Stenotrophomonas maltophilia: A Gram-Negative Bacterium Useful for Transformations of Flavanone and Chalcone
Abstract
:1. Introduction
2. Results and Discussion
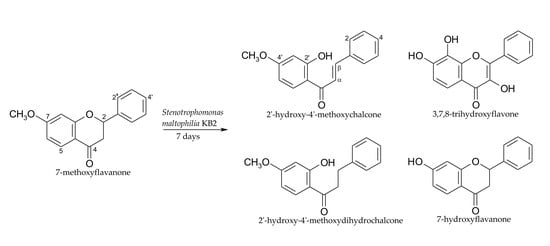
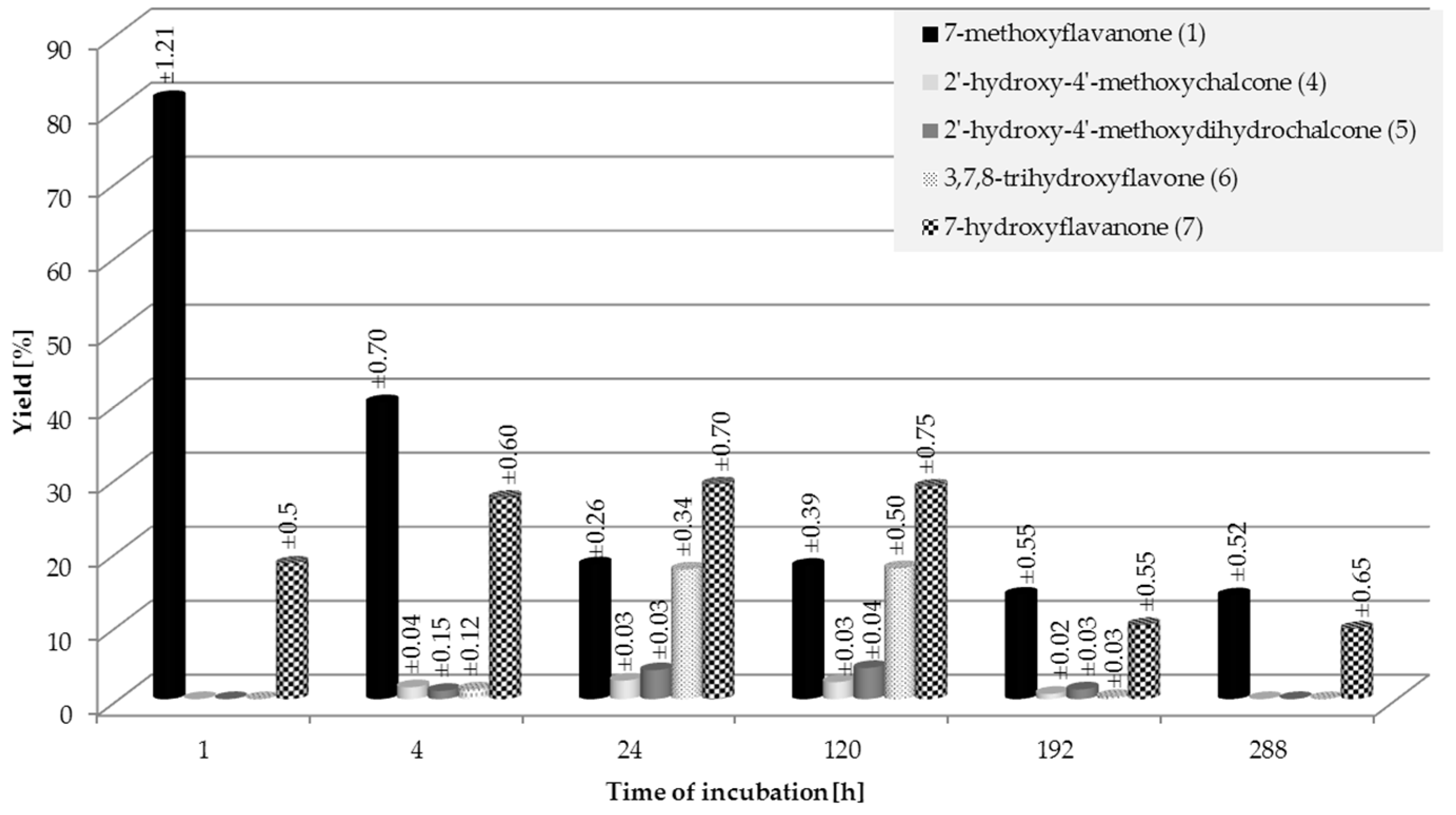
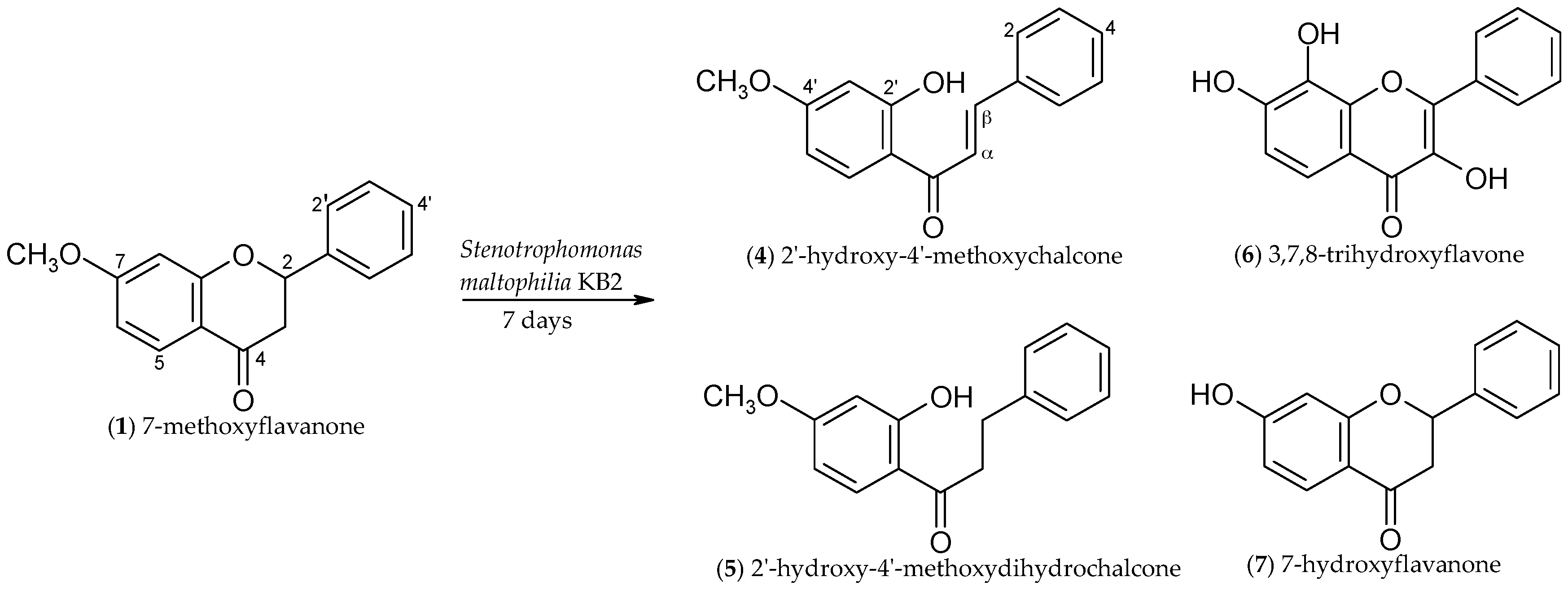
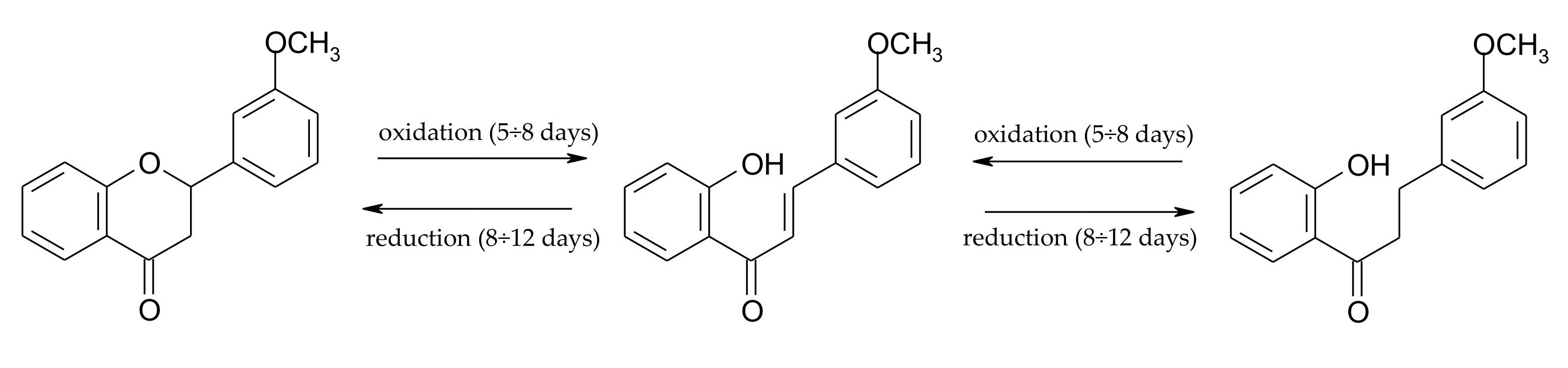
2.1. Biotransformations of 7-Methoxyflavanone (1)
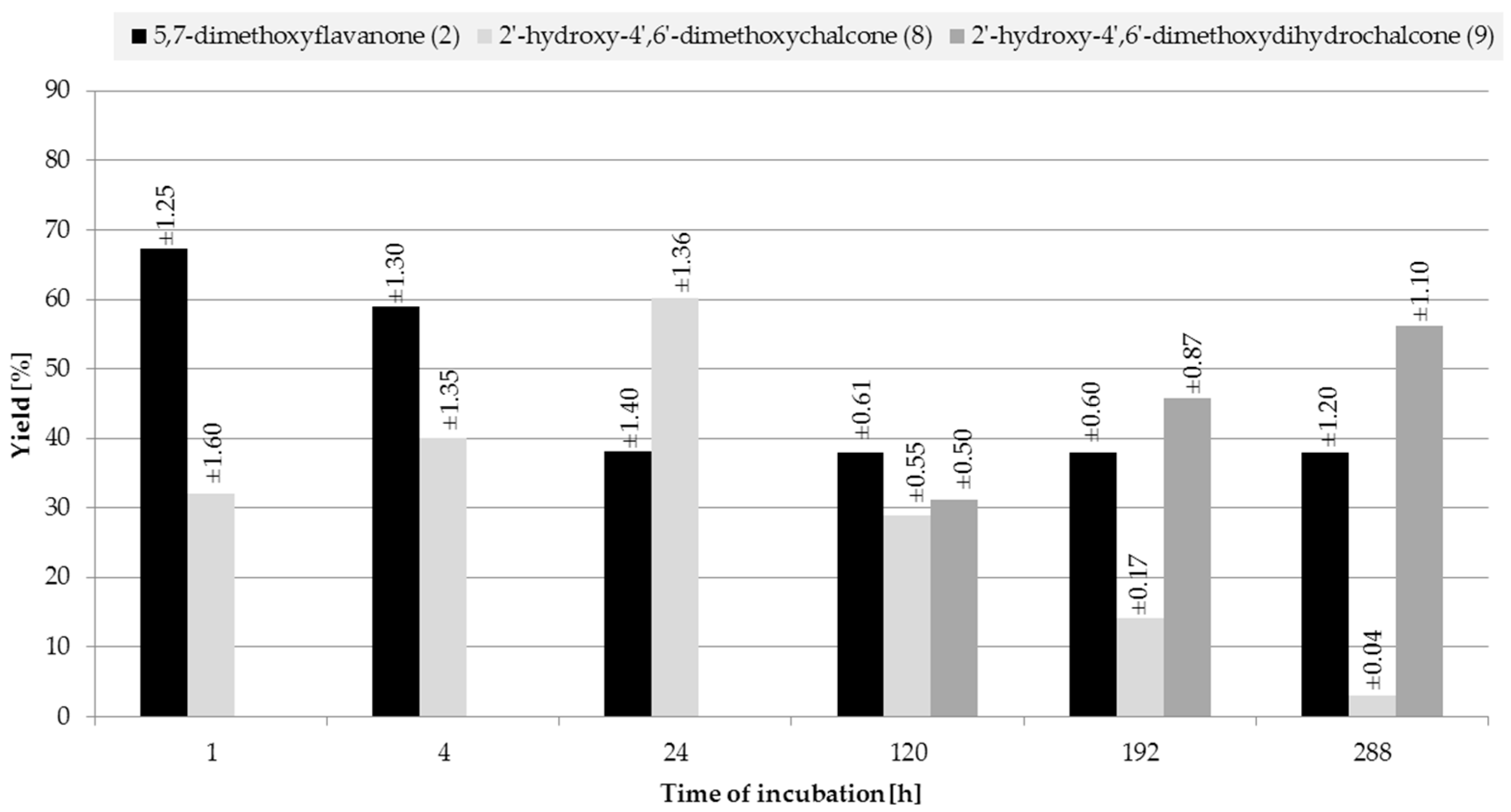
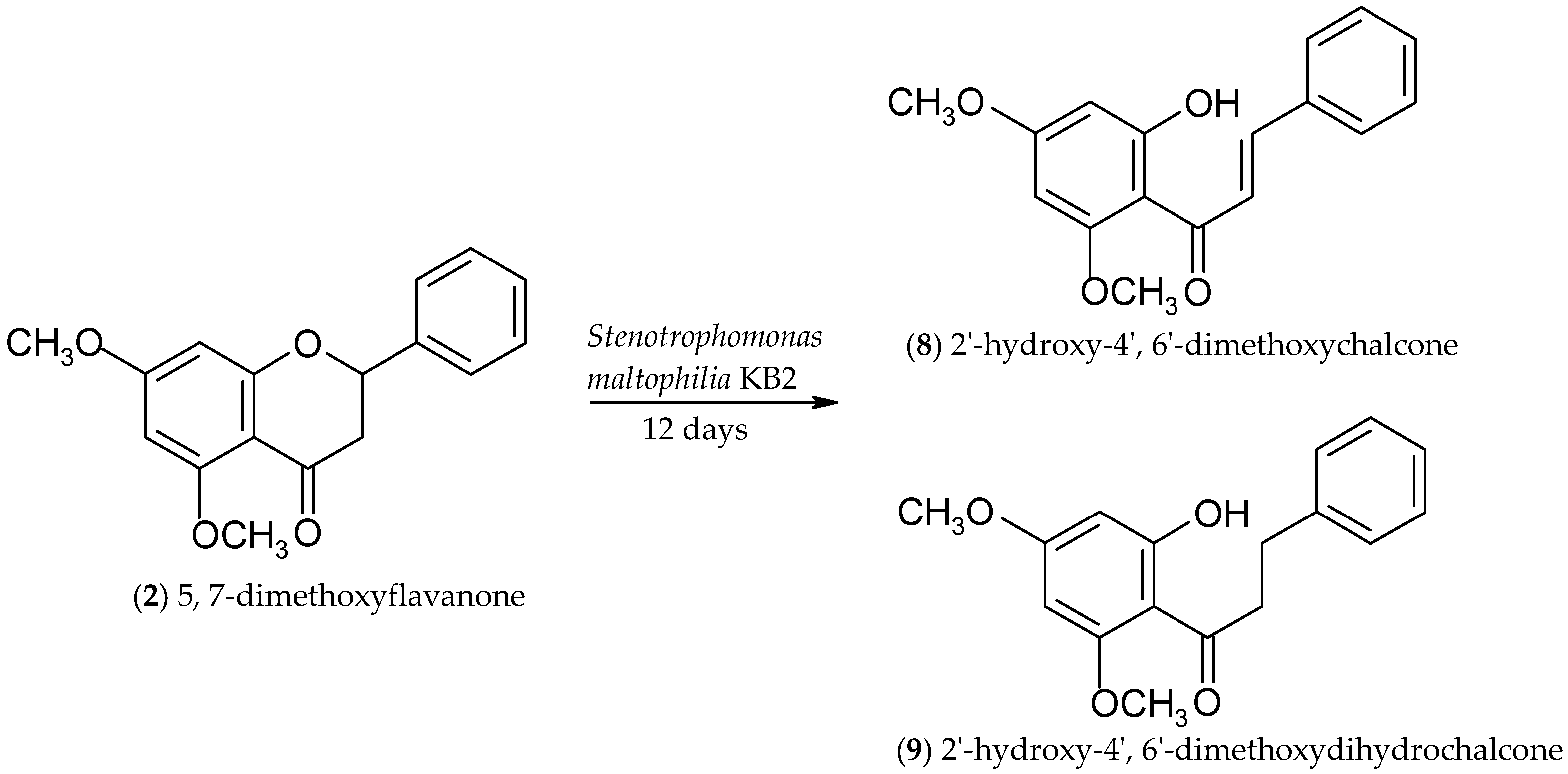
2.2. Biotransformations of 5,7-Dimethoxyflavanone (2)
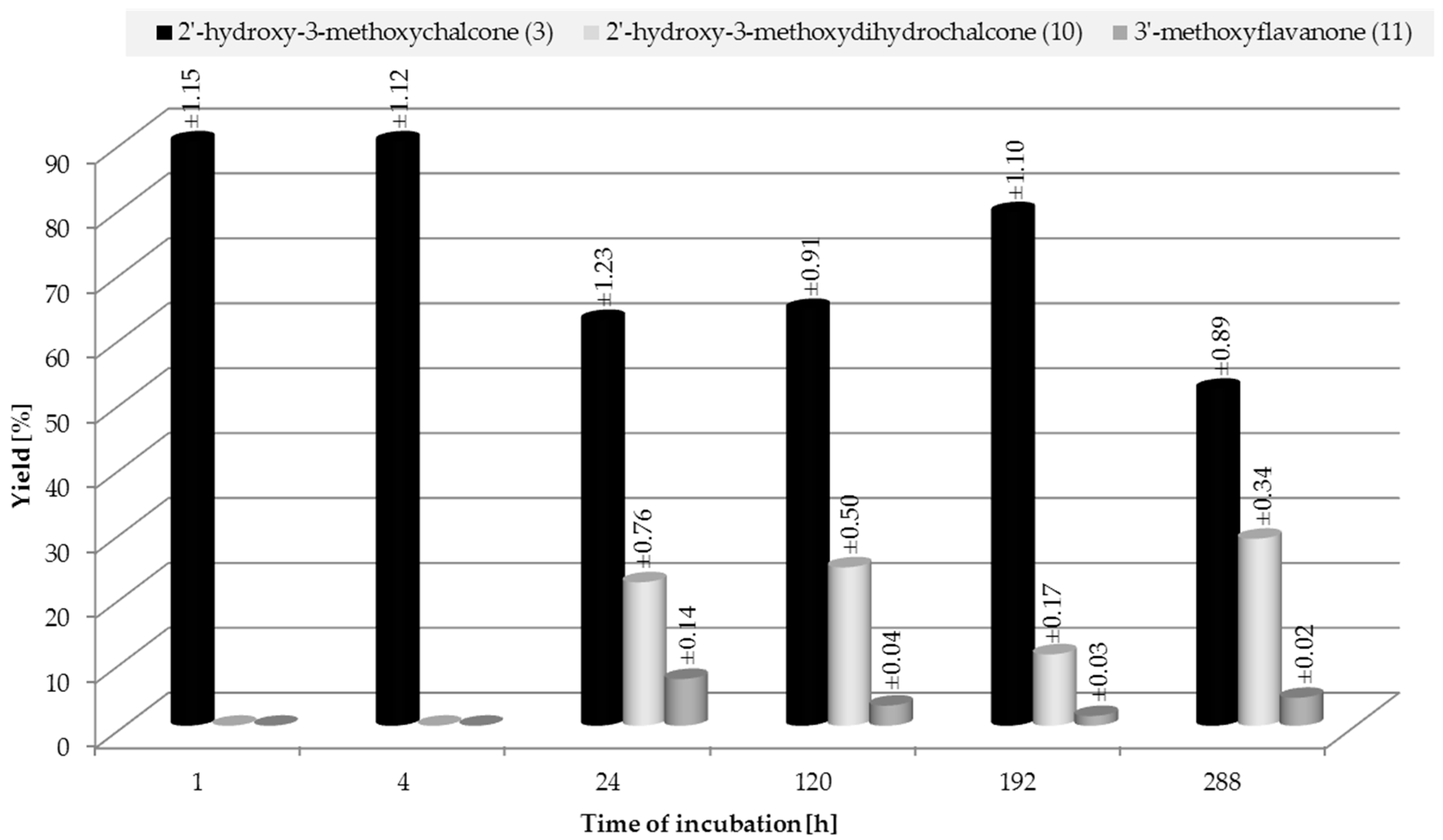
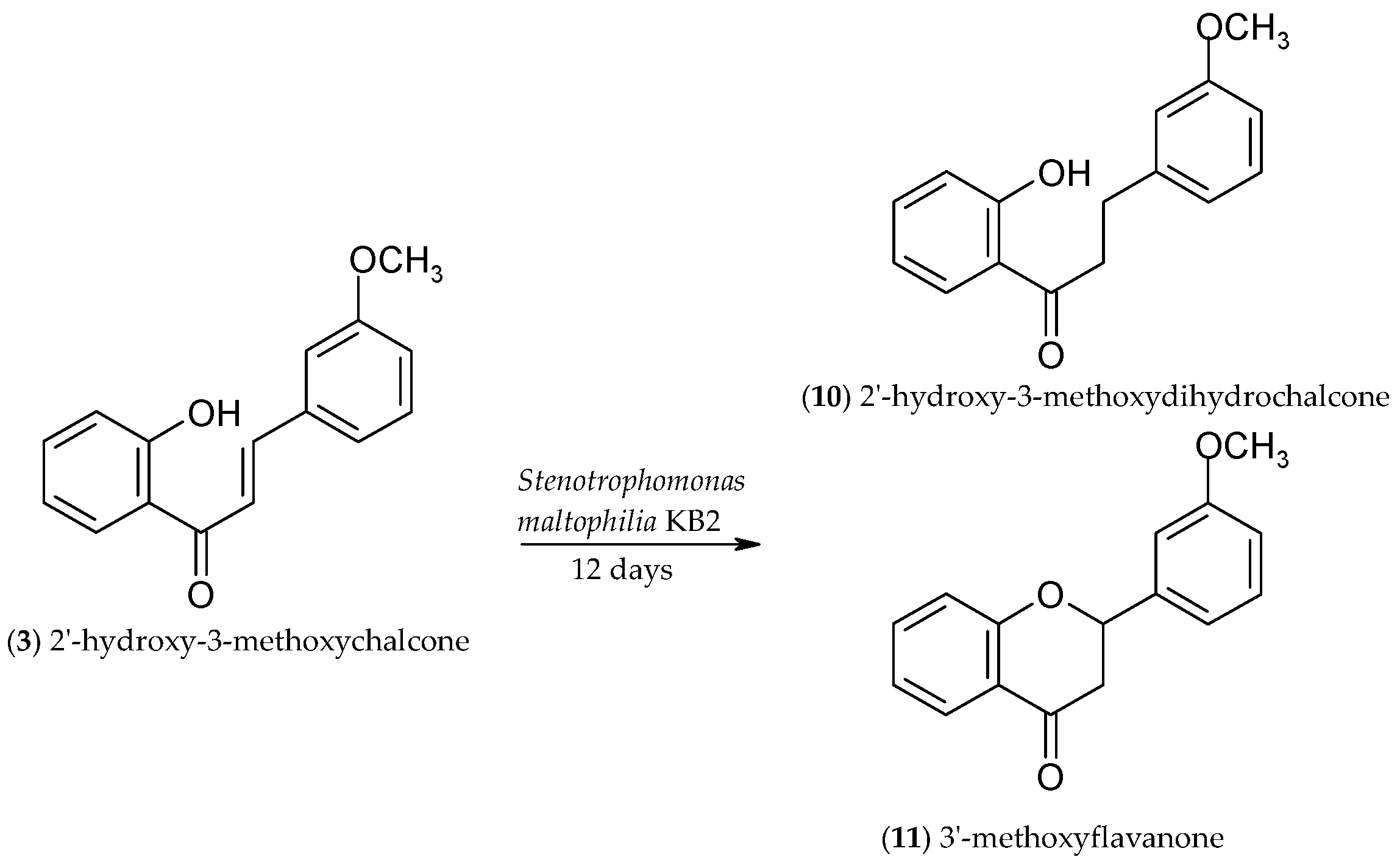
2.3. Biotransformation of 2′-Hydroxy-3-methoxychalcone (3)
3. Materials and Methods
3.1. Analysis
3.2. Materials
3.2.1. 7-Methoxyflavanone (1)
3.2.2. 5,7-Dimethoxyflavanone (2)
3.2.3. 2′-Hydroxy-3-methoxychalcone (3)
3.3. Microorganism
3.4. Biotransformations
3.4.1. 2′-Hydroxy-4′-methoxychalcone (4)
3.4.2. 2’-Hydroxy-4’-methoxydihydrochalcone (5)
3.4.3. 3,7,8-Trihydroxyflavone (6)
3.4.4. 7-Hydroxyflavanone (7)
3.4.5. 2′-Hydroxy-4′,6′-dimethoxychalcone (8)
3.4.6. 2′-Hydroxy-4′,6′-dimethoxydihydrochalcone (9)
3.4.7. 2′-Hydroxy-3-methoxydihydrochalcone (10)
3.4.8. 3′-methoxyflavanone (11)
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kostrzewa-Susłow, E.; Dmochowska-Gładysz, J.; Białońska, A.; Ciunik, Z.; Rymowicz, W. Microbial transformations of flavanone and 6-hydroxyflavanone by Aspergillus niger strains. J. Mol. Catal. B 2006, 39, 18–23. [Google Scholar] [CrossRef]
- Xiao, J.; Muzashvili, T.; Georgiev, M. Advances in the biotechnological glycosylation of valuable flavonoids. Biotechnol. Adv. 2014, 32, 1145–1156. [Google Scholar] [CrossRef] [PubMed]
- Kostrzewa-Susłow, E.; Dmochowska-Gładysz, J.; Oszmiański, J. Microbial transformation of baicalin and baicalein. J. Mol. Catal. B 2007, 49, 113–117. [Google Scholar] [CrossRef]
- Kostrzewa-Susłow, E.; Dmochowska-Gładysz, J.; Białońska, A.; Ciunik, Z. Microbial transformations of flavanone by Aspergillus niger and Penicillium chermesinum cultures. J. Mol. Catal. B 2008, 52–53, 34–39. [Google Scholar] [CrossRef]
- Janeczko, T.; Gładkowski, W.; Kostrzewa-Susłow, E. Microbial transformations of chalcones to produce food sweetener derivatives. J. Mol. Catal. B 2013, 98, 55–61. [Google Scholar] [CrossRef]
- Janeczko, T.; Dymarska, M.; Siepka, M.; Gniłka, R.; Leśniak, A.; Popłoński, J.; Kostrzewa-Susłow, E. Enantioselective reduction of flavanone and oxidation of cis- and trans-flavan-4-ol by selected yeast cultures. J. Mol. Catal. B 2014, 109, 47–52. [Google Scholar] [CrossRef]
- Stompor, M.; Potaniec, B.; Szumny, A.; Zieliński, P.; Żołnierczyk, A.K.; Anioł, M. Microbial synthesis of dihydrochalcones using Rhodococcus and Gordonia species. J. Mol. Catal. B 2013, 97, 283–288. [Google Scholar] [CrossRef]
- Seo, J.; Kang, S.; Kim, M.; Han, J.; Hur, H. Flavonoids biotransformation by bacterial non-heme dioxygenases, biphenyl and naphthalene dioxygenase. Appl. Microbiol. Biotechnol. 2011, 91, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.J.; Krishnamurty, H.G.; Jones, G.A.; Simpson, F.J. Identification of products produced by the anaerobic degradation of naringenin by Butyrivibrio sp. Can. J. Microbiol. 1970, 17, 129–131. [Google Scholar]
- Shimoda, K.; Kubota, N.; Taniuchi, K.; Sato, D.; Nakajima, N.; Hamada, H.; Hamada, H. Biotransformation of naringin and naringenin by cultured Eucalyptus perriniana cells. Phytochemistry 2010, 71, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Rosazza, J.P.N. Microbial and enzymatic transformations of flavonoids. J. Nat. Prod. 2006, 69, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Szliszka, E.; Kostrzewa-Susłow, E.; Bronikowska, J.; Jaworska, D.; Janeczko, T.; Czuba, Z.; Król, W. Synthetic flavanones augment the anticancer effect of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). Molecules 2012, 17, 11693–11711. [Google Scholar] [CrossRef] [PubMed]
- Kostrzewa-Susłow, E.; Dymarska, M.; Janeczko, T. Microbial transformations of 3-methoxyflavone by strains of Aspergillus niger. Pol. J. Microbiol. 2014, 63, 111–114. [Google Scholar] [PubMed]
- Wang, A.; Zhang, F.; Huang, L.; Yin, X.; Li, H.; Wang, Q.; Zeng, Z.; Xie, T. New progress in biocatalysis and biotransformation of flavonoids. J. Med. Plant Res. 2010, 4, 847–856. [Google Scholar] [CrossRef]
- Kostrzewa-Susłow, E.; Dmochowska-Gładysz, J.; Janeczko, T. Microbial transformation of selected flavanones as a method of increasing the antioxidant properties. Z. Naturforsch. C 2010, 65, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Kostrzewa-Susłow, E.; Dmochowska-Gładysz, J.; Janeczko, T.; Środa, K.; Michalak, K.; Palko, A. Microbial transformations of 6- and 7-methoxyflavones in Aspergillus niger and Penicillium chermesinum cultures. Z. Naturforsch. C 2012, 67, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Kostrzewa-Susłow, E.; Dymarska, M.; Białońska, A.; Janeczko, T. Enantioselective conversion of certain derivatives of 6-hydroxyflavanone. J. Mol. Catal. B 2014, 102, 59–65. [Google Scholar] [CrossRef]
- Cao, H.; Chen, X.; Jassbi, A.; Xiao, J. Microbial biotransformation of bioactive flavonoids. Biotechnol. Adv. 2015, 33, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Hosny, M.; Rosazza, J. Novel oxidations of (+)-catechin by horseradish peroxidase and laccase. J. Agric. Food Chem. 2002, 50, 5539–5545. [Google Scholar] [CrossRef] [PubMed]
- Holland, H. Organic Synthesis with Oxidative Enzymes; VCH Publishers: Weinheim, Germany, 1992; pp. 5–40. [Google Scholar]
- Aktas, N.; Tanyolac, A. Kinetics of laccase-catalysed oxidative polymerization of catechol. J. Mol. Catal. B 2003, 22, 61–69. [Google Scholar] [CrossRef]
- Makris, D.; Rossiter, J. An investigation on structural aspects influencing product formation in enzymic and chemical oxidation of quercetin and related flavonols. Food Chem. 2002, 77, 177–185. [Google Scholar] [CrossRef]
- Mejias, L.; Reihmann, M.; Sepulveda-Boza, S.; Ritter, H. New polymers from natural phenols using horseradish or soybean peroxidase. Macromol. Biosci. 2002, 21, 24–32. [Google Scholar] [CrossRef]
- Guzik, U.; Greń, I.; Wojcieszyńska, D.; Łabużek, S. Isolation and characterization of a novel strain of Stenotrophomonas maltophilia possessing various dioxygenases for monocyclic hydrocarbon degradation. Braz. J. Microbiol. 2009, 40, 285–291. [Google Scholar] [CrossRef]
- Wojcieszyńska, D.; Guzik, U.; Greń, I.; Perkosz, M.; Hupert-Kocurek, K. Induction of aromatic ring: Cleavage dioxygenases in Stenotrophomonas maltophilia strain KB2 in cometabolic systems. World J. Microbiol. Biotechnol. 2011, 27, 805–811. [Google Scholar] [CrossRef] [PubMed]
- Greń, I.; Wojcieszyńska, D.; Guzik, U.; Perkosz, M.; Hupert-Kocurek, K. Enhanced biotransformation of mononitrophenols by Stenotrophomonas maltophilia KB2 in the presence of aromatic compounds of plant origin. World J. Microbiol. Biotechnol. 2010, 26, 289–295. [Google Scholar] [CrossRef]
- Stompor, M.; Kałużny, M.; Żarowska, B. Biotechnological methods for chalcone reduction using whole cells of Lactobacillus, Rhodococcus and Rhodotorula strains as a way to produce new derivatives. Appl. Microbiol. Biotechnol. 2016, 100, 8371. [Google Scholar] [CrossRef] [PubMed]
- Wojcieszyńska, D.; Greń, I.; Hupert-Kocurek, K.; Guzik, U. Modulation of FAD-dependent monooxygenase activity from aromatic compounds-degrading Stenotrophomonas maltophilia strain KB2. Acta Biochim. Pol. 2011, 58, 421–426. [Google Scholar] [PubMed]
- Wojcieszyńska, D.; Hupert-Kocurek, K.; Greń, I.; Guzik, U. High activity catechol 2,3-dioxygenase from the cresols—Degrading Stenotrophomonas maltophilia strain KB2. Int. Biodeterior. Biodegrad. 2011, 65, 853–858. [Google Scholar] [CrossRef]
- Krishnamurty, H.G.; Cheng, K.J.; Jones, G.A.; Simpson, F.J.; Watkin, J.E. Identification of products produced by the anaerobic degradation of rutin and related flavonoids by Butyrivibrio sp. C3. Can. J. Microbiol. 1970, 16, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Yeom, J.E.; Kumar, M.R.; Lee, S.; Lee, J.B.; Park, H.R. Synthesis of flavanol-4-ol and its spectroscopic properties in aqueous solution. Bull. Korean Chem. Soc. 2011, 32, 4092–4094. [Google Scholar] [CrossRef]
- Yadav, N.; Dixit, S.K.; Bhattacharya, A.; Mishra, L.C.; Sharma, M.; Awasthi, S.K.; Bhasin, V.K. Antimalarial activity of newly synthesized chalcone derivatives in vitro. Chem. Biol. Drug Des. 2012, 80, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Kostrzewa-Susłow, E.; Janeczko, T. Microbial transformations of 7-methoxyflavanone. Molecules 2012, 17, 14810–14820. [Google Scholar] [CrossRef] [PubMed]
- Kostrzewa-Susłow, E.; Janeczko, T. Microbial transformations of 7-hydroxyflavanone. Sci. World J. 2012. [Google Scholar] [CrossRef] [PubMed]


| Proton | Compound | ||||
|---|---|---|---|---|---|
| 1 a | 2 a | 7 a | 11 b | 6 b | |
| H-2 | 5.47 (dd) (J2,3ax = 13.4, J2,3eq = 2.9) | 5.41 (dd) (J2,3ax = 13.2, J2,3eq = 2.6) | 5.46 (dd) (J2,3ax = 12.9, J2,3eq = 3.0) | 5.46 (dd) (J2,3ax = 13.3, J2,3eq = 2.8) | - |
| H-3 | H-3ax 3.05 (dd) (J3ax,3eq = 16.9, J3ax,2 = 13.4) H-3eq 2.84 (dd) (J3eq,3ax = 16.9, J3eq,2 = 2.9) | H-3ax 3.02 (dd) (J3ax,3eq = 16.5, J3ax,2 = 13.3) H-3eq 2.80 (dd) (J3eq,3ax = 16.5, J3eq,2 = 2.8) | H-3ax 2.95 (dd) (J3ax,3eq = 16.9, J3ax,2 = 12.8) H-3eq 2.69 (dd) (J3eq,3ax = 16.9, J3eq,2 = 3.1) | H-3ax 3.08 (dd) (J3ax,3eq = 16.8, J3ax,2 = 13.4) H-3eq 2.90 (dd) (J3eq,3ax = 16.5, J3eq,2 = 2.9) | - |
| H-5 | 7.87 (d) (J5,6 = 8.8) | - | 7.72 (d) (J5.6 = 8.7) | 7.94 (dd) (J = 8.2, J = 1.6) | 7.07 (d) (J = 8.1) |
| H-6 | 6.63 (dd) (J6,5 = 8.8, J6,8 = 2.4) | 6.16 (d) (J6,8 = 2.0) | 6.46 (dd) (J6.5 = 8.7, J6.8 = 2.2) | 7.06 (m) | 6.81 (d) (J = 8.2) |
| H-7 | - | - | - | 7.52 (m) | - |
| H-8 | 6.51 (d) (J8,6 = 2.4) | 6.10 (d) (J8,6 = 1.9) | 6.35 (d) (J8.6 = 2.2) | 7.06 (m) | - |
| H-2’ | 7.48 (d) (J2′,3′ = 6.4) | 7.46 (d) (J = 7.5) | 7.49 (d) (J2’,3’ = 7.6) | 7.06 (m) | 7.23 (d) (J = 7.5) |
| H-3’ | 7.39 (m) | 7.42 (t) (J = 7.5) | 7.38 (m) | - | 7.35 (m) |
| H-4’ | 7.39 (m) | 7.37 (t) (J = 7.2) | 7.32 (m) | 6.92 (m) | 7.29 (m) |
| H-5’ | 7.39 (m) | 7.42 (t) (J = 7.5) | 7.38 (m) | 7.35 (t) (J = 8.2) | 7.35 (m) |
| H-6’ | 7.48 (d) (J6′,5′ = 6.4) | 7.46 (d) (J = 7.5) | 7.49 (d) (J6′,5′ = 7.6) | 7.06 (m) | 7.23 (d) (J = 7.5) |
| 3-OH | - | - | - | - | 6.13 (s) |
| 8-OH | - | - | - | - | 6.09 (s) |
| 7-OH | - | - | 9.70 (s) | - | - |
| 7-OCH3 | 3.84 (s) | 3.82 (s) | - | - | - |
| 5-OCH3 | - | 3.90 (s) | - | - | - |
| 3’-OCH3 | - | - | - | 3.85 (s) | - |
| Carbon | Flavonoids | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 a | 2 a | 3 b | 4 b | 5 b | 6 b | 7 a | 8 b | 9 b | 10 b | 11 b | |
| C-1 | - | - | 136.1 | 135.0 | 141.0 | - | - | 135.7 | 142.1 | 142.5 | - |
| C-2 | 80.2 | 79.4 | 113.9 | 128.6 | 128.5 | 169.5 | 81.0 | 128.7 | 128.7 | 114.4 | 79.6 |
| C-3 | 44.5 | 45.7 | 160.2 | 129.1 | 128.7 | 135.9 | 45.4 | 129.0 | 128.6 | 159.9 | 44.9 |
| C-4 | 190.7 | 189.3 | 116.8 | 130.7 | 126.4 | 170.3 | 193.1 | 130.2 | 126.1 | 111.7 | 192.1 |
| C-5 | 129.1 | 162.4 | 130.2 | 129.1 | 128.7 | 130.4 | 129.6 | 129.0 | 128.6 | 129.8 | 127.2 |
| C-6 | 110.4 | 93.3 | 121.4 | 128.6 | 128.5 | 116.1 | 111.4 | 128.7 | 128.7 | 120.9 | 118.5 |
| C-7 | 166.4 | 166.1 | - | - | - | 165.1 | 166.9 | - | - | - | 136.4 |
| C-8 | 101.1 | 93.7 | - | - | - | 166.2 | 103.7 | - | - | - | 118.3 |
| C-9 | 163.7 | 165.1 | - | - | - | 165.1 | 165.4 | - | - | - | 160.1 |
| C-10 | 115.0 | 106.1 | - | - | - | 116.1 | 115.4 | - | - | - | 121.1 |
| C-1′ | 139.0 | 138.9 | 120.2 | - | 113.6 | - | 140.7 | - | 141.9 | 119.4 | 140.4 |
| C-2′ | 126.3 | 126.2 | 163.8 | 167.0 | 165.6 | 128.5 | 127.2 | 168.6 | 167.8 | 162.6 | 121.8 |
| C-3′ | 129.0 | 128.9 | 129.8 | 107.9 | 107.8 | 129.1 | 129.4 | 93.9 | 93.8 | 118.7 | 161.6 |
| C-4′ | 128.9 | 128.8 | 119.0 | 166.4 | 166.2 | 127.6 | 129.2 | 107.8 | 162.9 | 136.5 | 114.2 |
| C-5′ | 129.0 | 128.9 | 136.6 | 101.2 | 101.1 | 129.1 | 129.4 | 91.4 | 91.0 | 119.1 | 130.1 |
| C-6′ | 126.3 | 126.2 | 118.8 | 131.4 | 131.6 | 128.5 | 127.2 | 111.9 | 166.1 | 130.0 | 112.0 |
| C-α | - | - | 145.6 | 144.7 | 39.8 | - | - | 127.7 | 45.8 | 40.1 | - |
| C-β | - | - | 120.6 | 120.6 | 30.5 | - | - | 142.5 | 30.8 | 30.2 | - |
| C=O | - | - | 193.9 | - | 203.7 | - | - | 202.1 | 204.8 | 205.5 | - |
| 3-OCH3 | - | - | 55.6 | - | - | - | - | - | - | 55.3 | - |
| 5-OCH3 | - | 56.3 | - | - | - | - | - | - | - | - | - |
| 7-OCH3 | 55.8 | 55.7 | - | - | - | - | - | - | - | - | - |
| 3′-OCH3 | - | - | - | - | - | - | - | - | - | - | 55.5 |
| 4′-OCH3 | - | - | - | 55.8 | 55.7 | - | - | 55.8 | 55.8 | - | - |
| 6′-OCH3 | - | - | - | - | - | - | - | 56.0 | 55.7 | - | - |
| Proton | Chalcones | Dihydrochalcones | ||||
|---|---|---|---|---|---|---|
| 3 | 4 | 8 | 5 | 9 | 10 | |
| H-α | 7.89 (d) (J = 15.5) | 7.89 (d) (J = 15.5) | 7.90 (d) (J = 15.6) | 3.24 (m) | 3.32 (m) | 3.33 (m) |
| H-β | 7.65 (d) (J = 15.5) | 7.59 (d) (J = 15.5) | 7.79 (d) (J = 15.6) | 3.06 (m) | 3.03 (m) | 3.05 (m) |
| H-3’ | 7.93 (d) (J = 8.0) | 6.51 (m) | 6.12 (s) | 6.41 (d) (J = 2.3) | 6.10 (d) (J3′,5′ = 2.3) | 6.99 (d) (J = 8.4) |
| H-4’ | 6.96 (t) (J = 7.6) | - | - | - | - | 7.47 (t) (J = 7.7) |
| H-5’ | 7.51 (t) (J = 7.8) | 6.49 (m) | 5.97 (s) | 6.43 (m) | 5.92 (d) (J5′,3′ = 2.2) | 6.88 (t) (J = 7.6) |
| H-6’ | 7.04 (d) (J = 8.4) | 7.84 (d) (J = 8.8) | - | 7.64 (d) (J = 8.6) | - | 7.75 (d) (J = 7.8) |
| H-2 | 7.18 (s) | 7.66 (m) | 7.61 (d) (J = 7.4) | 7.25 (d) (J2,3(6,5) = 7.5) | 7.24 (d) (J2,3(6,5) = 7.7) | 6.80 (s) |
| H-3 | - | 7.43 (m) | 7.41 (m) | 7.30 (t) (J = 7.5) | 7.30 (t) (J = 7.5) | - |
| H-4 | 7.00 (dd) (J = 8.2, J = 2.0) | 7.43 (m) | 7.41 (m) | 7.22 (t) (J = 7.4) | 7.20 (t) (J = 7.2) | 6.77 (dd) (J = 8.3, J = 2.1) |
| H-5 | 7.36 (t) (J = 7.9) | 7.43 (m) | 7.41 (m) | 7.30 (t) (J = 7.5) | 7.30 (t) (J = 7.5) | 7.23 (t) (J = 7.9) |
| H-6 | 7.27 (d) (J = 7.7) | 7.66 (m) | 7.61 (d) (J = 7.4) | 7.25 (d) (J2,3(6,5) = 7.5) | 7.24 (d) (J2,3(6,5) = 7.7) | 6.84 (d) (J = 7.5) |
| 2′-OH | 12.79 (s) | 13.43 (s) | 14.28 (s) | 12.79 (s) | 14.05 (s) | 12.29 (s) |
| 3-OCH3 | 3.88 (s) | - | - | - | - | 3.80(s) |
| 4′-OCH3 | - | 3.87 (s) | 3.84 (s) | 3.84 (s) | 3.82 (s) | - |
| 6′-OCH3 | - | - | 3.93 (s) | - | 3.84 (s) | - |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kostrzewa-Susłow, E.; Dymarska, M.; Guzik, U.; Wojcieszyńska, D.; Janeczko, T. Stenotrophomonas maltophilia: A Gram-Negative Bacterium Useful for Transformations of Flavanone and Chalcone. Molecules 2017, 22, 1830. https://doi.org/10.3390/molecules22111830
Kostrzewa-Susłow E, Dymarska M, Guzik U, Wojcieszyńska D, Janeczko T. Stenotrophomonas maltophilia: A Gram-Negative Bacterium Useful for Transformations of Flavanone and Chalcone. Molecules. 2017; 22(11):1830. https://doi.org/10.3390/molecules22111830
Chicago/Turabian StyleKostrzewa-Susłow, Edyta, Monika Dymarska, Urszula Guzik, Danuta Wojcieszyńska, and Tomasz Janeczko. 2017. "Stenotrophomonas maltophilia: A Gram-Negative Bacterium Useful for Transformations of Flavanone and Chalcone" Molecules 22, no. 11: 1830. https://doi.org/10.3390/molecules22111830