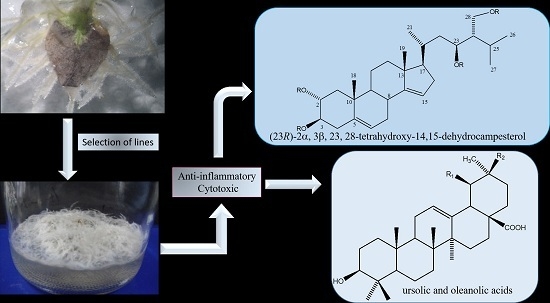
A Cytotoxic and Anti-inflammatory Campesterol Derivative from Genetically Transformed Hairy Roots of Lopezia racemosa Cav. (Onagraceae)
Abstract
:1. Introduction
2. Results and Discussion
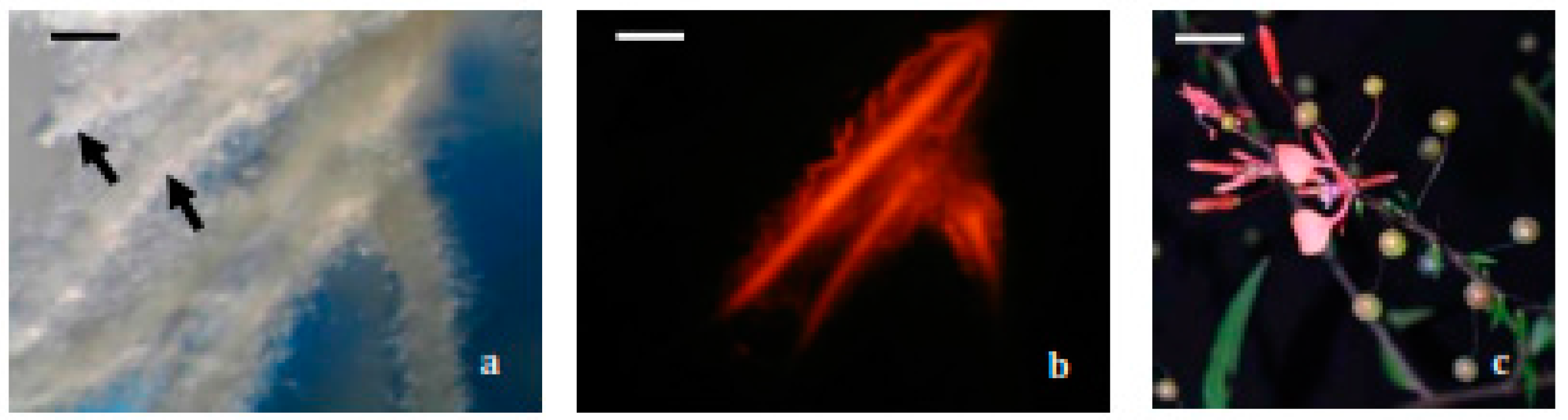
2.1. Hairy Roots Obtaining and Selection
2.2. Anti-Inflammatory and Cytotoxic Activity of the Transformed Line LRT 7.31
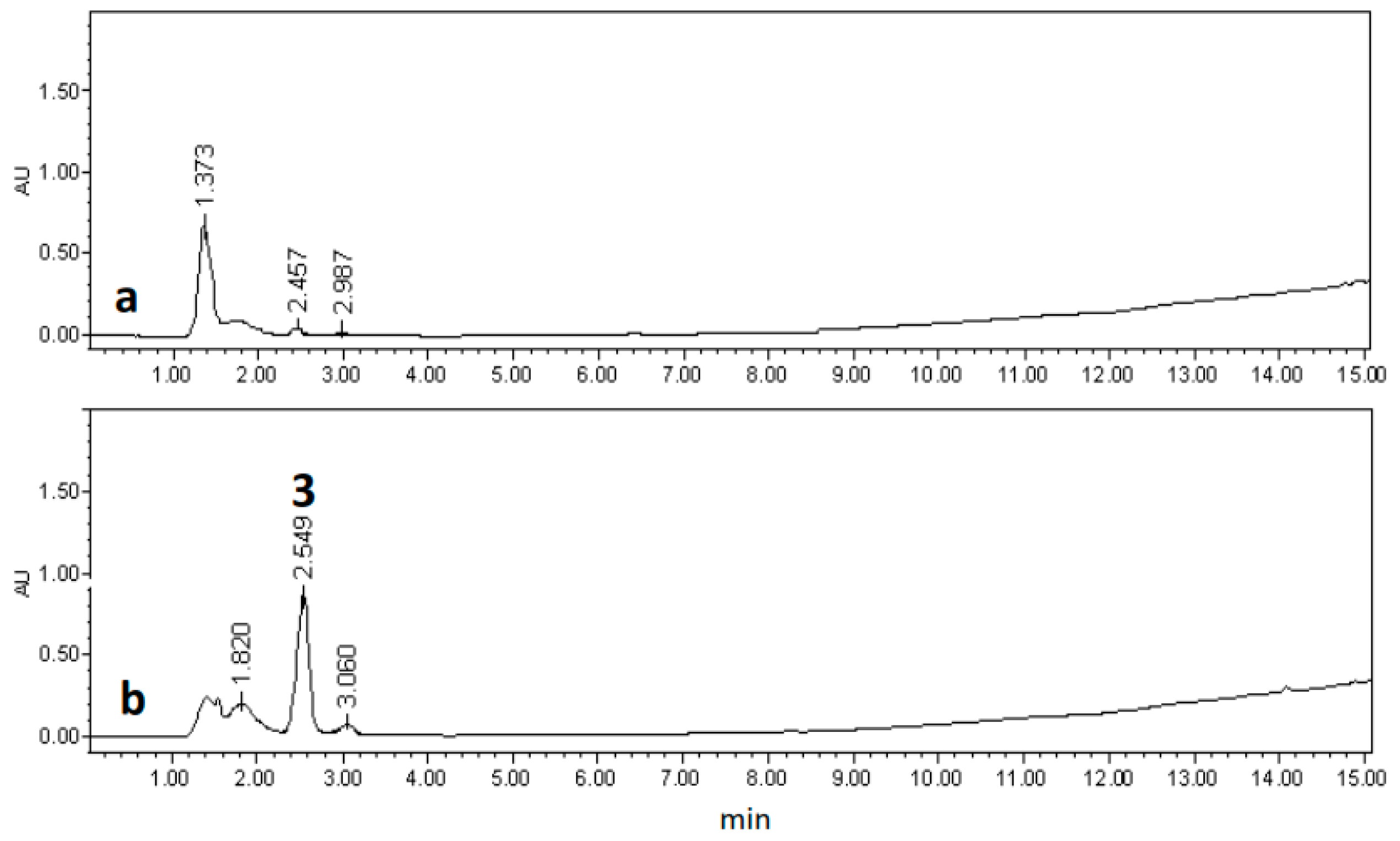
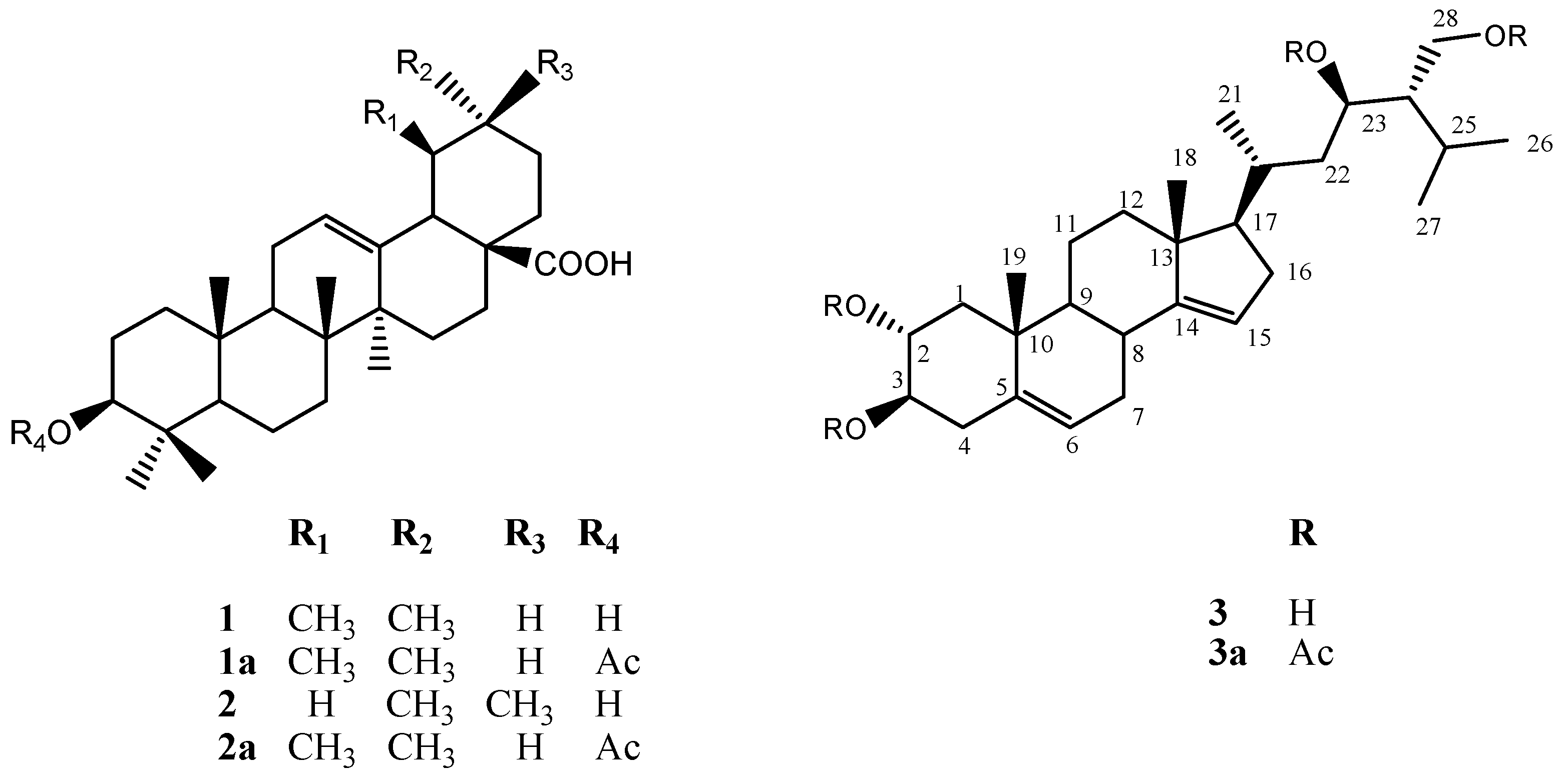
2.3. Chemical Characterization
3. Experimental Section
3.1. General Procedures
3.2. Bacterial Strain
3.3. Plant Material
3.4. Seed Germination and Axenic Seedling Obtaining
3.5. Genetic Transformation
3.5.1. Hairy Root Induction
3.5.2. Selection and Establishment of Hairy Root Lines
3.6. DNA Isolation and PCR Analysis
3.6.1. DNA Isolation
3.6.2. PCR Analysis
3.7. Extraction and Isolation of Chemicals Compounds from of Selected Hairy Root Line LTR-7.31
3.8. Antiinflammatory and Cytotoxic Activities
3.8.1. TPA Induced Mice Ear Inflammation Model
3.8.2. Cytotoxic Activity
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Tian, L. Using Hairy Roots for Production of Valuable Plant Secondary Metabolites. In Filaments in Bioprocesses; Advances in Biochemical Engineering/Biotechnology; Krull, R., Bley, T., Eds.; Springer: New York, NY, USA, 2000; Volume 149, pp. 275–324. [Google Scholar]
- Rahnama, H.; Hasanloo, T.; Shams, M.R.; Sepehrifar, R. Silymarin production by hairy root culture of Silybum marianum (L.) Gaertn. Iran. J. Biotechnol. 2008, 3, 113–118. [Google Scholar]
- Gunjan, S.K.; Lutz, J.; Bushong, A.; Rogers, D.T.; Littleton, J. Hairy Root Cultures and Plant Regeneration in Solidago nemoralis Transformed with Agrobacterium rhizogenes. Am. J. Plant Sci. 2013, 4, 1675–1678. [Google Scholar] [CrossRef]
- Samadi, A.; Jafari, M.; Nejhad, N.M.; Hossenian, F. Podophyllotoxin and 6-methoxy podophyllotoxin production in hairy root cultures of Linum mucronatum ssp. mucronatum. Pharmacogn. Mag. 2014, 10, 154–160. [Google Scholar] [PubMed]
- Cequier-Sánchez, E.; Rodríguez, C.; Dorta-Guerra, R.; Ravelo, A.G.; Zárate, R. Echium acanthocarpum hairy root cultures, a suitable system for polyunsaturated fatty acid studies and production. BMC Biotechnol. 2011, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Weathers, P.J.; Bunk, G.; McCoy, M.C. The effect of phytohormones on growth and artemisinin production in Artemisia annua hairy roots. In Vitro Cell. Dev. Biol.-Plant 2005, 41, 47–53. [Google Scholar] [CrossRef]
- Sharma, P.; Padh, H.; Shrivastava, N. Hairy root cultures: A suitable biological system for studying secondary metabolic pathways in plants. Eng. Life Sci. 2013, 13, 62–75. [Google Scholar] [CrossRef]
- Alonso-Castro, A.J.; Villarreal, M.L.; Salazar-Olivo, L.A.; Gomez-Sanchez, M.; Dominguez, F.; García-Carranca, A. Mexican medicinal plants used for cancer treatment: Pharmacological, phytochemical and ethnobotanical studies. J. Ethnopharm. 2011, 133, 945–972. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Paredes, C.; Bolívar, B.P.; Gómez-Velasco, A.; Juárez, Z.N.; Sánchez, A.E.; Hernández, L.R.; Bach, H. Antimicrobial, Antiparasitic, Anti-Inflammatory, and Cytotoxic Activities of Lopezia racemosa. Sci. World J. 2013, 2013, 237438. [Google Scholar] [CrossRef] [PubMed]
- Salinas, R.; Arellano-García, J.; Perea-Arango, I.; Álvarez, L.; Garduño-Ramírez, M.L.; Marquina, S.; Zamilpa, A.; Castillo-España, P. Production of the Anti-inflammatory Compound 6-O-Palmitoyl-3-O-β-d-glucopyranosylcampesterol by Callus Cultures of Lopezia racemosa Cav. (Onagraceae). Molecules 2014, 19, 8679–8690. [Google Scholar] [CrossRef] [PubMed]
- Rubnov, S.; Kashman, Y.; Rabinowitz, R.; Schlesinger, M.; Mechoulam, R. Suppressors of Cancer Cell Proliferation from Fig (Ficus carica) Resin: Isolation and Structure Elucidation. J. Nat. Prod. 2001, 64, 993–996. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.F.; Zain, C.R.C.M.; Zainal, Z.; Noor, N.M.; Anuar, N.; Markom, M.; Ismail, I. Establishment of Persicaria minor hairy roots and analysis of secreted β-caryophyllene in medium broth. Plant Cell Tissue Organ Cult. 2015, 121, 11–20. [Google Scholar] [CrossRef]
- Wahyuni, D.K.; Vidianti, F.; Purnobasuki, H.; Ermayanti, T.M.; Prajoga, B.; Utami, E.S.W. Agrobacterium rhizogenes Mediated Hairy Root Induction in Justicia gendarussa Burm.f. J. Appl. Environ. Biol. Sci. 2015, 5, 87–93. [Google Scholar]
- Schmulling, T.; Schell, J.; Spena, A. Single genes from Agrobacterium rhizogenes influence plant development. EMBO J. 1988, 7, 2621–2629. [Google Scholar] [PubMed]
- Gheysen, G.; Montagu, M.V.; Zambryski, P. Integration of Agrobacterium tumefaciens transfer DNA (T-DNA) involves rearrangements of target plant DNA sequences. Proc. Natl. Acad. Sci. USA 1987, 84, 6169–6173. [Google Scholar] [CrossRef] [PubMed]
- Peerbolte, R.; Ruigrok, P.; Gullems, G.; Schilperoort, R. T-DNA Rearrangements due to tissue culture: Somaclonal variation in crown gall tissues. Plant Mol. Biol. 1987, 9, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Meza, T.J.; Stangeland, B.; Mercy, I.S.; Skarn, M.; Nymoen, D.A.; Berg, A.; Butenko, M.A.; Hakelien, A.M.; Camilla Hasleka, C.; Meza-Zepeda, L.A.; et al. Analyses of single-copy Arabidopsis T-DNA-transformed lines show that the presence of vector backbone sequences, short inverted repeats and DNA methylation is not suficient or necessary for the induction of transgene silencing. Nucleic Acids Res. 2002, 30, 4556–4566. [Google Scholar] [CrossRef] [PubMed]
- Gelvin, S.B. Agrobacterium-Mediated Plant Transformation: The Biology behind the “Gene-Jockeying” Tool. Microbiol. Mol. Biol. Rev. 2003, 67, 16–37. [Google Scholar] [CrossRef] [PubMed]
- Jaki, B.U.; Franzblau, S.G.; Chadwick, L.R. Purity-Activity Relationships of Natural Products: The case of Anti-TB Active Ursolic Acid. J. Nat. Prod. 2008, 71, 1742–1748. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, L.; Núñez, M.; Pérez, M.C.; Villarreal, M.L.; Delgado, G. Chemical and Biological Study of Astianthus viminalis. Planta Med. 1994, 60, 98. [Google Scholar] [CrossRef] [PubMed]
- Misharin, A.Y.; Mehtiev, A.R.; Morozevich, G.E.; Tkachev, Y.V.; Timofeev, V.P. Synthesis and cytotoxicity evaluation of 22,23-oxygenated stigmastane derivatives. Bioorg. Med. Chem. 2008, 16, 1460–1473. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Watanabe, B.; Otsuki, J.; Nakagawa, Y.; Akamatsu, M.; Miyagawa, H. Synthesis of 26,27-bisnorcastasterone analogs and analysis of conformation-activity relationship for brassinolide-like activity. Bioorg. Med. Chem. 2006, 14, 1761–1770. [Google Scholar] [CrossRef] [PubMed]
- Michelini, F.M.; Ramírez, J.A.; Berra, A.; Galagovsky, L.R.; Alché, L.E. In vitro and in vivo antiherpetic activity of three new synthetic brassinosteroid analogues. Steroids 2004, 69, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Bazaldúa, C.; Cardoso-Taketa, A.; Arellano, J.; Camacho-Díaz, B.; Ventura-Zapata, E.; Villarreal, M.L. Podophyllotoxin-like lignans production through hairy roots of Hyptis suaveolens. JCPBS 2014, 4, 37–47. [Google Scholar]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Gamborg, O.L.; Miller, R.A.; Ojima, K. Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell. Res. 1968, 50, 151–158. [Google Scholar] [CrossRef]
- Bonhomme, V.; Laurain-Mattara, D.; Lacoux, J.; Fliniaux, M.A.; Jacquin-Dubreuil, A. Tropane alkaloid production by hairy roots of Atropa belladonna obtained after transformation with Agrobacterium rhizogenes 15834 and Agrobacterium tumefaciens containing rol A, B, C genes only. J. Biotechnol. 2000, 81, 151–158. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds are not available from the authors.


| Sample Origin | Cancer Cell Lines | |||
|---|---|---|---|---|
| HeLa (μg/mL) | HCT-15 (μg/mL) | OVCAR (μg/mL) | KB (μg/mL) | |
| Crude extract | 63.97 ± 0.09 | 3.14 ± 0.03 | 0.57 ± 0.01 | >100 |
| C1F3 | 0.00089 ± 0.000098 | 3.32 ± 0.03 | 3.069 ± 0.01 | 5.39 ± 0.12 |
| Wild plants | >100 | 5.6 ± 0.11 | 0.08 ± 0.03 | >100 |
| Position | δH | δC | Type |
|---|---|---|---|
| 1 | 1.31, 2H, m | 39.92 | CH2 |
| 2 | 5.14, 1H, ddd (4.4, 10.4, 11.6) | 70.29 | CH |
| 3 | 5.05, 1H, ddd (3.2, 10.4) | 75.55 | CH |
| 4 | 1.76, 2H, m | 31.43 | CH2 |
| 5 | ------------ | 139.52 | C |
| 6 | 5.24, 1H, t (6.8) | 122.72 | CH |
| 7 | 1.74, 2H, m | 28.83 | CH2 |
| 8 | 1.82, 1H, m | 38.63 | CH |
| 9 | 1.68, 1H, m | 48.45 | CH |
| 10 | ------------ | 31.38 | C |
| 11 | 1.38, 2H, m | 23.86 | CH2 |
| 12 | 1.32, 2H,m | 34.53 | CH2 |
| 13 | ---------- | 46.84 | C |
| 14 | ---------- | 145.19 | C |
| 15 | 5.22, 1H,t (7.2) | 125.89 | CH |
| 16 | 1.72, 2H, m | 33.46 | CH2 |
| 17 | 1.72, 1H, m | 53.96 | CH |
| 18 | 1.16, 3H, s | 24.14 | CH3 |
| 19 | 1.21, 3H, s | 24.24 | CH3 |
| 20 | 1.44, 1H, c | 33.14 | CH |
| 21 | 0.81, 3H, d (6.4) | 17.72 | CH3 |
| 22 | 1.38, 2H, m | 37.65 | CH2 |
| 23 | 5.05, 1H, ddd (3.2, 10.4) | 75.55 | CH |
| 24 | 1.24, 1H, m | 44.41 | CH |
| 25 | 1.76, 1H, m | 23.79 | CH |
| 26 | 0.87, 3H, d, (7.6) | 17.43 | CH3 |
| 27 | 0.87, 3H, d (7.6) | 17.58 | CH3 |
| 28 | 3.80, 1H, dd, (3.6, 11.6) 3.55, 1H, dd, (1.2, 12) | 65.93 | CH2 |
| CH3-CO- | 1.93, 3H, s | 20.77 | CH3 |
| 1.93, 3H, s | 20.80 | CH3 | |
| 1.99, 3H, s | 21.02 | CH3 | |
| 2.02, 3H, s | 21.53 | CH3 | |
| CH3-CO- | 170.42 | C | |
| 170.54 | C | ||
| 170.54 | C | ||
| 170.72 | C |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreno-Anzúrez, N.E.; Marquina, S.; Alvarez, L.; Zamilpa, A.; Castillo-España, P.; Perea-Arango, I.; Torres, P.N.; Herrera-Ruiz, M.; Díaz García, E.R.; García, J.T.; et al. A Cytotoxic and Anti-inflammatory Campesterol Derivative from Genetically Transformed Hairy Roots of Lopezia racemosa Cav. (Onagraceae). Molecules 2017, 22, 118. https://doi.org/10.3390/molecules22010118
Moreno-Anzúrez NE, Marquina S, Alvarez L, Zamilpa A, Castillo-España P, Perea-Arango I, Torres PN, Herrera-Ruiz M, Díaz García ER, García JT, et al. A Cytotoxic and Anti-inflammatory Campesterol Derivative from Genetically Transformed Hairy Roots of Lopezia racemosa Cav. (Onagraceae). Molecules. 2017; 22(1):118. https://doi.org/10.3390/molecules22010118
Chicago/Turabian StyleMoreno-Anzúrez, Norma Elizabeth, Silvia Marquina, Laura Alvarez, Alejandro Zamilpa, Patricia Castillo-España, Irene Perea-Arango, Pilar Nicasio Torres, Maribel Herrera-Ruiz, Edgar Rolando Díaz García, Jaime Tortoriello García, and et al. 2017. "A Cytotoxic and Anti-inflammatory Campesterol Derivative from Genetically Transformed Hairy Roots of Lopezia racemosa Cav. (Onagraceae)" Molecules 22, no. 1: 118. https://doi.org/10.3390/molecules22010118