidin-2-ones and Thiazolidin-4-ones. Indian J. Pharm. Sci. 2011, 73, 478–482. [Google Scholar] [PubMed]
- El-masry, A.H.; Fahmy, H.H.; Abdelwahed, S.H.A. Synthesis and Antimicrobial Activity of Some New Benzimidazole Derivatives. Molecules 2000, 5, 1429–1438. [Google Scholar] [CrossRef]
- Mariappan, G.; Korim, R.; Joshi, N.M.; Alam, F.; Hazarika, R.; Kumar, D.; Uriah, T. Synthesis and biological evaluation of formazan derivatives. J. Adv. Pharm. Technol. Res. 2010, 1, 396–400. [Google Scholar] [CrossRef] [PubMed]
- Sah, P.; Bidawat, P.; Seth, M.; Gharu, C.P. Synthesis of formazans from Mannich base of 5-(4-chlorophenyl amino)-2-mercapto-1,3,4-thiadiazole as antimicrobial agents. Arabian J. Chem. 2014, 7, 181–187. [Google Scholar] [CrossRef]
- Rollas, S.; Küçükgüzel, S.G. Biological Activities of Hydrazone Derivatives. Molecules 2007, 12, 1910–1939. [Google Scholar] [CrossRef] [PubMed]
- Melnyk, P.; Leroux, V.; Sergheraert, C.; Grellier, P. Design, synthesis and in vitro antimalarial activity of an acylhydrazone library. Bioorg. Med. Chem. Lett. 2006, 16, 31–35. [Google Scholar] [CrossRef] [PubMed]
- El-Faham, A.; Farooq, M.; Khattab, S.N.; Elkayal, A.M.; Ibrahim, M.F.; Abutaha, N.; Wadaan, M.A.; Hamed, E.A. Synthesis and Biological Activity of Schiff Base Series of Valproyl, N-Valproyl Glycinyl, and N-Valproyl-4-aminobenzoyl Hydrazide Derivatives. Chem. Pharm. Bull. 2014, 62, 591–599. [Google Scholar] [CrossRef]
- Vicini, P.; Incerti, M.; LaColla, P.; Loddo, R. Anti-HIV evaluation of benzo[d]isothiazole hydrazones. Eur. J. Med. Chem. 2009, 44, 1801–1807. [Google Scholar] [CrossRef] [PubMed]
- El-Sabbagh, O.I.; Rady, H.M. Synthesis of new acridines and hydrazones derived from cyclic β-diketone for cytotoxic and antiviral evaluation. Eur. J. Med. Chem. 2009, 44, 3680–3688. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Judge, V.; Narang, R.; Sangwan, S.; Clercq, E.D.; Balzarini, J.; Narasimhan, B. Benzylidene/2-chlorobenzylidene hydrazides: Synthesis, antimicrobial activity, QSAR studies and antiviral evaluation. Eur. J. Med. Chem. 2010, 45, 2806–2816. [Google Scholar] [CrossRef] [PubMed]
- Čačić, M.; Molnar, M.; Bojan Šarkanj, B.; Has-Schön, E.; Rajković, V. Synthesis and Antioxidant Activity of Some New Coumarinyl-1,3-Thiazolidine-4-ones. Molecules 2010, 15, 6795–6809. [Google Scholar] [CrossRef] [PubMed]
- Rajitha, G.; Saideepa, N.; Praneetha, P. Synthesis and evaluation of N-(α-benzamido cinnamoyl) aryl hydrazone derivatives for anti-inflammatory and antioxidant activities. Indian J. Chem. 2011, 50B, 729–733. [Google Scholar]
- Alafeefy, A.M.; Bakht, M.A.; Ganaie, M.A.; Ansarie, M.N.; El-Sayed, N.N.; Awaad, A.S. Synthesis, analgesic, anti-inflammatory and anti-ulcerogenic activities of certain novel Schiff’s bases as fenamate isosteres. Bioorg. Med. Chem. Lett. 2015, 25, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Gelb, M.H.; Jain, M.K.; Hanel, A.M.; Berg, O.G. Interfacial enzymology of glycerolipid hydrolases: Lessons from secreted phospholipases A2. Annu. Rev. Biochem. 1995, 64, 653–688. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Kudo, I. Diversity and regulatory functions of mammalian secretory phospholipase A2s. Adv. Immunol. 2001, 77, 163–194. [Google Scholar] [PubMed]
- Valentin, E.; Lambeau, G. Increasing molecular diversity of secreted phospholipases A(2) and their receptors and binding proteins. Biochim. Biophys. Acta 2000, 1488, 59–70. [Google Scholar] [CrossRef]
- Hanasaki, K.; Arita, H. Phospholipase A2 receptor: A regulator of biological functions of secretory phospholipase A2. Prostaglandins Lipid Mediat. 2002, 68–69, 71–82. [Google Scholar] [CrossRef]
- Yagami, T.; Ueda, K.; Askura, K.; Hayasaki-Kajiwara, Y.; Nakazato, H.; Sakaeda, T.; Hata, S.; Kuroda, T.; Takasu, N.; Hori, Y. Group IB secretory phospholipase A2 induces neuronal cell death via apoptosis. J. Neurochem. 2002, 81, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Peuravuori, H.J.; Funatomi, H.; Nevalainen, T.J. Group I and group II phospholipases A2 in serum in uraemia. Eur. J. Clin. Chem. Clin. Biochem. 1993, 31, 491–494. [Google Scholar] [CrossRef] [PubMed]
- Rae, D.; Porter, J.; Beechey- Newman, N.; Sumar, N.; Bennett, D.; Hermon-Tylor, J. Type I phospholipase A2 propeptide in acute lung injury. Lancet 1994, 344, 1472–1473. [Google Scholar] [CrossRef]
- Pruzanski, W.; Vasdas, P.; Stefanski, E.; Urowitz, M.B. Phospholipase A2 activity in sera and synovial fluids in rheumatoid arthritis and osteoarthritis. Its possible role as a proinflammatory enzyme. J. Rheumatol. 1985, 12, 211–216. [Google Scholar] [PubMed]
- Kitsiouli, E.; Nakos, G.; Lekka, M.E. Phospholipase A2 subclasses in acute respiratory distress syndrome. Biochim. Biophys. Acta 2009, 1792, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Chen, C.; Wang, W.-X.; Chen, X.-Y. Crucial role of group IIA phospholipase A2 in pancreatitis-associated adrenal injury in acute necrotizing pancreatitis. Pathol. Res. Pract. 2009, 206, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Abe, T.; Sakamoto, K.; Kamohara, H.; Hirano, Y.; Kuwahara, N.; Ogwa, M. Group II phospholipase A2 is increased in peritoneal and pleural effusions in patients with various types of cancer. Int. J. Cancer 1997, 74, 245–250. [Google Scholar] [CrossRef]
- Garcia-Carreno, F.; Toro, M.N. Classification of Proteases without Tears. Biochem. Educ. 1997, 25, 161–167. [Google Scholar] [CrossRef]
- Krishnaswamy, S. Exosite-driven substrate specificity and function in coagulation. J. Thromb. Haemost. 2005, 3, 54–67. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Van den Steen, P.E.; Sang, Q.; Opdenakker, G. Matrix metalloproteinase inhibitors as therapy for inflammatory and vascular diseases. Nat. Rev. Drug Discov. 2007, 6, 480–498. [Google Scholar] [CrossRef] [PubMed]
- Overall, C.M.; Dean, R.A. Degradomics: Systems biology of the protease web. Pleiotropic roles of MMPs in cancer. Cancer Metastasis Rev. 2006, 25, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Dusing, R.; Sellers, F. ACE inhibitors, angiotensin receptor blockers and direct renin inhibitors in combination: A review of their role after the ONTARGET trial. Curr. Med. Res. Opin. 2009, 25, 2287–2301. [Google Scholar] [CrossRef] [PubMed]
- Brik, A.; Wong, C.-H. HIV-1 protease: Mechanism and drug discovery. Org. Biomol. Chem. 2003, 1, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Patick, A.K.; Potts, K.E. Protease Inhibitors as Antiviral Agents. Clin. Microbiol. Rev. 1998, 11, 614–627. [Google Scholar] [PubMed]
- El-Sayed, N.N. E.; AL-Balawi, N.A.; Alafeefy, A.M.; Al-AlShaikh, M.A.; Khan, K.M. Synthesis, Characterization and Antimicrobial Evaluation of some Thiazole-Derived Carbamates, Semicarbazones, Amides and Carboxamide. J. Chem. Soc. Pak. 2016, 38, 358–368. [Google Scholar]
- El-Sayed, N.N.E.; Abdelaziz, M.A.; Wardakhan, W.W.; Mohareb, R.M. The Knoevenagel reaction of cyanoacetylhydrazine with pregnenolone, Synthesis of thiophene, thieno[2,3-d]pyrimidine, 1,2,4-triazole, pyran and pyridine derivatives with anti-inflammatory and anti-ulcer activities. Steroids 2016, 107, 98–111. [Google Scholar] [CrossRef] [PubMed]
- Wardakhan, W.W.; El-Sayed, N.N. New approaches for the synthesis of 1,3,4-thiadiazole and 1,2,4-triazole derivatives with antimicrobial activity. Phosphorous Slufur Silicon Relat. Elem. 2009, 184, 790–804. [Google Scholar] [CrossRef]
- Alafeefy, A.M.; Awaad, A.S.; Abdel-Aziz, H.A.; El-Meligy, R.M.; Zain, M.E.; Al-Outhman, M.R.; Becha, A.B. Synthesis and biological evaluation of certain 3-substituted benzylideneamino-2-(4-nitrophenyl)quinazolin-4(3H)-one derivatives. J. Enzyme Inhib. Med. Chem. 2015, 30, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Vanden Berghe, D.A.; Vlietinck, A.J. Screening methods for antibacterial and antiviral agents from higher plants. In Methods in Plant Biochemistry-Assay for Bioactivity; Dey, P.M., Harborne, J.B., Hostettman, K., Eds.; Academic Press: London, UK, 1991; pp. 47–69. [Google Scholar]
- Mouchlis, V.D.; Magrioti, V.; Barbyianni, E.; Cermak, N.; Oslund, R.C.; Mavromoustakos, T.M.; Gelb, M.H.; Kokotos, G. Inhibition of secreted phospholipases A2 by 2-oxoamides based on α-amino acids: Synthesis, in vitro evaluation and molecular docking calculations. Bioorg. Med. Chem. 2011, 19, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Jany, K.-D.; Lederer, G.; Maye, B. Amino acid sequence of proteinase K from the mold Tritirachium album Limber: Proteinase K—A subtilisin-related enzyme with disulfide bonds. FEBS Lett. 1986, 199, 139–144. [Google Scholar] [CrossRef]
- Milewski, S. Glucosamine-6-phosphate synthase—The multi-facets enzyme. Biochim. Biophys. Acta 2002, 1597, 173–192. [Google Scholar] [CrossRef]
- Durand, P.; Golinelli-Pimpaneau, B.; Mouilleron, S.; Badet, B.; Badet-Denisot, M.A. Highlights of glucosamine-6P synthase catalysis. Arch. Biochem. Biophys. 2008, 474, 302–317. [Google Scholar] [CrossRef] [PubMed]
- Hollenhorst, M.A.; Ntai, I.; Badet, B.; Kelleher, N.L.; Walsh, C.T. A Head-to-Head Comparison of Eneamide and Epoxyamide Inhibitors of Glucosamine-6-Phosphate Synthase from the Dapdiamide Biosynthetic Pathway. Biochemistry 2011, 50, 3859–3861. [Google Scholar] [CrossRef] [PubMed]
- Wojciechowski, M.A.; Milewski, S.; Mazerski, J.; Borowski, E. Glucosamine-6-phosphate synthase, a novel target for antifungal agents. Molecular modelling studies in drug design. Acta Biochim. Pol. 2005, 52, 647–653. [Google Scholar] [PubMed]
- De Araújo, A.L.; Radvanyi, F. Determination of phospholipase A2 activity by a colorimetric assay using a pH indicator. Toxicon 1987, 25, 1181–1188. [Google Scholar] [CrossRef]
- Kunitz, M.J. Crystalline soyabean trypsin inhibitor II. General properties. Gen. Physiol. 1947, 30, 291–310. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds are available from the authors.


| Comp # | Gram-Negative Bacteria | Gram-Positive Bacteria | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli | p. aeruginosa | K. Pneumoniae | Salmonella | Serratia | S. aureus | S. aureus ALA1 | MRSA ATCC3 | S. mutans | E. feacalis | B. subtilis | |
| 6a | 0 ± 0 | 17 ± 1 | 0 ± 0 | 8 ± 1 | 15.5 ± 0 | 0 ± 0 | 10 ± 0 | 12 ± 0 | 12 ± 0.5 | 18.5 ± 0.5 | 0 ± 0 |
| 6b | 14.5 ± 1.5 | 12 ± 0 | 0 ± 0 | 12.5 ± 0.5 | 12.5 ± 0.5 | 8 ± 0.5 | 0 ± 0 | 6 ± 0 | 0 ± 0 | 9.5 ± 0.5 | 0 ± 0 |
| 6c | 1 ± 0 | 1.5 ± 0 | 0 ± 0 | 10.25 ± 1 | 1.4 ± 1 | 0 ± 0 | 0 ± 0 | 8 ± 0.5 | 0.6 ± 1.2 | 1.3 ± 1 | 0 ± 0 |
| 6d | 14.5 ± 0.5 | 10 ± 0 | 12.2 ± 1 | 6.5 ± 0 | 12 ± 0 | 8.5 ± 0.5 | 0 ± 0 | 0 ± 0 | 10 ± 0.0 | 18 ± 1 | 0 ± 0 |
| 6e | 10 ± 0 | 12.5 ± 0.1 | 12.5 ± 0.5 | 4 ± 0 | 14 ± 1 | 8.5 ± 0.5 | 0 ± 0 | 14 ± 1 | 8 ± 1 | 18.5 ± 0.5 | 0 ± 0 |
| 6f | 0 ± 0 | 10.5 ± 0.1 | 4.5 ± 0.1 | 10 ± 0 | 14 ± 0.1 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 8 ± 1 | 0 ± 0 | 0 ± 0 |
| 6g | 0 ± 0 | 10.5 ± 0.5 | 8 ± 1 | 0 ± 0 | 10.5 ± 0.5 | 8.5 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| 6h | 8 ± 0 | 12.5 ± 0.7 | 0 ± 0 | 0 ± 0 | 10 ± 0 | 8.5 ± 0.7 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| 6i | 8.5 ± 0.5 | 10.5 ± 0.5 | 0 ± 0 | 0 ± 0 | 8.5 ± 0.5 | 10.5 ± 1.5 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| 6j | 8 ± 1 | 10 ± 0 | 0 ± 0 | 0 ± 0 | 8.5 ± 0.5 | 19.5 ± 1.5 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| 6k | 10 ± 0 | 13.5 ± 1.5 | 14.5 ± 1.5 | 0 ± 0 | 10 ± 0 | 10.5 ± 1.5 | 0 ± 0 | 0 ± 0 | 24.5 ± 0.5 | 0 ± 0 | 8.5 ± 0.5 |
| 6l | 14 ± 1 | 11 ± 0 | 11 ± 0 | 8 ± 1 | 13 ± 0.1 | 18.5 ± 0.5 | 0 ± 0 | 0 ± 0 | 14 ± 2 | 0 ± 0 | 0 ± 0 |
| 6m | 10 ± 1 | 18.5 ± 0.5 | 0 ± 0 | 4 ± 0 | 14.5 ± 1.5 | 8.5 ± 0.5 | 0 ± 0 | 8.5 ± 0.5 | 0 ± 0 | 13.5 ± 1.5 | 0 ± 0 |
| 6n | 10 ± 0 | 0 ± 0 | 13 ± 1 | 0 ± 0 | 12.5 ± 0.5 | 14 ± 1 | 0 ± 0 | 10 ± 0 | 15 ± 1 | 18.5 ± 0.5 | 0 ± 0 |
| 6o | 10 ± 0 | 18.5 ± 0.5 | 10 ± 0 | 0 ± 0 | 12.5 ± 0.5 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 9 ± 0.6 | 0 ± 0 | 0 ± 0 |
| TCN | 19 ± 0.1 | 17 ± 0.2 | 17 ± 0.5 | 16 ± 0.1 | 12 ± 0.3 | 31 ± 0.6 | 29 ± 0.4 | 18 ± 0.3 | 27 ± 1.2 | 0 ± 0 | 15 ± 0.5 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Sayed, N.N.E.; Alafeefy, A.M.; Bakht, M.A.; Masand, V.H.; Aldalbahi, A.; Chen, N.; Fan, C.; Ben Bacha, A. Synthesis, Antiphospholipase A2, Antiprotease, Antibacterial Evaluation and Molecular Docking Analysis of Certain Novel Hydrazones. Molecules 2016, 21, 1664. https://doi.org/10.3390/molecules21121664
El-Sayed NNE, Alafeefy AM, Bakht MA, Masand VH, Aldalbahi A, Chen N, Fan C, Ben Bacha A. Synthesis, Antiphospholipase A2, Antiprotease, Antibacterial Evaluation and Molecular Docking Analysis of Certain Novel Hydrazones. Molecules. 2016; 21(12):1664. https://doi.org/10.3390/molecules21121664
Chicago/Turabian StyleEl-Sayed, Nahed N. E., Ahmed M. Alafeefy, Mohammed A. Bakht, Vijay H. Masand, Ali Aldalbahi, Nan Chen, Chunhai Fan, and Abir Ben Bacha. 2016. "Synthesis, Antiphospholipase A2, Antiprotease, Antibacterial Evaluation and Molecular Docking Analysis of Certain Novel Hydrazones" Molecules 21, no. 12: 1664. https://doi.org/10.3390/molecules21121664







