Isolation of CHS Gene from Brunfelsia acuminata Flowers and Its Regulation in Anthocyanin Biosysthesis
Abstract
:1. Introduction
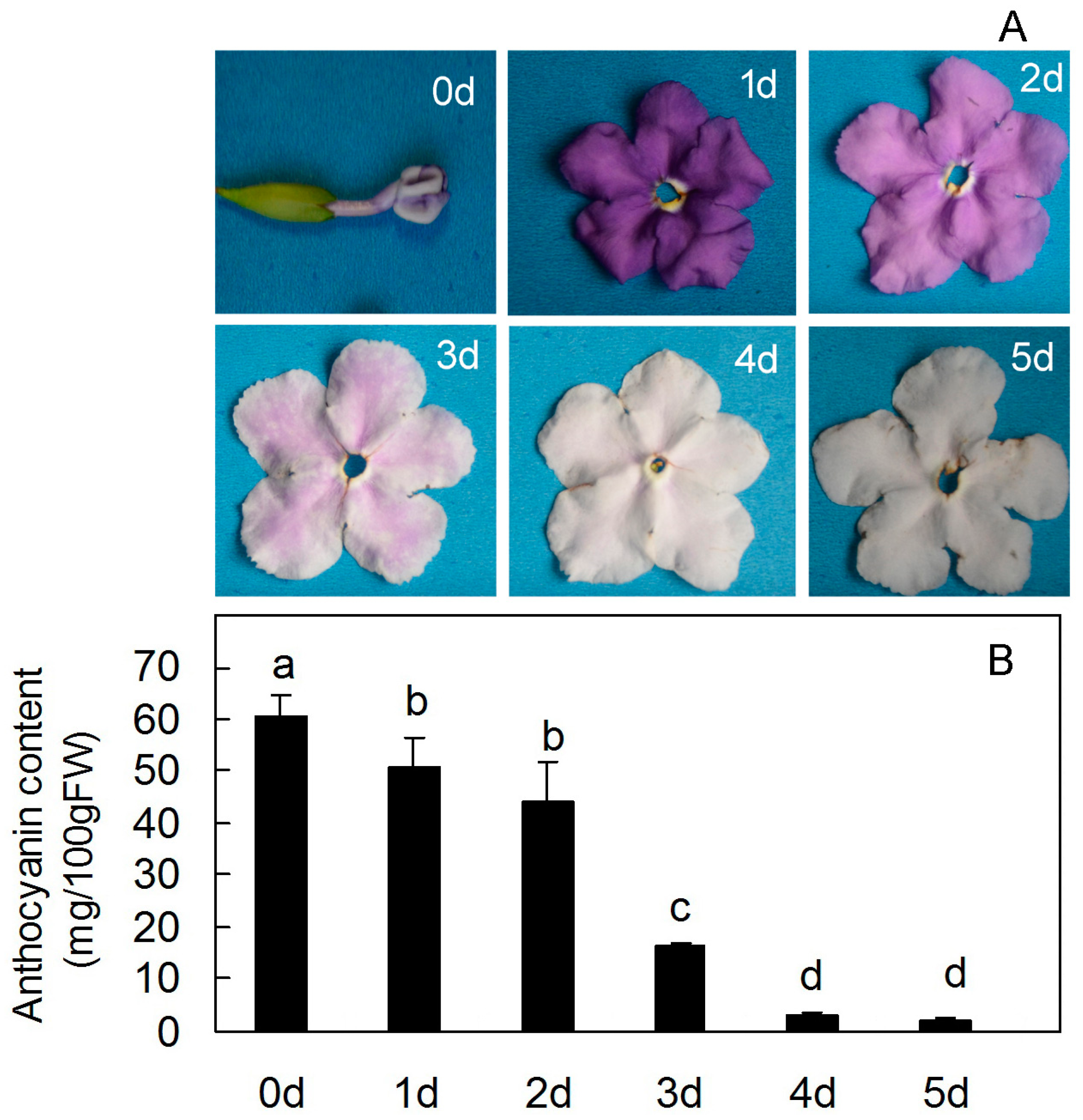
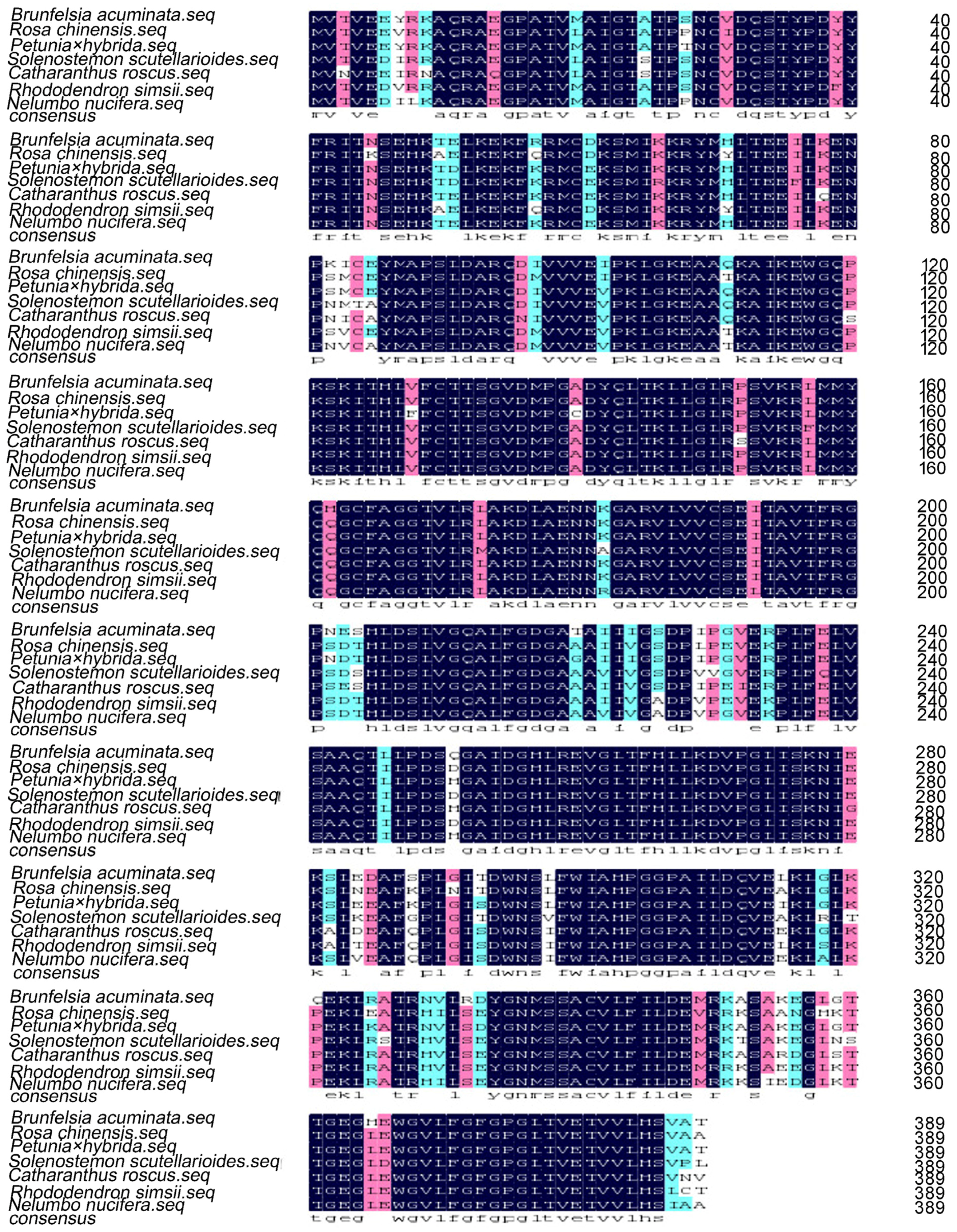
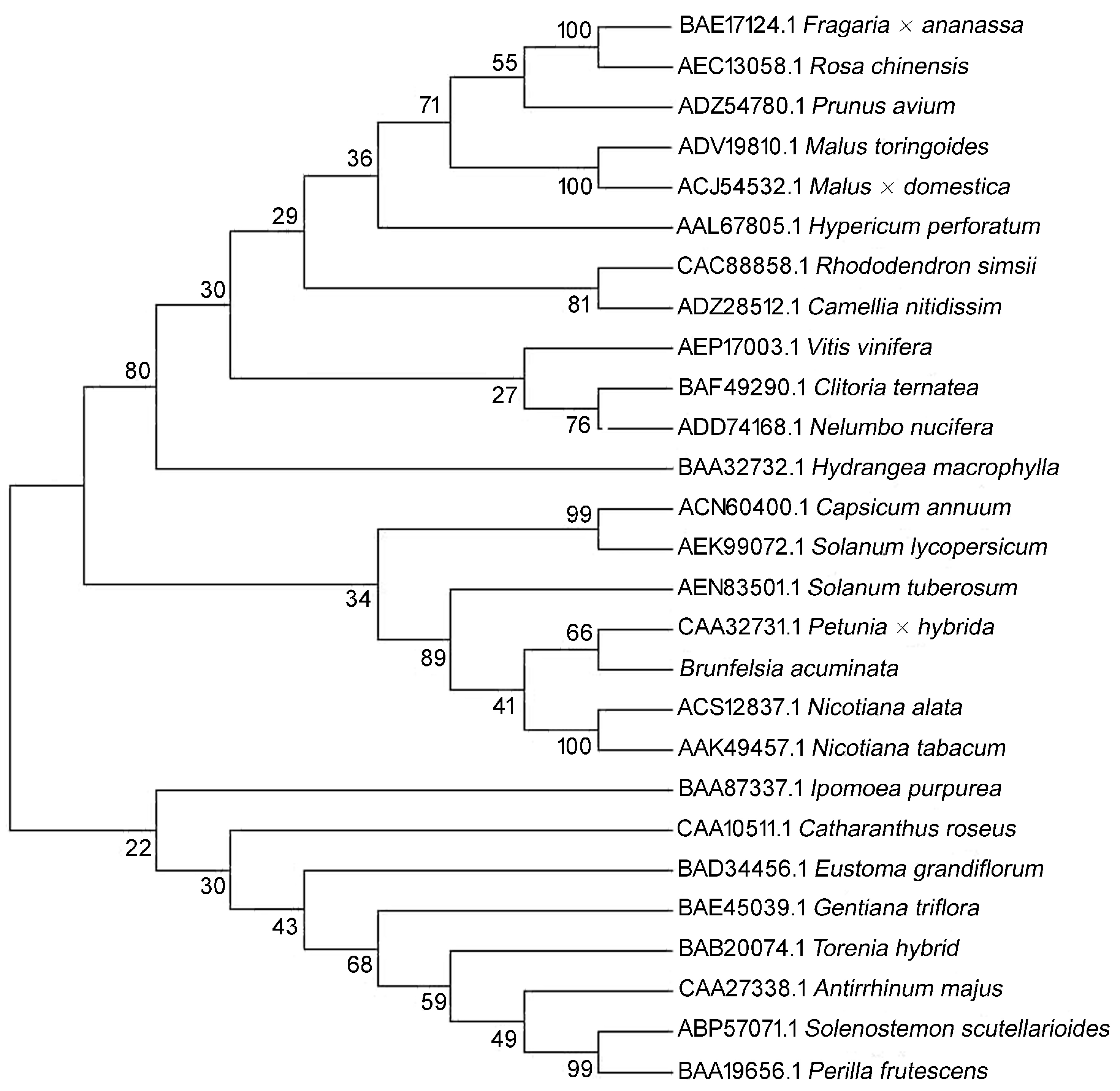
2. Results
3. Discussion
4. Materials and Methods
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Claudot, A.C.; Ernst D, H.S., Jr.; Drouet, A. Cloning and characterization of two members of the chalcone synthase gene family from walnut. Plant Physiol. Biochem. 1999, 37, 721–730. [Google Scholar] [CrossRef]
- Schmelzer, E.; Jahnen, W.; Hahlbrock, K. In situ localization of light-induced chalcone synthase mRNA, chalcone synthase, and flavonoid end products in epidermal cells of parsley leaves. Proc. Nat. Acad. Sci. USA 1988, 85, 2989–2993. [Google Scholar] [CrossRef] [PubMed]
- Schulze-Lefert, P.; Becker-Andre, M.; Schulz, W. Functional architecture of the light-responsive chalcone synthase promoter from parsley. Plant Cell 1989, 1, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Holton, T.A.; Cornish, E.C. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 1995, 7, 1071–1083. [Google Scholar] [CrossRef] [PubMed]
- Franken, P.; Niesbach-Klosgen, U.; Weydemann, U.; Marechal-Drouard, L.; Heinz, S.; Wienand, U. The duplicated chalcone synthase genes C2 and Whp. (white pollen) of Zea mays are independently regulated; evidence for translational control of Whp. expression by the anthocyanin intensifying gene. EMBO J. 1991, 10, 2605–2612. [Google Scholar] [PubMed]
- Saslowsky, D.E.; Dana, C.D.; Winkel-Shirley, B. An allelic series for the chalcone synthase locus in Arabidopsis. Gene 2000, 255, 127–138. [Google Scholar] [CrossRef]
- Sommer, H.; Saedler, H. Structure of the chalcone synthase gene of Antirrhinum majus. Mol. Gen. Genet. 1986, 202, 429–434. [Google Scholar] [CrossRef]
- Helariutta, Y.; Kotilainen, M.; Elomaa, P. Duplication and functional divergence in the chalcone synthase gene family of Asteraceae: Evolution with substrate change and catalytic simplification. Proc. Nat. Acad. Sci. USA 1996, 3, 9033–9038. [Google Scholar] [CrossRef]
- Sparvoli, F.; Martin, C.; Scienza, A.; Gavazzi, G.; Tonelli, C. Cloning and molecular analysis of structural genes involved in flavonoid and stilbene biosynthesis in grape (Vitis. vinifera L.). Plant Mol. Biol. 1994, 24, 743–755. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Ma, X.H.; Shi, Y.M.; Tang, D.Q. Isolation and expression analysis of six putative structural genes involved in anthocyanin biosynthesis in Tulipa fosteriana. Sci. Hort. 2013, 153, 93–102. [Google Scholar] [CrossRef]
- Koes, R.E.; Spelt, C.E.; Mol, J.N.M. The chalcone synthase multigene family of Petunia hybrid (V30): Differential, light-regulated expression during flower development and UV light induction. Plant Mol. Biol. 1989, 12, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Durbin, M.L.; McCaig, B.; Clegg, M.T. Molecular evolution of the chalcone synthase multigene family in the morning glory genome. Plant Mol. Biol. 2000, 42, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Elomaa, P.; Honkanen, J.; Puska, R.; Seppanen, P.; Helariutta, Y.; Mehto, M.; Kotilainen, M.; Nevalainen, L.; Teeri, T.H. Agrobacterium-mediated transfer of antisense chalcone synthase cDNA to Gerbera hybrida inhibits flower pigmentation. Biotechnology 1993, 11, 508–511. [Google Scholar] [CrossRef]
- Li, J.; Lü, R.H.; Zhao, A.C.; Wang, X.L.; Liu, C.Y.; Zhang, Q.Y.; Wang, X.H.; Umuhoza, D.; Jin, X.Y.; Lu, C.; et al. Isolation and expression analysis of anthocyanin biosynthetic genes in Morus. alba L. Biol. Plant 2014, 58, 618–626. [Google Scholar] [CrossRef]
- Grotewold, E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006, 57, 761–780. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.T.; Qiu, D.L. Cloning and sequence analysis of chalcone isomerase Gene (CHI) from Brunfelsia acuminata flowers. J. Trop. Subtrop. Bot. 2012, 20, 475–481. [Google Scholar]
- Zipor, G.; Duarte, P.; Carqueijeiro, I.; Shahar, L.; Ovadia, R.; Teper-Bamnolker, P.; Eshel, D.; Levin, Y.; Doron-Faigenboim, A.; Sottomayor, M.; et al. In planta anthocyanin degradation by a vacuolar class III peroxidase in Brunfelsia calycina flowers. New Phytol. 2015, 205, 653–665. [Google Scholar] [CrossRef] [PubMed]
- Vaknin, H.; Bar-Akiva, A.; Ovadia, R.; Nissim-Levi, A.; Forer, I.; Weiss, D.; Oren-Shamir, M. Active anthocyanin degradation in Brunfelsia calycina (yesterday–today–tomorrow) flowers. Planta 2005, 222, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.J.; Li, H.; Bai, B.B.; Yu, H.Q.; You, J. Effect of Light on the Anthocyanin Biosythesis and Expression of CHS and DFR in Rosa chinensis ‘Spectra’. Mol. Plant Breed. 2013, 11, 126–131. [Google Scholar]
- Ohto, M.; Onai, K.; Furukawa, Y.; Aoki, E.; Araki, T.; Nakamura, K. Effects of sugar on vegetative development and floral transition in Arabidopsis. Plant Physiol. 2001, 127, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhao, M.Z.; Hong-Mei, Y.U. Semilethal Temperatures for Flowers of Two Strawberry Varieties under Low Temperature Stress. Jiangxi Nongye Daxue Xuebao 2012, 34, 255–258. [Google Scholar]
- Tian, J.; Shen, H.X.; Zhang, J.; Song, T.T.; Yao, Y.C. Characteristics of chalcone synthase promoters from different leaf-color malus crabapple cultivars. Sci. Hort. 2011, 129, 449–453. [Google Scholar] [CrossRef]
- Azuma, A.; Ban, Y.; Sato, A.; Kono, A.; Shiraishi, M.; Yakushiji, H.; Kobayashi, S. MYB diplotypes at the color locus affect the ratios of tri/di-hydroxylated and methylated non-methylated anthocyanins in grape berry skin. Tree Genet. Genoms 2015, 11, 1–13. [Google Scholar] [CrossRef]
- González, C.V.; Fanzone, M.L.; Cortés, L.E.; Bottini, R.; Lijavetzky, D.C.; Ballaré, C.L.; Boccalandro, H.E. Fruit-localized photoreceptors increase phenolic compounds in berry skins of field-grown Vitis. vinifera L. cv. Malbec. Phytochemistry 2015, 110, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.N.; Yao, G.F.; Zheng, D.M.; Zhang, S.L.; Wang, C.; Zhang, M.Y.; Wu, J. Expression differences of anthocyanin biosynthesis genes reveal regulation patterns for red pear coloration. Plant Cell Rep. 2015, 34, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Kayesh, E.; Shangguan, L.F.; Korir, N.K.; Sun, X.; Bilkish, N.; Zhang, Y.P.; Han, J.; Song, C.N.; Cheng, Z.M.; Fang, J.G. Fruit skin color and the role of anthocyanin. Acta Physiol. Plant 2013, 35, 2879–2890. [Google Scholar] [CrossRef]
- Ju, Z.G.; Liu, C.L.; Yuan, Y.B. Activities of chalcone synthase and UDPGal: Xavonoid-3-O-glycosyltransferase in relation to anthocyanin synthesis in apple. Sci. Hort. 1995, 63, 175–185. [Google Scholar] [CrossRef]
- Mori, T.; Sakurai, M.; Sakuta, M. Effects of conditioned medium on activities of PAL, CHS, DAHP synthase (DS-Co and DS-Mn) and anthocyanin production in suspension cultures of Fragaria ananassa. Plant Sci. 2001, 160, 355–360. [Google Scholar] [CrossRef]
- Muir, S.R.; Collins, G.J.; Robinson, S.; Hughes, S.; Bovy, A.; Vos, C.H.R.D.; Tunen, A.J.; Martine, E.; Verhoeyen, M.E. Over expression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nat. Bioteehnol. 2001, 19, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Tunen, A.J.V.; Koes, R.E.; Mol, J.N.M. Coning of the two chalcone flavanone isomerase genes form Petunia hybrid co-ordinate, light-regulated and differential expression of flavonoid genes. EMBO J. 1988, 7, 1257–1263. [Google Scholar] [PubMed]
- Weiss, D.; Halevy, A.H. Stamens and gibberellin in the regulation of corolla pigmentation and growth in Petunia hybrida. Planta 1989, 179, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.C.; Chen, X.Y.; Wang, J.; Du, C.; Zhang, K.; Cai, Y.M.; Gao, X.; Tang, D.B.; Zhao, Y. Anthocyanin extraction from purple sweet potato by citric acid-disodium hydrogen phosphate buffer. J. South. Agric. 2013, 44, 653–656. [Google Scholar]
- Bar-Akiva, A.; Ovadia, R.; Rogachev, I.; Bar-Or, C.; Bar, E.; Freiman, Z.; Nissim-Levi, A.; Gollop, N.; Lewinsohn, E.; Aharoni, A. Metabolic networking in Brunfelsia calycina petals after flower opening. J. Exp. Bot. 2010, 61, 1393–1403. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds are available from the authors.
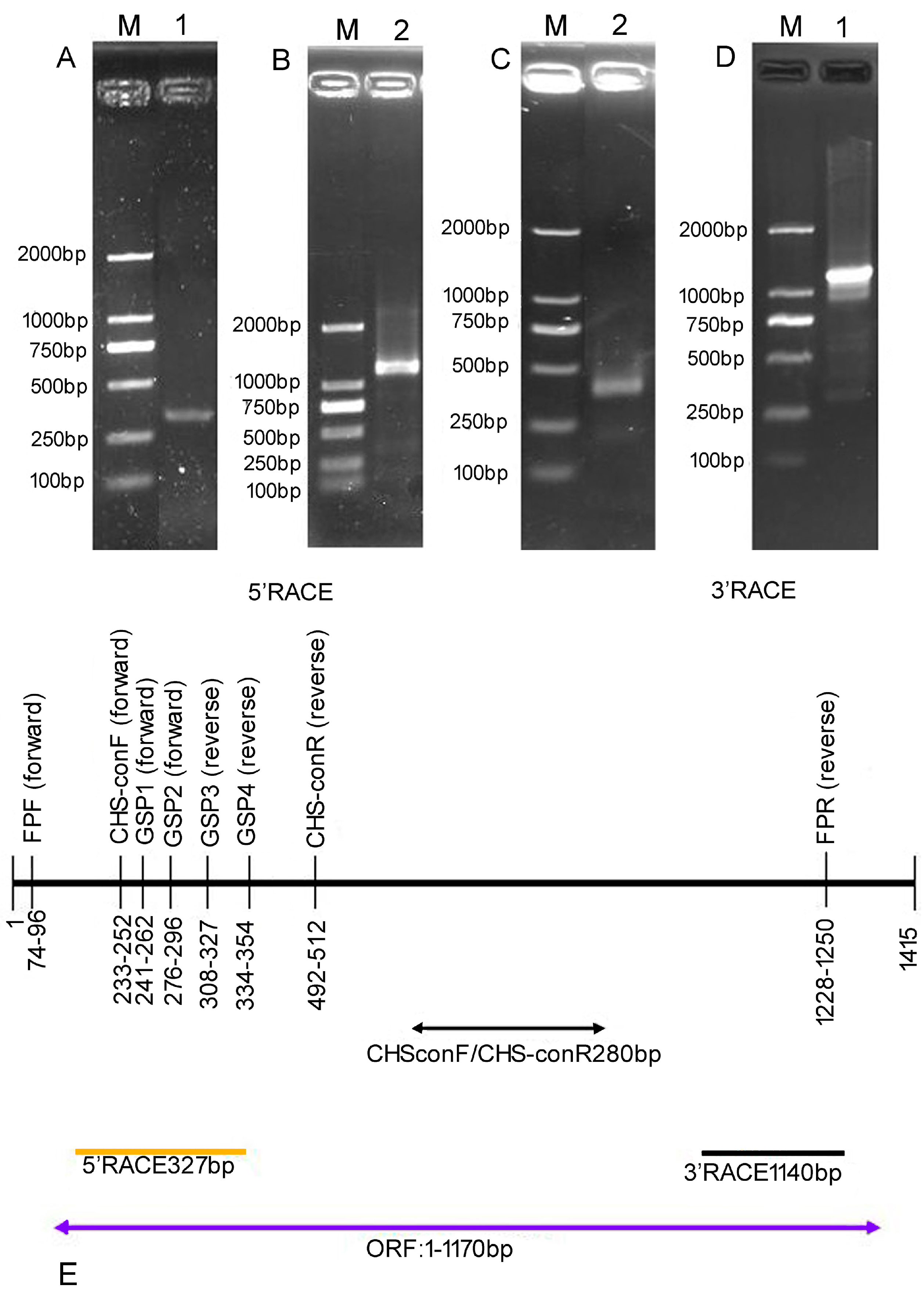


| Primer | Primer Sequence | Use |
|---|---|---|
| CHS-conF(forward) | 5′-GAGAARTTCMRGCGCATGTGYG-3′ | cDNA |
| CHS-conR(reverse) | 5′-GCTTGGTGAGYTGRTAGTCRG-3′ | |
| GSP1(forward) | 5′-CGGCGCATGTGTGATAAATCA-3′ | 3′RACE-PCR |
| GSP2(forward) | 5′-GGTACATGCACTTAACAGAGG-3′ | |
| UPM | 5′-CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT-3′ | |
| GSP3(reverse) | 5′-TCACAAATCTTGGGGTTCTC-3′ | 5′RACE-PCR |
| GSP4(reverse) | 5′-CTAGCATCAAGGGAAGGAGCC-3′ | |
| UPM | 5′-CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT-3′ | |
| FPF(forward) | 5′-ATGGTGACCGTAGAGGAGTATCG-3′ | Full-length |
| FPR (reverse) | 5′-GAACCCACTAAGTAGCCACACTG-3′ | |
| CHS-F(forward) | 5′-CAAAGGAGGGCTTAGGAACTACT-3′ | RT-qPCR |
| CHS-R(reverse) | 5′-CAAATTCAACCCCAAGAACAAATGA-3′ | |
| BaActin-F(forward) | 5′-AACCATAAACGATNCCGACCAG-3′ | |
| BaActin-R(reverse) | 5′-NCTTGCGACCATACTCCC-3′ |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Cao, Y.-T.; Ye, S.-R.; Irshad, M.; Pan, T.-F.; Qiu, D.-L. Isolation of CHS Gene from Brunfelsia acuminata Flowers and Its Regulation in Anthocyanin Biosysthesis. Molecules 2017, 22, 44. https://doi.org/10.3390/molecules22010044
Li M, Cao Y-T, Ye S-R, Irshad M, Pan T-F, Qiu D-L. Isolation of CHS Gene from Brunfelsia acuminata Flowers and Its Regulation in Anthocyanin Biosysthesis. Molecules. 2017; 22(1):44. https://doi.org/10.3390/molecules22010044
Chicago/Turabian StyleLi, Min, Yu-Ting Cao, Si-Rui Ye, Muhammad Irshad, Teng-Fei Pan, and Dong-Liang Qiu. 2017. "Isolation of CHS Gene from Brunfelsia acuminata Flowers and Its Regulation in Anthocyanin Biosysthesis" Molecules 22, no. 1: 44. https://doi.org/10.3390/molecules22010044





