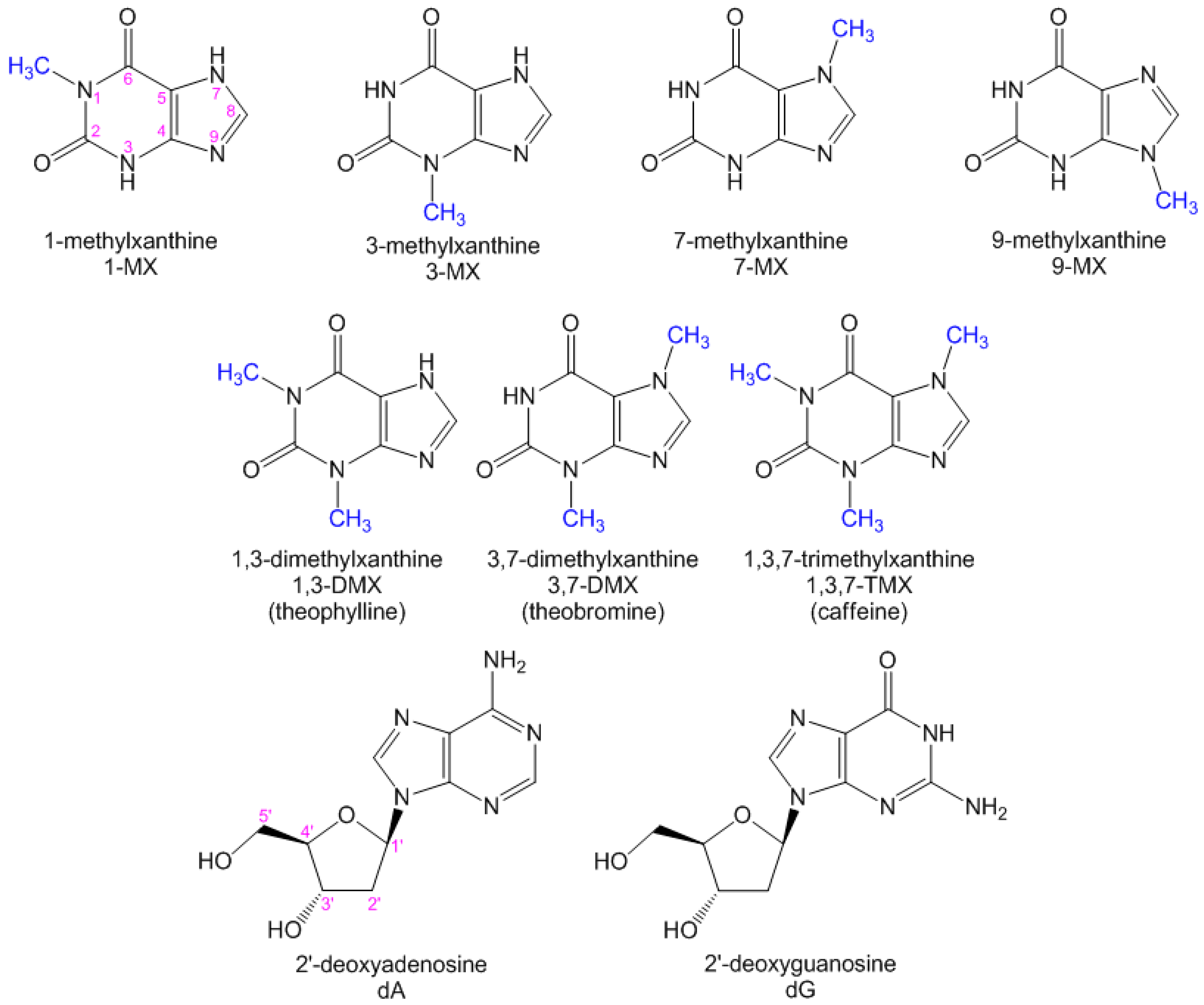
Xanthines Studied via Femtosecond Fluorescence Spectroscopy
Abstract
:1. Introduction
2. Results
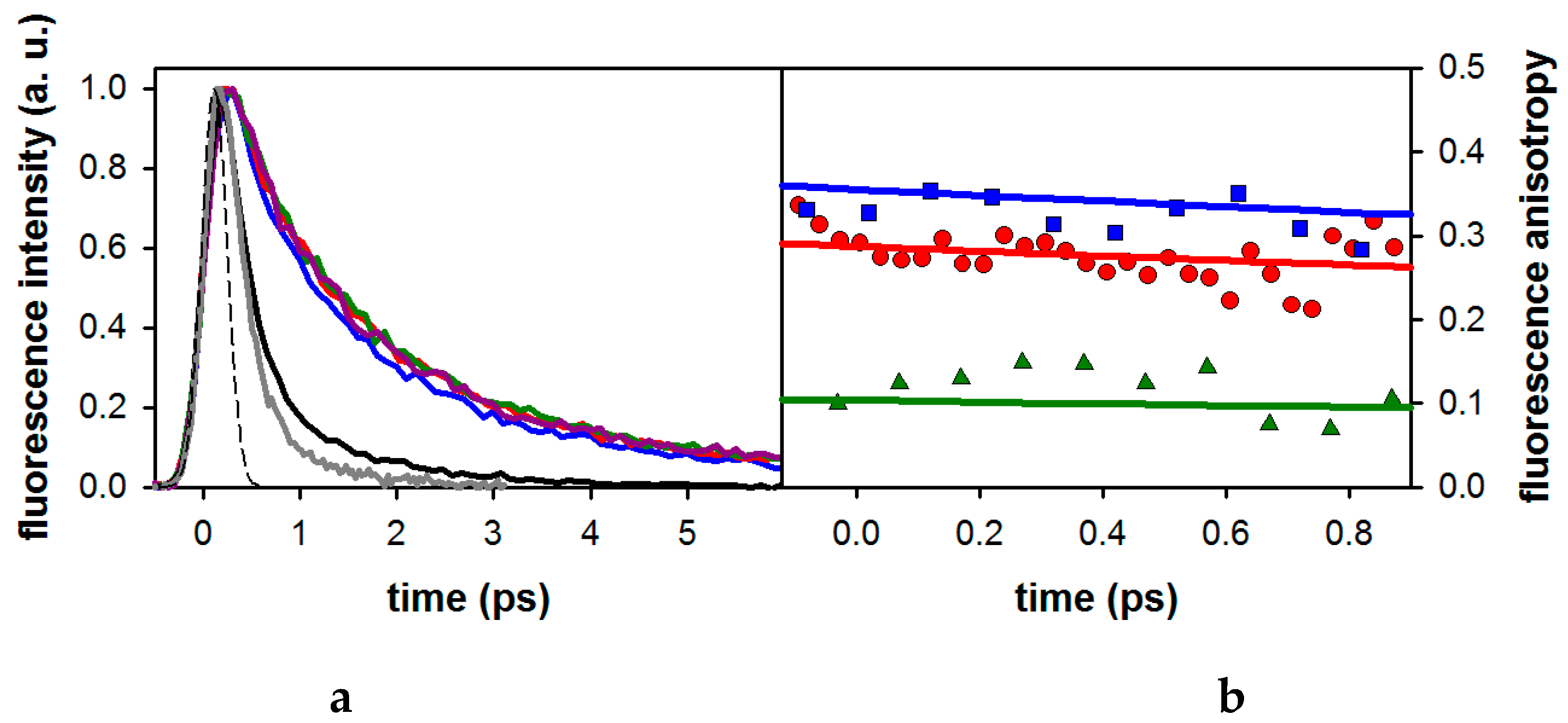
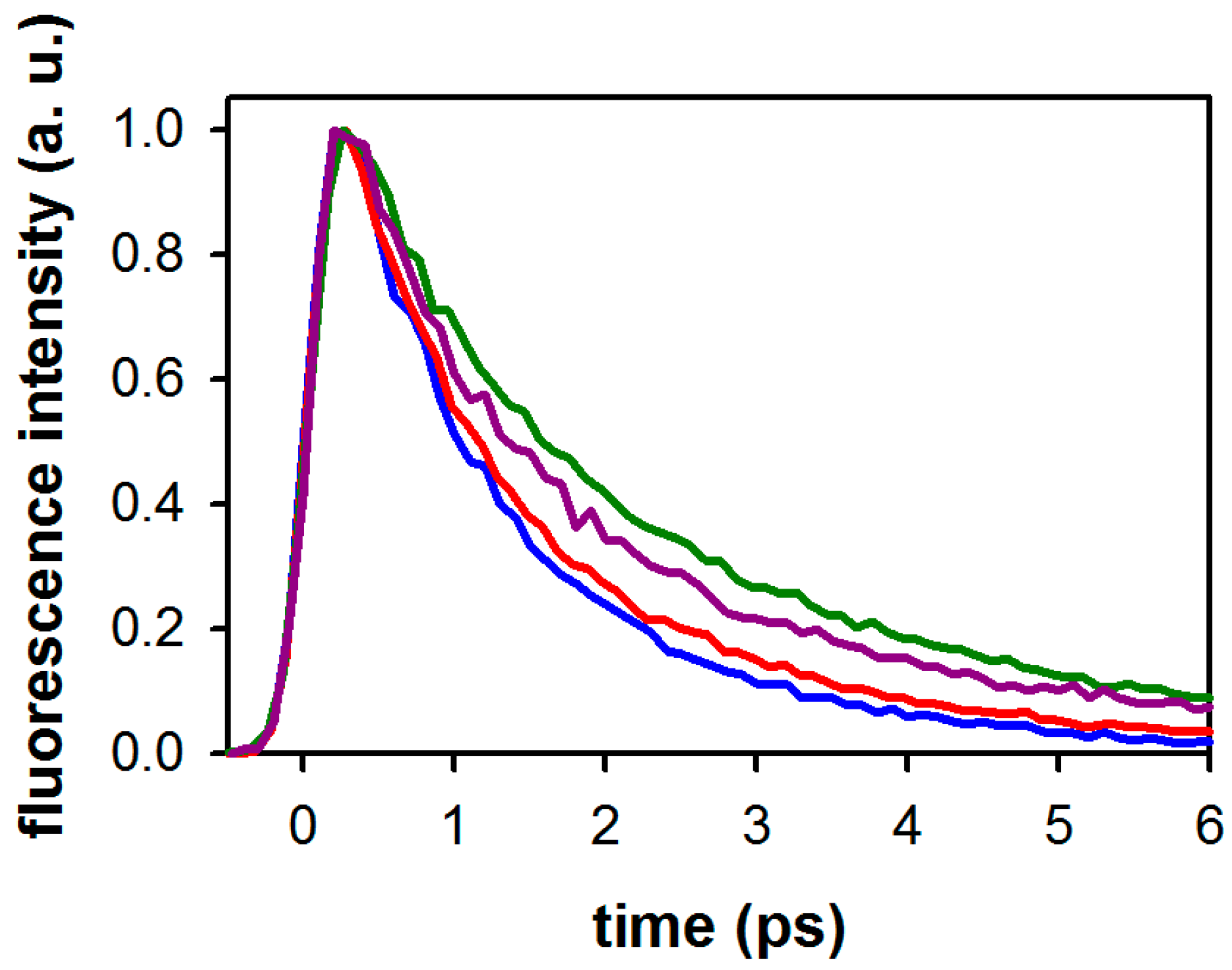
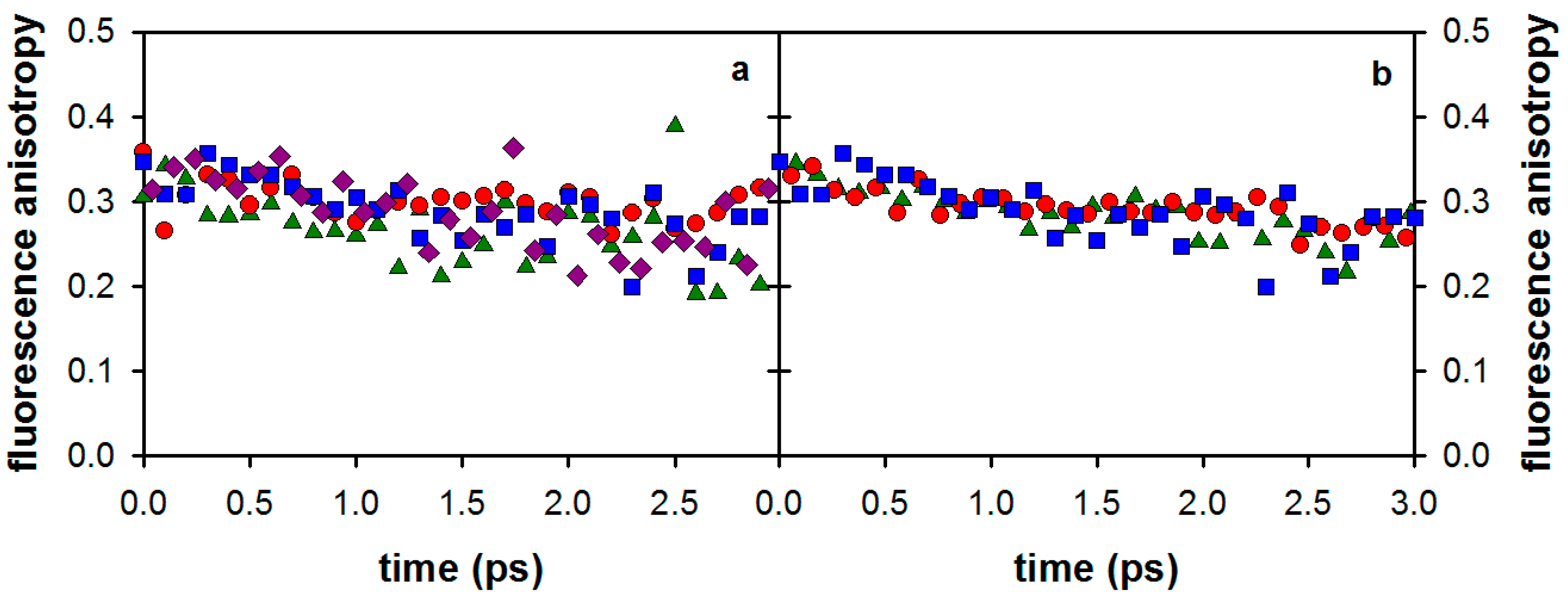
2.1. Fluorescence Properties of 3-Methylxanthine
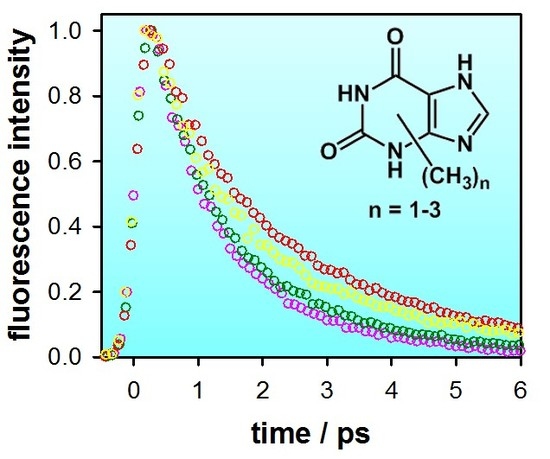
2.2. Substituent Effect on the Fluorescence Dynamics
3. Discussion
4. Materials and Methods
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Eritja, R.; Horowitz, D.M.; Walker, P.A.; Ziehlermartin, J.P.; Boosalis, M.S.; Goodman, M.F.; Itakura, K.; Kaplan, B.E. Synthesis and properties of oligonucleotides containing 2′-deoxynebularine and 2′-deoxyxanthosine. Nucleic Acids Res. 1986, 14, 8135–8153. [Google Scholar] [CrossRef] [PubMed]
- Szolomájer, J.; Paragi, G.; Batta, G.; Guerra, C.F.; Bickelhaupt, F.M.; Kele, Z.; Pádár, P.; Kupihár, Z.; Kovács, L. 3-substituted xanthines as promising candidates for quadruplex formation: Computational, synthetic and analytical studies. New J. Chem. 2011, 35, 476. [Google Scholar] [CrossRef]
- Cheong, V.V.; Heddi, B.; Lech, C.J.; Phan, A.T. Xanthine and 8-oxoguanine in g-quadruplexes: Formation of a ggxo tetrad. Nucleic Acids Res. 2015, 43, 10506. [Google Scholar] [CrossRef] [PubMed]
- Cheong, V.V.; Lech, C.J.; Heddi, B.; Phan, A.T. Inverting the G-tetrad polarity of a G-quadruplex by using xanthine and 8-oxoguanine. Angew. Chem. Int. Ed. 2016, 55, 160–163. [Google Scholar] [CrossRef] [PubMed]
- Paragi, G.; Kovács, L.; Kupihár, Z.; Szolomájer, J.; Penke, B.; Fonseca Guerra, C.; Bickelhaupt, F.M. Neutral and positively charged new purine tetramer structures: A computational study of xanthine and uric acid derivatives. New J. Chem. 2011, 35, 119. [Google Scholar] [CrossRef]
- Novotny, J.; Kulhanek, P.; Marek, R. Biocompatible xanthine-quadruplex scaffold for ion-transporting DNA channels. J. Phys. Chem. Lett. 2012, 3, 1788–1792. [Google Scholar] [CrossRef] [PubMed]
- Novotny, J.; Yurenko, Y.P.; Kulhanek, P.; Marek, R. Tailoring the properties of quadruplex nucleobases for biological and nanomaterial applications. Phys. Chem. Chem. Phys. 2014, 16, 15241–15248. [Google Scholar] [CrossRef] [PubMed]
- Yurenko, Y.P.; Novotny, J.; Sklenar, V.; Marek, R. Exploring non-covalent interactions in guanine- and xanthine-based model DNA quadruplex structures: A comprehensive quantum chemical approach. Phys. Chem. Chem. Phys. 2014, 16, 2072–2084. [Google Scholar] [CrossRef] [PubMed]
- Livshits, G.I.; Stern, A.; Rotem, D.; Borovok, N.; Eidelshtein, G.; Migliore, A.; Penzo, E.; Wind, S.J.; di Felice, R.; Skourtis, S.S.; et al. Long-range charge transport in single G-quadruplex DNA molecules. Nat. Nanotechnol. 2014, 9, 1040–1046. [Google Scholar] [CrossRef] [PubMed]
- Fredholm, B.B. Methylxanthines; Springer: Berlin/Heidelberg, Germany, 2011; Volume 200. [Google Scholar]
- Monteiro, J.P.; Alves, M.G.; Oliveira, P.F.; Silva, B.M. Structure-bioactivity relationships of methylxanthines: Trying to make sense of all the promises and the drawbacks. Molecules 2016, 21, 974. [Google Scholar] [CrossRef] [PubMed]
- Lutz, M.J.; Held, H.A.; Hottiger, M.; Hubscher, U.; Benner, S.A. Differential discrimination of DNA polymerases for variants of the non-standard nucleobase pair between xanthosine and 2,4-diaminopyrimidine, two components of an expanded genetic alphabet. Nucleic Acids Res. 1996, 24, 1308–1313. [Google Scholar] [CrossRef] [PubMed]
- Shukla, M.K.; Mishra, P.C. Electronic spectra and structures of some biologically important xanthines. J. Mol. Struct. 1994, 324, 241–249. [Google Scholar] [CrossRef]
- McKemy, D.D.; Welch, W.; Airey, J.A.; Sutko, J.L. Concentrations of caffeine greater than 20 mm increase the indo-1 fluorescence ratio in a Ca2+-independent manner. Cell Calcium 2000, 27, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Karim, M.M.; Jeon, C.W.; Lee, H.S.; Alam, S.M.; Lee, S.H.; Choi, J.H.; Jin, S.O.; Das, A.K. Simultaneous determination of acetylsalicylic acid and caffeine in pharmaceutical formulation by first derivative synchronous fluorimetric method. J. Fluoresc. 2006, 16, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.L.; Dong, C.; Shuang, S.M.; Liu, D.S. Study for luminescence performance of three methyl xanthine derivatives. Spectrochim. Acta Part A 2005, 61, 2584–2589. [Google Scholar] [CrossRef] [PubMed]
- Onidas, D.; Markovitsi, D.; Marguet, S.; Sharonov, A.; Gustavsson, T. Fluorescence properties of DNA nucleosides and nucleotides: A refined steady-state and femtosecond investigation. J. Phys. Chem. B 2002, 106, 11367–11374. [Google Scholar] [CrossRef]
- Gustavsson, T.; Improta, R.; Markovitsi, D. DNA/RNA: Building blocks of life under UV irradiation. J. Phys. Chem. Lett. 2010, 1, 2025–2030. [Google Scholar] [CrossRef] [Green Version]
- Miannay, F.A.; Gustavsson, T.; Banyasz, A.; Markovitsi, D. Excited-state dynamics of dgmp measured by steady-state and femtosecond fluorescence spectroscopy. J. Phys. Chem. A 2010, 114, 3256–3263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gustavsson, T.; Sharonov, A.; Onidas, D.; Markovitsi, D. Adenine, deoxyadenosine and deoxyadenosine 5′-monophosphate studied by femtosecond fluorescence upconversion spectroscopy. Chem. Phys. Lett. 2002, 356, 49–54. [Google Scholar] [CrossRef]
- Yamazaki, S.; Sobolewski, A.L.; Domcke, W. Photophysics of xanthine: Computational study of the radiationless decay mechanisms. Phys. Chem. Chem. Phys. 2009, 11, 10165–10174. [Google Scholar] [CrossRef] [PubMed]
- Nachtigallova, D.; Aquino, A.J.A.; Horn, S.; Lischka, H. The effect of dimerization on the excited state behavior of methylated xanthine derivatives: A computational study. Photochem. Photobiol. Sci. 2013, 12, 1496–1508. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Q.; Kohler, B. Ultrafast nonradiative decay by hypoxanthine and several methylxanthines in aqueous and acetonitrile solution. Phys. Chem. Chem. Phys. 2012, 14, 10677–10682. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Y.; Chen, J.Q.; Kohler, B. Hydrogen bond donors accelerate vibrational cooling of hot purine derivatives in heavy water. J. Phys. Chem. A 2013, 117, 6771–6780. [Google Scholar] [CrossRef] [PubMed]
- Gustavsson, T.; Sarkar, N.; Vayá, I.; Jiménez, M.C.; Markovitsi, D.; Improta, R. A joint experimental/theoretical study of the ultrafast excited state deactivation of deoxyadenosine and 9-methyladenine in water and acetonitrile. Photochem. Photobiol. Sci. 2013, 12, 1375–1386. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Plenum Press: New York, NY, USA, 2006. [Google Scholar]
- Gustavsson, T.; Sharonov, A.; Markovitsi, D. Thymine, thymidine and thymidine 5′-monophosphate studied by femtosecond fluorescence upconversion spectroscopy. Chem. Phys. Lett. 2002, 351, 195–200. [Google Scholar] [CrossRef]
- Middleton, C.T.; de la Harpe, K.; Su, C.; Law, Y.K.; Crespo-Hernandez, C.E.; Kohler, B. DNA excited-state dynamics: From single bases to the double helix. Ann. Rev. Phys. Chem. 2009, 60, 217–239. [Google Scholar] [CrossRef] [PubMed]
- Improta, R.; Santoro, F.; Blancafort, L. Quantum mechanical studies on the photophysics and the photochemistry of nucleic acids and nucleobases. Chem. Rev. 2016, 116, 3540–3593. [Google Scholar] [CrossRef] [PubMed]
- Gustavsson, T.; Banyasz, A.; Lazzarotto, E.; Markovitsi, D.; Scalmani, G.; Frisch, M.J.; Barone, V.; Improta, R. Singlet excited-state behavior of uracil and thymine in aqueous solution: A combined experimental and computational study of 11 uracil derivatives. J. Am. Chem. Soc. 2006, 128, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Gustavsson, T.; Sarkar, N.; Lazzarotto, E.; Markovitsi, D.; Barone, V.; Improta, R. Solvent effect on the singlet excited state dynamics of 5-fluorouracil in acetonitrile as compared to water. J. Phys. Chem. B 2006, 110, 12843–12847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gustavsson, T.; Sarkar, N.; Lazzarotto, E.; Markovitsi, D.; Improta, R. Singlet excited state dynamics of uracil and thymine derivatives. A femtosecond fluorescence upconversion study in acetonitrile. Chem. Phys. Lett. 2006, 429, 551–557. [Google Scholar] [CrossRef] [Green Version]
- Santoro, F.; Barone, V.; Gustavsson, T.; Improta, R. Solvent effect on the singlet excited state lifetimes of nucleic acid bases: A computational study of 5-fluorouracil and uracil in acetonitrile and water. J. Am. Chem. Soc. 2006, 128, 16312–16322. [Google Scholar] [CrossRef] [PubMed]
- Santoro, F.; Gustavsson, T.; Lami, S.; Barone, V.; Improta, R. Towards the understanding of the excited state dynamics of nucleic acids: Solvent and stacking effect on the photophysical behavior of nucleobases. AIP Conf. Proc. 2007, 963, 631–634. [Google Scholar]
- Zgierski, M.Z.; Patchkovskii, S.; Fujiwara, T.; Lim, E.C. On the origin of the ultrafast internal conversion of electronically excited pyrimidine bases. J. Phys. Chem. A 2005, 109, 9384–9387. [Google Scholar] [CrossRef] [PubMed]
- Zgierski, M.Z.; Fujiwara, T.; Kofron, W.G.; Lim, E.C. Highly effective quenching of the ultrafast radiationless decay of photoexcited pyrimidine bases by covalent modification: Photophysics of 5,6-trimethylenecytosine and 5,6-trimethyleneuracil. Phys. Chem. Chem. Phys. 2007, 9, 3206–3209. [Google Scholar] [CrossRef] [PubMed]
- Karunakaran, V.; Kleinermanns, K.; Improta, R.; Kovalenko, S.A. Photoinduced dynamics of guanosine monophosphate in water from broad-band transient absorption spectroscopy and quantum-chemical calculations. J. Am. Chem. Soc. 2009, 131, 5839–5850. [Google Scholar] [CrossRef] [PubMed]
- Bridson, P.K.; Richmond, G.; Yeh, F. Preparation of xanthine-3-acetic acid and some derivatives. Synth. Commun. 1990, 20, 2459–2467. [Google Scholar] [CrossRef]
- Muller, C.E.; Thorand, M.; Qurishi, R.; Diekmann, M.; Jacobson, K.A.; Padgett, W.L.; Daly, J.W. Imidazo [2,1-i]purin-5-ones and related tricyclic water-soluble purine derivatives: Potent A2A− and A3− adenosine receptor antagonists. J. Med. Chem. 2002, 45, 3440–3450. [Google Scholar] [CrossRef] [PubMed]
- Miannay, F.A.; Banyasz, A.; Gustavsson, T.; Markovitsi, D. Excited states and energy transfer in G-quadruplexes. J. Phys. Chem. C 2009, 113, 11760–11765. [Google Scholar] [CrossRef] [Green Version]
- Changenet-Barret, P.; Hua, Y.; Markovitsi, D. Electronic excitations in guanine quadruplexes. Top. Curr. Chem. 2015, 356, 183–201. [Google Scholar] [PubMed]
- Changenet-Barret, P.; Hua, Y.; Gustavsson, T.; Markovitsi, D. Electronic excitations in g-quadruplexes formed by the human telomeric sequence: A time-resolved fluorescence study. Photochem. Photobiol. 2015, 91, 759–765. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Not available.
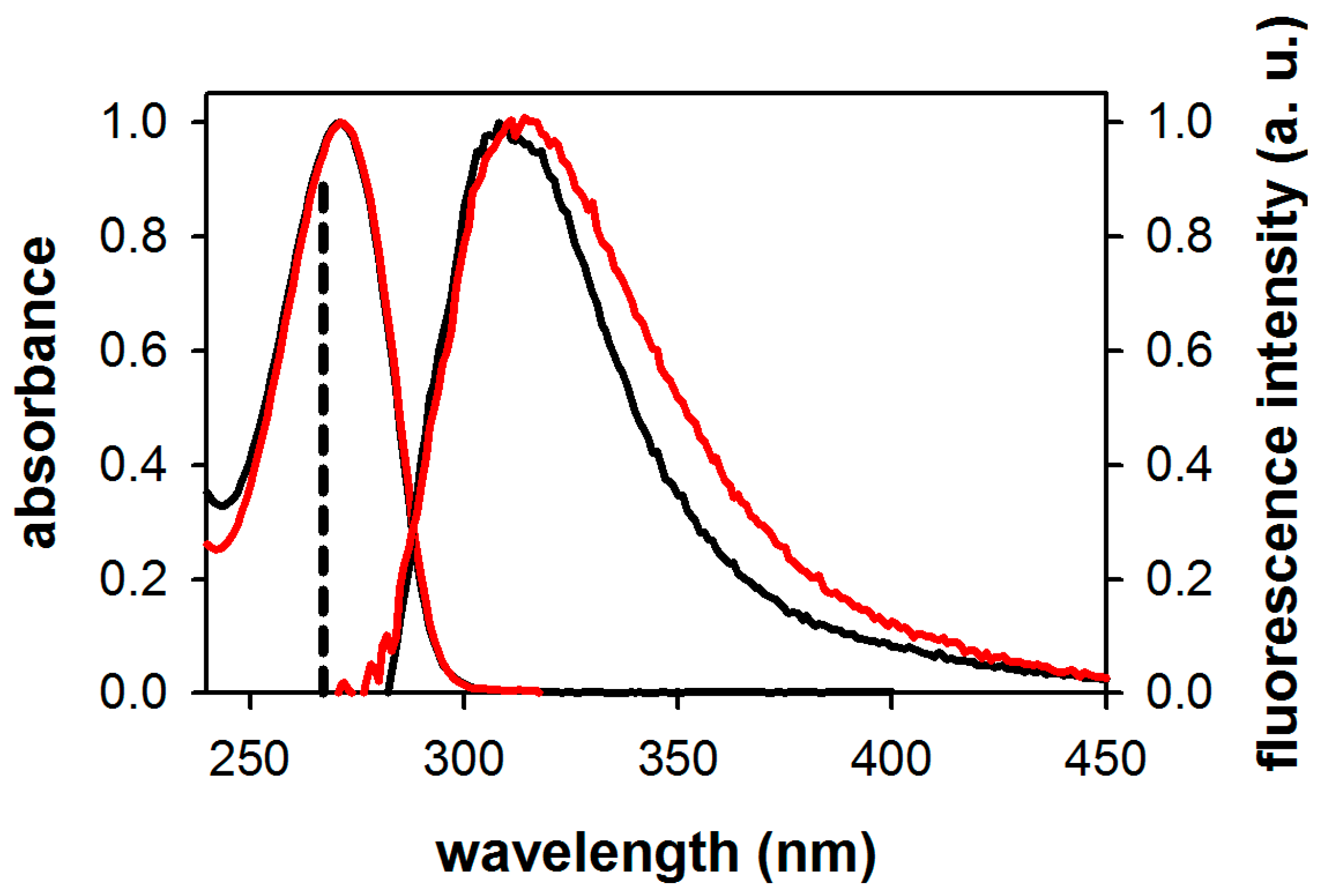

| Compound/Solvent | λfl (nm) | φf × 104 | Δν (cm−1) |
|---|---|---|---|
| 3-MX/MeOH | 311 | 4.5 | 5200 |
| 3-MX/H2O | 315 | 2.4 | 6150 |
| dA/H2O | 307 1 | 0.9 1 | 10,150 |
| dG/H2O | 334 1 | 1.0 1 | 11,390 |
| Compound/Solvent | <τ> (ps) 1 | τrad (ns) |
|---|---|---|
| 3-MX/MeOH | 1.67 | 3.7 |
| dA/H2O | 0.13 2 | 1.4 |
| dA/MeOH | 0.27 3 | nd |
| dG/H2O | 0.33 4 | 3.3 |
| dG/MeOH | 0.42 3 | nd |
| Compound | α | τ1 (ps) | τ2 (ps) | <τ> (ps) 1 | r0 | τTA (ps) 2 |
|---|---|---|---|---|---|---|
| 1-MX | 0.82 | 0.54 | 2.62 | 0.91 | 0.34 | |
| 3-MX | 0.49 | 0.66 | 2.65 | 1.67 | 0.33 | |
| 7-MX | 0.49 | 0.44 | 1.72 | 1.10 | 0.32 | |
| 9-MX | 0.68 | 0.52 | 2.50 | 1.15 | 0.31 | |
| 1,3-DMX | 0.51 | 0.60 | 1.69 | 1.13 | 0.33 | 1.7 |
| 3,7-DMX | 0.35 | 0.71 | 2.70 | 2.01 | 0.32 | 2.0 |
| 1,3,7-TMX | 0.65 | 0.85 | 2.35 | 1.37 | 0.33 | 1.3 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Changenet-Barret, P.; Kovács, L.; Markovitsi, D.; Gustavsson, T. Xanthines Studied via Femtosecond Fluorescence Spectroscopy. Molecules 2016, 21, 1668. https://doi.org/10.3390/molecules21121668
Changenet-Barret P, Kovács L, Markovitsi D, Gustavsson T. Xanthines Studied via Femtosecond Fluorescence Spectroscopy. Molecules. 2016; 21(12):1668. https://doi.org/10.3390/molecules21121668
Chicago/Turabian StyleChangenet-Barret, Pascale, Lajos Kovács, Dimitra Markovitsi, and Thomas Gustavsson. 2016. "Xanthines Studied via Femtosecond Fluorescence Spectroscopy" Molecules 21, no. 12: 1668. https://doi.org/10.3390/molecules21121668