Quality Evaluation of Pseudostellariae Radix Based on Simultaneous Determination of Multiple Bioactive Components Combined with Grey Relational Analysis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Optimization of Extraction Conditions
2.2. Optimization of UFLC Conditions
2.3. Optimization of MS Conditions
2.4. UFLC Method Validation
2.5. Quantification of Cyclopeptides, Nucleosides, and Amino Acids
2.6. GRA of the Samples
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Plant Materials
3.3. Preparation of Standard Solution
3.4. Preparation of Sample Solutions
3.5. Chromatographic and Mass Spectrometric Conditions
3.6. Validation of the Method
3.7. Statistical Analysis
3.7.1. Normalization Treatment of Raw Data
3.7.2. Calculation of the Correlation Coefficient
3.7.3. Calculation of the Correlation Degree and Weight Value
3.7.4. The Grey Comprehensive Evaluation Value
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Liu, X.H.; Han, L.; Wang, L.J.; Fu, X.S. Study on Quality Standards of Pseudostellaria heterophylla. Chin. Pharm. 2010, 21, 1769–1771. [Google Scholar]
- Shen, X.C.; Tao, L.; Bo, S.; Gan, H.R.; Duan, J.A. A meliorated effects of Radix Pseudostellariaeon oxidative stress in rat chronic heart failure induced by acute cardiac infarction. West Chin. J. Pharm. Sci. 2008, 23, 413–416. [Google Scholar]
- Fu, X.S.; Liu, X.H.; Xu, H.; Zhou, Y.Z.; Chen, F. Research status and trends of pseudostellariae radix. Chin. J. New Grugs 2012, 21, 757–760. [Google Scholar]
- Sheng, R.; Xu, X.; Tang, Q.; Bian, D.; Li, Y.; Qian, C.; He, X.; Gao, X.; Pan, R.; Wang, C.; et al. Polysaccharide of radix pseudostellariae improves chronic fatigue syndrome induced by poly I:C in mice. Evid. Based Complement. Altern. Med. 2011, 2011, 840516. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.S.; Liu, X.H.; Xu, H.; Zhou, Y.Z.; Chen, F. Effect of pseudostellaria polysaccharides in diabetic mice by alloxan. Anhui Med. Pharm. J. 2010, 14, 521–522. [Google Scholar]
- Pang, W.; Lin, S.; Dai, Q.; Zhang, H.; Hu, J. Antitussive activity of Pseudostellaria heterophylla (Miq.) Pax extracts and improvement in lung function via adjustment of multi-cytokine levels. Molecules 2011, 16, 3360–3370. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Shen, Y.; Chen, J.; Lee, F.S.; Wang, X. HPLC fingerprinting and LC-TOF-MS analysis of the extract of Pseudostellaria heterophylla (Miq.) Pax root. J. Chromatogr. B 2008, 862, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Hou, Y.; Zou, L.S.; Liu, X.H.; Xu, L.; Yuan, J.D. QTRAP LC-MS/MS analytical study on nucleosides and nucleobases of Pseudostellariae Radix cultivated in different idioplasm resources. J. Chin. Med. Mater. 2015, 38, 711–714. [Google Scholar]
- An, K.; He, J.; Wan, Z.M.; Shao, H.W.; Yang, X.C.; Jiao, W. Determination and multivariate statistical analysis of amino acid in Pseudostellaria Heterophylla from different producing areas. Nat. Prod. Res. Dev. 2012, 24, 594–598. [Google Scholar]
- Morita, H.; Kayashita, T.; Kobata, H.; Gonda, A.; Takeya, K.; Itokawa, H. Pseudostellarins A-C, new tyrosinase inhibitory cyclic peptides from Pseudostellaria heterophylla. Tetrahedron 1994, 50, 9975–9982. [Google Scholar] [CrossRef]
- Han, B.X.; Zhu, Z.X.; Yao, Y.; Li, Y.Y.; Chen, J. A comparative analysis of amino acids and polysaccharides of Pseudostellaria heterophylla from different areas. Chin. Tradit. Pat. Med. 2010, 32, 513–514. [Google Scholar]
- Ma, Y.; Hou, Y.; Zou, L.S.; Xu, L.; Liu, X.H.; Lan, C.W.; Luo, Y.Y.; Liu, J.X. Quantitative Determination of Thirteen Nucleosides and Nucleobases in Pseudostellariae Radix in Different Harvest Periods by QTRAP LC-MS/MS. Chin. Pharm. J. 2015, 50, 75–79. [Google Scholar]
- Liu, W.X.; Hu, H.; Liu, X.C.; Duan, Q. Fingerprint of Radix Pseudostellariae by HPLC. Chin. Tradit. Herb. Drugs 2007, 38, 761–764. [Google Scholar]
- Liu, X.H.; Wang, M.; Cai, B.C.; Wang, Y.X.; Lin, X.Y. GC-MS Fingerprint of root tuber of Pseudostellaria heterophylla. Chin. Tradit. Herb. Drugs 2007, 38, 113–116. [Google Scholar]
- Liu, X.H.; Li, W.; Cai, B.C.; Huang, M.H. Study of capillary electrophoresis fingerprint spectra of Pseudostellariae Radix. J. Nanjing Univ. TCM 2007, 23, 238–240. [Google Scholar]
- Zhang, L.L.; Bai, Y.L.; Shu, S.L.; Qian, D.W.; Ou-yang, Z.; Liu, L.; Duan, J.A. Simultaneous quantitation of nucleosides, nucleobases, amino acids, and alkaloids in mulberry leaf by ultra high performance liquid chromatography with triple quadrupole tandem mass spectrometry. J. Sep. Sci. 2014, 37, 1265–1275. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.G.; Yang, H.; Cheng, X.L.; Liu, L.; Qin, Y.; Wang, Q.; Qi, L.W.; Li, P. Direct analysis of 18 flavonol glycosides, aglycones and terpene trilactones in Ginkgo biloba tablets by matrix solid phase dispersion coupled with ultra-high performance liquid chromatography tandem triple quadrupole mass spectrometry. J. Pharm. Biomed. Anal. 2014, 97, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Guo, L.; Duan, L.; Dong, X.; Zhou, P.; Liu, E.H.; Li, P. Simultaneous determination of 16 phenolic constituents in Spatholobi Caulis by high performance liquid chromatography/electrospray ionization triple quadrupole mass spectrometry. J. Pharm. Biomed. Anal. 2015, 102, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Chen, X.F.; Yang, W.J.; Li, Z.F.; Zhang, Y.; Song, J.H.; Yang, X.W. Evaluation of Bupleuri Radix resources in Qingchuan based on DTOPSIS and grey related degree. Chin. J. Chin. Mater. Med. 2014, 39, 433–437. [Google Scholar]
- Liu, W.; Zhang, Y.N.; Pei, J.; Kang, Y.L.; Luo, J.; Chen, C.P.; Ma, Y.T. Grey Relational Analysis of the Relationship between Variety and Quality of Radix Cyathulae. Chin. Pharm. J. 2014, 49, 1796–1801. [Google Scholar]
- Sample Availability: Samples of the compounds 1–30 are available from the authors.
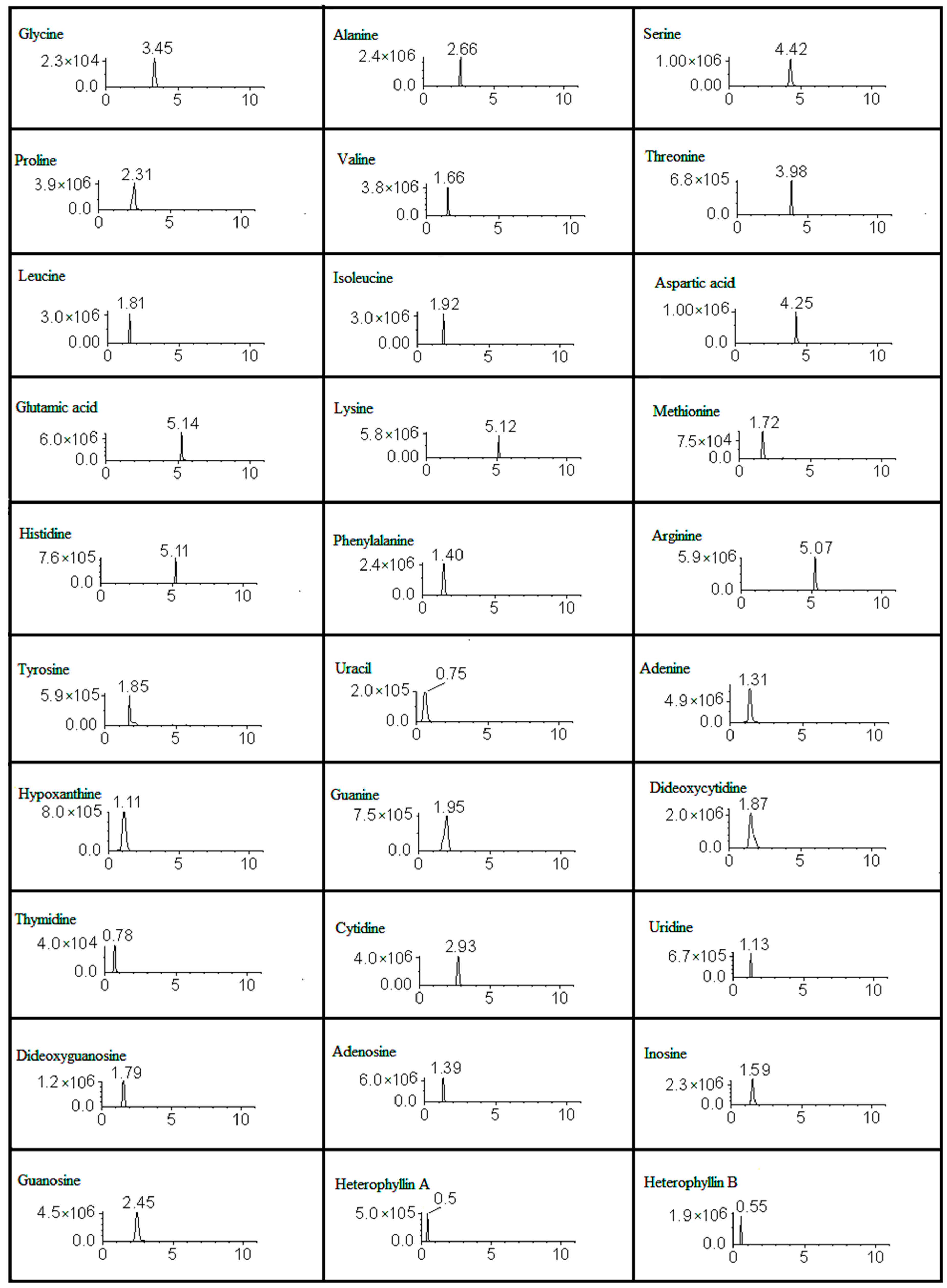
| Coumpounds | RT (min) | [M + H]+ (m/z) | Precursor Ion | Product Ion | FV | CE |
|---|---|---|---|---|---|---|
| Heterophyllin A | 0.50 | 728.43 | 728.4 | 70.05 | 211 | 119 |
| Heterophyllin B | 0.55 | 779.44 | 779.39 | 70.05 | 226 | 117 |
| Uracil | 0.75 | 112.09 | 113.04 | 70 | 111 | 21 |
| Thymidine | 0.78 | 242.23 | 243.1 | 127.07 | 61 | 13 |
| Hypoxanthine | 1.11 | 136.11 | 137.05 | 137.05 | 51 | 24 |
| Uridine | 1.13 | 244.2 | 244.9 | 113 | 103 | 13 |
| Adenine | 1.31 | 135.13 | 136.06 | 136 | 51 | 24 |
| Adenosine | 1.39 | 267.24 | 268.1 | 136.07 | 86 | 23 |
| Phenylalanine | 1.4 | 165.19 | 166.1 | 120.05 | 56 | 14 |
| Inosine | 1.59 | 268.23 | 269 | 137.07 | 46 | 15 |
| Valine | 1.66 | 117.15 | 118.09 | 72.06 | 54 | 10 |
| Methionine | 1.72 | 149.21 | 150.06 | 104.03 | 91 | 10 |
| Dideoxyguanosine | 1.79 | 267.2 | 268.1 | 152.1 | 61 | 15 |
| Leucine | 1.81 | 131.18 | 132.1 | 86.05 | 98 | 10 |
| Tyrosine | 1.85 | 182.1 | 182.16 | 136.08 | 46 | 17 |
| Dideoxycytidine | 1.87 | 227.3 | 228.2 | 112.05 | 76 | 13 |
| Isoleucine | 1.92 | 131.18 | 132.1 | 86.05 | 64 | 10 |
| Guanine | 1.95 | 151.12 | 152 | 135 | 51 | 25 |
| Proline | 2.31 | 115.13 | 116.07 | 70.02 | 68 | 10 |
| Guanosine | 2.45 | 283.24 | 284.3 | 152 | 62 | 15 |
| Alanine | 2.66 | 89.09 | 90.06 | 44.02 | 79 | 10 |
| Cytidine | 2.93 | 243.22 | 244.09 | 112 | 61 | 10 |
| Glycine | 3.45 | 75.07 | 76.04 | 30 | 73 | 6 |
| Threonine | 3.98 | 119.12 | 120.07 | 74 | 93 | 20 |
| Aspartic acid | 4.25 | 133.1 | 134.05 | 87.96 | 59 | 10 |
| Serine | 4.42 | 105.09 | 106.05 | 59.99 | 67 | 8 |
| Arginine | 5.07 | 174.2 | 175.12 | 70.02 | 88 | 18 |
| Histidine | 5.11 | 155 | 156.08 | 110.03 | 95 | 16 |
| Lysine | 5.12 | 146.19 | 147.11 | 83.91 | 66 | 14 |
| Glutamic acid | 5.14 | 147.13 | 147.08 | 83.92 | 83 | 14 |
| No. | Compounds | Regression Equation | r2 | Liner Range (ng/mL) | LOQ (ng/mL) | LOD (ng/mL) | Precision RSD (%) | Repeatability RSD (%) (n = 6) | Stability RSD (%) | Recovery (%) (n = 3) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Intar-Day (n = 6) | Inter-Day (n = 6) | Low | Medium | High | ||||||||||||
| Mean | RSD | Mean | RSD | Mean | RSD | |||||||||||
| 1 | Glycine | Y = 138X + 4880 | 0.9991 | 20.04–2004 | 10.02 | 4.01 | 2.09 | 3.47 | 2.66 | 3.70 | 98.75 | 1.4 | 99.7 | 1.81 | 100.79 | 1.19 |
| 2 | Alanine | Y = 1860X + 222000 | 0.9999 | 186.84–18684 | 93.40 | 31.13 | 2.49 | 2.59 | 1.45 | 1.02 | 100.78 | 0.92 | 102 | 1.11 | 101.32 | 1.2 |
| 3 | Serine | Y = 872X + 52100 | 0.9996 | 4.93–4930 | 4.93 | 1.64 | 2.10 | 3.48 | 3.31 | 2.67 | 97.89 | 1.72 | 102.76 | 2.55 | 101.3 | 1.52 |
| 4 | Proline | Y = 10100X − 459000 | 0.9995 | 30.06–6012 | 15.03 | 7.52 | 1.82 | 2.54 | 1.38 | 3.67 | 103.53 | 1.06 | 102.22 | 2.72 | 99.34 | 2.3 |
| 5 | Valine | Y = 10100X + 136000 | 0.9992 | 10.06–2012 | 0.40 | 0.13 | 2.96 | 3.07 | 2.12 | 3.20 | 99.05 | 2.24 | 101.64 | 2.35 | 102.28 | 1.84 |
| 6 | Threonine | Y = 1250X + 70400 | 0.9991 | 81.28–4064 | 40.64 | 20.32 | 2.01 | 2.61 | 3.14 | 1.33 | 101.68 | 1.79 | 101.08 | 1.76 | 102.58 | 2.39 |
| 7 | Leucine | Y = 27700X + 403000 | 0.9979 | 4.94–988 | 1.98 | 0.66 | 2.1 | 3.79 | 2.89 | 3.77 | 103.04 | 1.09 | 102.19 | 2.11 | 99.17 | 2.75 |
| 8 | Isoleucine | Y = 27900X + 403000 | 0.9979 | 4.91–982 | 1.96 | 0.65 | 2.88 | 4.07 | 1.14 | 1.11 | 104.36 | 2.31 | 100.87 | 4.72 | 100.44 | 4.18 |
| 9 | Aspartic acid | Y = 1120X + 36200 | 0.9998 | 29.7–5940 | 11.88 | 3.57 | 1.55 | 1.93 | 1.83 | 3.57 | 98.92 | 2.62 | 98.97 | 2.34 | 99.89 | 1.71 |
| 10 | Glutamic acid | Y = 4740X + 99900 | 1.0000 | 20.28–4056 | 4.06 | 0.81 | 1.02 | 2.57 | 2.46 | 3.12 | 101.76 | 2.39 | 100.2 | 1.73 | 99.86 | 1.58 |
| 11 | Lysine | Y = 3040X + 43100 | 0.9996 | 24.70–4940 | 4.94 | 2.47 | 0.61 | 2.98 | 2.53 | 1.34 | 98.74 | 4.23 | 101.59 | 2.56 | 99.41 | 1.79 |
| 12 | Methionine | Y = 4160X + 3600 | 0.9998 | 0.996–99.6 | 0.50 | 0.20 | 1.75 | 3.37 | 1.01 | 3.84 | 98.09 | 4.76 | 102.97 | 4.52 | 99.74 | 3.04 |
| 13 | Histidine | Y = 12400X + 91600 | 1.0000 | 19.76–1976 | 9.88 | 3.95 | 1.55 | 2.43 | 2.55 | 3.58 | 97.8 | 2.31 | 100.08 | 1.52 | 100.83 | 1.47 |
| 14 | Phenylalanine | Y = 18800X + 102000 | 0.9999 | 4.97–994 | 1.99 | 0.50 | 1.31 | 3.58 | 1.47 | 2.40 | 99.03 | 3.06 | 101.54 | 3.63 | 100.81 | 4.04 |
| 15 | Arginine | Y = 5690X + 430000 | 0.9998 | 19.88–7952 | 1.08 | 0.32 | 0.71 | 1.95 | 2.36 | 2.24 | 99.27 | 1.51 | 99.84 | 1.61 | 99.03 | 1.25 |
| 16 | Tyrosine | Y = 9780X + 179000 | 0.9998 | 20.04–2004 | 2.00 | 0.40 | 2.16 | 3.16 | 1.62 | 2.70 | 99.29 | 2.44 | 99.55 | 2.39 | 101.53 | 2.17 |
| 17 | Uracil | Y = 299X + 35600 | 0.9990 | 1.97–3936 | 0.79 | 0.32 | 3.3 | 4.39 | 2.89 | 2.11 | 104.85 | 2.67 | 99.53 | 3.43 | 97.94 | 3.18 |
| 18 | Adenine | Y = 5810X + 461000 | 0.9947 | 1.96–196 | 1.96 | 0.98 | 1.03 | 2.49 | 3.93 | 3.87 | 104.3 | 1.66 | 101.64 | 4.63 | 98.17 | 2.67 |
| 19 | Hypoxanthine | Y = 2470X + 59300 | 0.9993 | 19.84–1984 | 9.92 | 3.97 | 3.34 | 4.08 | 3.71 | 1.71 | 99.67 | 1.59 | 99.93 | 1.02 | 99.99 | 1.05 |
| 20 | Guanine | Y = 3780X + 98000 | 0.9992 | 14.76–1476 | 5.90 | 2.36 | 1.05 | 3.17 | 3.65 | 3.71 | 103.61 | 1.61 | 101.93 | 2.16 | 101.35 | 2.14 |
| 21 | Dideoxycytidine | Y = 19900X + 46500 | 0.9999 | 4.97–497 | 1.99 | 0.50 | 2.01 | 3.44 | 2.81 | 2.12 | 98.96 | 2.77 | 101.58 | 4.83 | 98.45 | 2.01 |
| 22 | Thymidine | Y = 2940X + 3200 | 0.9994 | 2.00–200 | 1.00 | 0.50 | 1.92 | 3.12 | 3.13 | 3.34 | 100.21 | 3.99 | 103.16 | 3.58 | 95.98 | 1.72 |
| 23 | Cytidine | Y = 17600X + 361000 | 0.9999 | 15.15–3030 | 1.52 | 0.61 | 2.98 | 3.61 | 3.44 | 2.43 | 100.04 | 3.26 | 99.82 | 2.82 | 99.49 | 2.04 |
| 24 | Uridine | Y = 1260X + 28000 | 0.9992 | 30.48–3048 | 15.24 | 3.05 | 2.18 | 3.32 | 3.34 | 3.99 | 97.69 | 2.53 | 100.95 | 2.14 | 99.81 | 2.95 |
| 25 | Dideoxyguanosine | Y = 13000X + 143000 | 0.9997 | 4.95–990 | 0.99 | 0.25 | 1.94 | 4.28 | 3.52 | 2.67 | 103.88 | 2.35 | 103.01 | 1.51 | 101.45 | 4.16 |
| 26 | Adenosine | Y = 25400X + 944000 | 0.9994 | 19.72–1972 | 0.99 | 0.39 | 2.54 | 3.39 | 2.53 | 2.66 | 100.91 | 1.36 | 99.72 | 1.08 | 99.76 | 1.17 |
| 27 | Inosine | Y = 16900X + 127000 | 0.9994 | 10.16–1016 | 5.08 | 2.03 | 2.14 | 2.96 | 2.09 | 3.69 | 101.97 | 2.02 | 102.79 | 4.81 | 95.68 | 2.02 |
| 28 | guanosine | Y = 15600X + 346000 | 0.9999 | 14.70–2940 | 0.74 | 0.29 | 3.25 | 4.18 | 2.86 | 1.20 | 100.14 | 2.05 | 99.25 | 1.49 | 100.56 | 1.66 |
| 29 | Heterophyllin A | Y = 4190X + 10700 | 0.9996 | 1.25–750 | 0.75 | 0.38 | 2.1 | 3.35 | 2.67 | 3.28 | 101.52 | 3.32 | 97.57 | 3.34 | 99.96 | 1.96 |
| 30 | Heterophyllin B | Y = 3480X − 40800 | 0.9996 | 5.4–5400 | 0.54 | 0.22 | 3.28 | 4.37 | 2.55 | 2.87 | 100.42 | 3.6 | 101.35 | 1.99 | 102.45 | 1.1 |
| Analyte | Different Harvesting Times | Different Habitats | Different Processing Methods | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 a | S2 | S3 | S4 | S5 | S6 | S7 | S8 | S9 | S10 | S11 | S12 | S13 | S14 | S15 | |
| 1 b | 104.09 ± 5.85 | 77.34 ± 4.21 | 39.23 ± 9.39 | 117.41 ± 7.27 | 64.98 ± 1.63 | 114.33 ± 3.56 | 76.467 ± 4.05 | 67.99 ± 19.17 | 42.87 ± 2.64 | 100.42 ± 6.25 | 36.00 ± 3.41 | 41.267 ± 2.19 | 33.73 ± 5.66 | 45.53 ± 1.33 | 56.20 ± 1.71 |
| 2 | 1028.67 ± 23.35 | 910.67 ± 12.86 | 824.00 ± 20.88 | 2468.06 ± 81.89 | 1296.67 ± 27.15 | 1872.67 ± 19.73 | 978.00 ± 44.14 | 1250.67 ± 27.30 | 1130.00 ± 6.93 | 1077.33 ± 22.30 | 568.00 ± 7.21 | 531.33 ± 36.02 | 237.33 ± 3.06 | 696.00 ± 5.29 | 634.00 ± 10.58 |
| 3 | 118.40 ± 3.42 | 107.80 ± 11.72 | 129.53 ± 15.60 | 281.33 ± 11.72 | 131.00 ± 14.91 | 211.13 ± 28.46 | 139.67 ± 7.18 | 143.53 ± 6.45 | 87.93 ± 12.07 | 106.93 ± 13.06 | 52.60 ± 1.31 | 40.53 ± 6.35 | 6.10 ± 4.09 | 27.33 ± 2.66 | 21.20 ± 1.59 |
| 4 | 157.67 ± 5.83 | 148.60 ± 3.70 | 109.00 ± 0.87 | 9.29 ± 0.042 | 300.67 ± 6.43 | 443.33 ± 4.16 | 136.53 ± 3.45 | 195.13 ± 5.16 | 114.33 ± 0.76 | 240.67 ± 3.06 | 106.87 ± 2.32 | 80.73 ± 7.92 | 21.67 ± 0.46 | 106.47 ± 1.33 | 218.00 ± 4.00 |
| 5 | 118.93 ± 1.51 | 139.53 ± 2.80 | 84.13 ± 1.81 | 156.55 ± 1.60 | 74.73 ± 0.64 | 114.067 ± 2.37 | 78.2 ± 1.56 | 85.07 ± 2.00 | 75.00 ± 1.64 | 101.00 ± 2.31 | 71.00 ± 1.60 | 75.80 ± 3.94 | 41.67 ± 0.95 | 70.27 ± 2.08 | 74.80 ± 3.29 |
| 6 | 107.33 ± 2.91 | 141.13 ± 4.23 | 124.27 ± 3.06 | 250.00 ± 2.71 | 134.60 ± 1.91 | 206.67 ± 2.31 | 98.60 ± 7.32 | 127.00 ± 2.27 | 132.33 ± 5.25 | 126.47 ± 3.14 | 122.67 ± 2.00 | 125.13 ± 2.27 | 111.87 ± 2.20 | 146.60 ± 2.09 | 169.20 ± 2.31 |
| 7 | 89.93 ± 0.64 | 80.87 ± 1.01 | 39.00 ± 0.20 | 108.4 ± 1.91 | 53.13 ± 0.95 | 83.73 ± 2.20 | 65.93 ± 1.17 | 65.73 ± 0.76 | 53.20 ± 1.04 | 56.33 ± 0.83 | 27.67 ± 1.14 | 31.27 ± 3.72 | 9.48 ± 0.087 | 24.067 ± 0.12 | 31.80 ± 1.91 |
| 8 | 89.40 ± 0.53 | 80.40 ± 0.92 | 38.80 ± 0.20 | 107.67 ± 1.81 | 52.73 ± 0.95 | 83.27 ± 2.14 | 65.53 ± 1.17 | 65.33 ± 0.76 | 52.87 ± 0.99 | 56.00 ± 0.92 | 27.47 ± 1.14 | 31.07 ± 3.72 | 9.39 ± 0.075 | 23.87 ± 0.12 | 31.53 ± 1.90 |
| 9 | 223.33 ± 13.01 | 240.67 ± 14.05 | 210.53 ± 11.25 | 513.33 ± 19.63 | 276.00 ± 4.00 | 395.33 ± 9.02 | 173.73 ± 5.82 | 250.67 ± 16.29 | 160.6 ± 0.60 | 202.00 ± 2.00 | 90.80 ± 6.77 | 80.33 ± 0.42 | 39.67 ± 2.80 | 79.40 ± 2.42 | 58.13 ± 1.81 |
| 10 | 115.07 ± 2.02 | 116.4 ± 6.51 | 94.27 ± 4.02 | 211.2 ± 12.24 | 98.4 ± 2.46 | 154.07 ± 2.84 | 62.67 ± 0.81 | 104.80 ± 4.73 | 82.53 ± 4.31 | 108.20 ± 1.73 | 54.80 ± 1.78 | 50.00 ± 2.42 | 29.93 ± 1.50 | 53.27 ± 0.76 | 50.33 ± 0.42 |
| 11 | 123.20 ± 4.04 | 126.80 ± 4.19 | 98.80 ± 6.45 | 224.00 ± 0.00 | 104.40 ± 3.27 | 169.20 ± 1.91 | 67.60 ± 2.31 | 112.60 ± 2.42 | 86.07 ± 2.48 | 121.80 ± 0.69 | 59.33 ± 3.11 | 54.40 ± 4.06 | 33.33 ± 1.67 | 59.07 ± 1.30 | 54.80 ± 2.60 |
| 12 | 3.76 ± 0.18 | 0.61 ± 0.20 | 1.00 ± 0.080 | 6.01 ± 1.72 | 4.79 ± 0.012 | 8.69 ± 0.69 | 2.33 ± 0.11 | 5.41 ± 0.59 | 0.37 ± 0.071 | 1.32 ± 0.31 | 0.27 ± 0.11 | 0.49 ± 0.00 | 0.29 ± 0.096 | 0.30 ± 0.12 | 0.23 ± 0.030 |
| 13 | 75.13 ± 1.86 | 108.00 ± 2.27 | 85.93 ± 2.14 | 216.67 ± 4.16 | 124.13 ± 4.22 | 177.80 ± 2.51 | 83.80 ± 2.88 | 295.33 ± 7.02 | 189.33 ± 4.00 | 45.73 ± 0.99 | 107.00 ± 1.25 | 103.00 ± 1.60 | 100.53 ± 2.54 | 130.67 ± 2.87 | 110.27 ± 1.67 |
| 14 | 33.80 ± 13.56 | 46.47 ± 0.61 | 27.07 ± 0.50 | 33.53 ± 4.11 | 41.73 ± 0.76 | 55.67 ± 18.25 | 39.53 ± 19.15 | 35.00 ± 2.62 | 43.87 ± 0.23 | 27.53 ± 11.68 | 32.53 ± 0.31 | 38.27 ± 1.29 | 22.93 ± 0.70 | 33.33 ± 0.50 | 57.00 ± 0.72 |
| 15 | 1352.67 ± 22.03 | 1230.67 ± 160.81 | 1288.00 ± 119.31 | 1205.33 ± 1.15 | 1422.67 ± 90.56 | 1140.67 ± 15.28 | 1172.00 ± 36.39 | 1196.00 ± 23.07 | 1179.33 ± 134.23 | 1304.00 ± 55.46 | 1257.33 ± 79.10 | 1285.33 ± 41.05 | 1366.00 ± 58.41 | 1357.33 ± 71.06 | 1319.33 ± 18.04 |
| 16 | 82.37 ± 35.21 | 55.13 ± 25.84 | 27.49 ± 0.66 | 250.17 ± 5.93 | 67.53 ± 3.19 | 119.56 ± 59.61 | 77.45 ± 5.86 | 101.79 ± 1.82 | 39.07 ± 3.89 | 61.25 ± 0.78 | 45.60 ± 25.46 | 82.40 ± 4.53 | 60.87 ± 0.42 | 68.60 ± 0.58 | 73.53 ± 0.76 |
| Total | 3823.75 ± 37.07 c | 3611.08 ± 185.73 c | 3221.06 ± 84.97 d | 6158.95 ± 139.62 a | 4248.17 ± 51.22 b | 5350.18 ± 33.29 a | 3318.05 ± 11.07 d | 4102.06 ± 75.32 b | 3469.71 ± 125.79 d | 3736.99 ± 40.10 c | 2659.93 ± 93.99 b | 2651.36 ± 77.58 b | 2124.80 ± 43.94 c | 2922.10 ± 73.45 a | 2960.37 ± 24.80 a |
| 17 | 66.07 ± 2.95 | 17.5 ± 4.50 | 9.16 ± 5.42 | 11.97 ± 2.43 | 2.18 ± 2.11 | 12.98 ± 9.39 | 8.46 ± 2.51 | 11.44 ± 2.97 | 20.12 ± 27.33 | 42.4 ± 8.46 | 3.42 ± 0.34 | 3.74 ± 1.06 | 2.78 ± 1.30 | 8.70 ± 1.50 | 2.58 ± 1.30 |
| 18 | 10.74 ± 1.22 | 6.76 ± 2.06 | 9.13 ± 0.60 | 15.35 ± 0.32 | 9.25 ± 0.85 | 15.09 ± 0.53 | 6.99 ± 0.84 | 13.60 ± 0.97 | 3.39 ± 0.55 | 10.31 ± 0.66 | 10.90 ± 0.53 | 4.90 ± 2.53 | 3.92 ± 1.98 | 6.63 ± 1.12 | 7.03 ± 2.51 |
| 19 | 213.33 ± 4.62 | 20.95 ± 7.56 | 10.64 ± 0.60 | 26.00 ± 0.69 | 17.36 ± 1.56 | 31.93 ± 15.37 | 13.79 ± 2.61 | 38.29 ± 16.70 | 15.59 ± 6.51 | 24.87 ± 2.83 | 22.87 ± 1.03 | 14.75 ± 6.28 | 16.27 ± 6.76 | 13.09 ± 8.25 | 32.93 ± 0.58 |
| 20 | 49.60 ± 1.59 | 13.29 ± 0.076 | 11.21 ± 0.49 | 50.40 ± 1.56 | 16.61 ± 1.02 | 33.40 ± 0.87 | 17.25 ± 0.56 | 30.47 ± 0.46 | 3.09 ± 0.00 | 22.13 ± 0.12 | 10.95 ± 0.74 | 11.59 ± 0.89 | 15.69 ± 1.15 | 16.61 ± 0.67 | 17.23 ± 0.63 |
| 21 | 3.02 ± 0.072 | 7.39 ± 0.095 | 6.97 ± 0.46 | 21.92 ± 0.23 | 7.50 ± 0.17 | 15.48 ± 0.060 | 7.61 ± 0.50 | 9.31 ± 0.067 | 1.56 ± 0.071 | 7.85 ± 0.77 | 7.47 ± 0.17 | 8.62 ± 0.63 | 7.99 ± 0.19 | 7.18 ± 0.21 | 6.77 ± 0.11 |
| 22 | 37.40 ± 10.86 | 15.54 ± 0.52 | 14.23 ± 13.89 | 41.80 ± 13.34 | 15.08 ± 2.48 | 30.33 ± 5.20 | 20.15 ± 1.37 | 23.47 ± 8.68 | 6.14 ± 7.43 | 20.80 ± 2.31 | 16.25 ± 1.40 | 19.67 ± 0.85 | 19.63 ± 1.61 | 16.64 ± 0.98 | 14.84 ± 0.26 |
| 23 | 64.27 ± 2.00 | 68.27 ± 1.75 | 57.18 ± 1.70 | 193.90 ± 1.10 | 68.20 ± 74.01 | 134.39 ± 1.57 | 130.13 ± 4.32 | 200.73 ± 4.61 | 60.60 ± 0.72 | 112.40 ± 1.51 | 148.27 ± 3.01 | 128.07 ± 3.92 | 123.60 ± 1.04 | 139.20 ± 2.84 | 156.33 ± 2.83 |
| 24 | 262.00 ± 6.93 | 160.73 ± 8.27 | 98.47 ± 0.99 | 234.00 ± 3.46 | 104.07 ± 3.35 | 174.87 ± 4.24 | 162.93 ± 10.00 | 218.67 ± 4.62 | 155.93 ± 6.33 | 155.07 ± 5.52 | 176.87 ± 2.42 | 167.20 ± 5.33 | 163.00 ± 7.41 | 176.20 ± 2.11 | 193.07 ± 4.00 |
| 25 | 31.20 ± 1.06 | 15.71 ± 0.47 | 14.13 ± 0.58 | 44.33 ± 1.10 | 12.53 ± 0.12 | 26.67 ± 0.61 | 18.11 ± 0.75 | 23.13 ± 0.31 | 3.34 ± 0.035 | 15.94 ± 0.20 | 12.85 ± 0.14 | 16.10 ± 0.53 | 16.01 ± 0.33 | 14.67 ± 0.10 | 13.57 ± 0.24 |
| 26 | 141.87 ± 2.19 | 162.47 ± 2.20 | 108.00 ± 5.64 | 259.33 ± 7.57 | 108.13 ± 2.72 | 163.60 ± 1.44 | 182.47 ± 3.97 | 241.33 ± 3.06 | 146.40 ± 5.05 | 157.53 ± 2.34 | 168.33 ± 4.10 | 171.53 ± 7.17 | 159.87 ± 2.60 | 166.20 ± 7.22 | 176.20 ± 3.47 |
| 27 | 34.27 ± 0.61 | 10.71 ± 0.25 | 6.55 ± 0.012 | 19.01 ± 0.32 | 6.38 ± 0.19 | 10.67 ± 0.076 | 12.42 ± 0.49 | 17.77 ± 0.095 | 9.55 ± 0.042 | 10.29 ± 0.061 | 11.31 ± 0.44 | 11.50 ± 0.41 | 10.65 ± 0.33 | 10.97 ± 0.14 | 11.71 ± 0.34 |
| 28 | 228.00 ± 9.17 | 199.47 ± 4.24 | 136.87 ± 4.71 | 366.67 ± 13.32 | 139.73 ± 1.79 | 252.67 ± 1.15 | 253.33 ± 12.22 | 349.33 ± 11.02 | 130.80 ± 0.60 | 193.87 ± 2.12 | 282.00 ± 3.46 | 247.33 ± 7.02 | 236.00 ± 2.00 | 251.33 ± 5.03 | 282.00 ± 2.00 |
| Total | 1141.76 ± 11.82 b | 698.87 ± 22.87 c | 482.54 ± 14.14 d | 1284.68 ± 22.44 a | 507.02 ± 2.10 d | 902.07 ± 13.08 b | 833.63 ± 30.40 c | 1177.45 ± 19.17 a | 556.51 ± 29.15 d | 773.46 ± 15.4 c | 871.48 ± 9.64 b | 805.00 ± 25.59 c,d | 775.40 ± 15.88 d | 827.41 ± 5.77 c | 914.27 ± 4.98 a |
| 29 | 19.20 ± 0.71 | 20.10 ± 0.97 | 13.43 ± 0.50 | 26.47 ± 0.58 | 12.21 ± 0.29 | 19.07 ± 0.70 | 104.07 ± 13.40 | 59.20 ± 3.30 | 26.60 ± 0.80 | 56.00 ± 1.78 | 20.87 ± 0.81 | 18.48 ± 1.08 | 18.38 ± 0.92 | 22.40 ± 1.22 | 24.67 ± 1.01 |
| 30 | 274.00 ± 5.29 | 296.00 ± 15.62 | 234.00 ± 6.93 | 388.00 ± 10.58 | 220.00 ± 0.00 | 309.33 ± 10.26 | 11.55 ± 0.84 | 4.98 ± 0.11 | 217.33 ± 5.77 | 102.67 ± 7.56 | 198.73 ± 8.25 | 174.07 ± 10.02 | 178.00 ± 1.74 | 220.00 ± 2.00 | 247.33 ± 3.06 |
| Total | 293.20 ± 5.92 b | 316.10 ± 16.58 b | 247.43 ± 6.53 c | 414.47 ± 10.41 a | 232.21 ± 0.29 c | 328.40 ± 9.81 a | 115.61 ± 14.15 d | 64.18 ± 3.29 e | 243.93 ± 5.83 b | 158.67 ± 7.14 c | 219.60 ± 8.99 c | 192.55 ± 10.88 d | 196.38 ± 0.90 d | 242.40 ± 1.04 b | 272.00 ± 4.06 a |
| Sample | Grey Comprehensive Evaluation Value (ri’) | Quality-Ranking |
|---|---|---|
| S1 | 0.0260 | 2 |
| S2 | 0.0231 | 3 |
| S3 | 0.0211 | 5 |
| S4 | 0.0311 | 1 |
| S5 | 0.0227 | 4 |
| S6 | 0.0300 | 1 |
| S7 | 0.0222 | 4 |
| S8 | 0.0262 | 2 |
| S9 | 0.0201 | 5 |
| S10 | 0.0228 | 3 |
| S11 | 0.0276 | 4 |
| S12 | 0.0278 | 3 |
| S13 | 0.0237 | 5 |
| S14 | 0.0281 | 2 |
| S15 | 0.0298 | 1 |
| Sample No. | Habitats | Harvesting Time | Processing Method |
|---|---|---|---|
| S1 | Jurong, Jiangsu | 15 June 2013 | sun drying |
| S2 | Jurong, Jiangsu | 9 July 2013 | sun drying |
| S3 | Jurong, Jiangsu | 15 July 2013 | sun drying |
| S4 | Jurong, Jiangsu | 6 August 2013 | sun drying |
| S5 | Jurong, Jiangsu | 12 September 2013 | sun drying |
| S6 | Jurong, Jiangsu | 10 August 2013 | sun drying |
| S7 | Zherong 1, Fujian | 10 August 2013 | sun drying |
| S8 | Zherong 2, Fujian | 10 August 2013 | sun drying |
| S9 | Shibing, Guizhou | 10 August 2013 | sun drying |
| S10 | Xuancheng, Anhui | 10 August 2013 | sun drying |
| S11 | Jurong, Jiangsu | 10 August 2013 | sun drying |
| S12 | Jurong, Jiangsu | 10 August 2013 | sun drying-twisting |
| S13 | Jurong, Jiangsu | 10 August 2013 | oven drying 40 °C |
| S14 | Jurong, Jiangsu | 10 August 2013 | oven drying 50 °C |
| S15 | Jurong, Jiangsu | 10 August 2013 | oven drying 60 °C |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hua, Y.; Wang, S.; Chai, C.; Liu, Z.; Liu, X.; Zou, L.; Wu, Q.; Zhao, H.; Ying, Y. Quality Evaluation of Pseudostellariae Radix Based on Simultaneous Determination of Multiple Bioactive Components Combined with Grey Relational Analysis. Molecules 2017, 22, 13. https://doi.org/10.3390/molecules22010013
Hua Y, Wang S, Chai C, Liu Z, Liu X, Zou L, Wu Q, Zhao H, Ying Y. Quality Evaluation of Pseudostellariae Radix Based on Simultaneous Determination of Multiple Bioactive Components Combined with Grey Relational Analysis. Molecules. 2017; 22(1):13. https://doi.org/10.3390/molecules22010013
Chicago/Turabian StyleHua, Yujiao, Shengnan Wang, Chuan Chai, Zixiu Liu, Xunhong Liu, Lisi Zou, Qinan Wu, Hui Zhao, and Yan Ying. 2017. "Quality Evaluation of Pseudostellariae Radix Based on Simultaneous Determination of Multiple Bioactive Components Combined with Grey Relational Analysis" Molecules 22, no. 1: 13. https://doi.org/10.3390/molecules22010013






