In-Silico UHPLC Method Optimization for Aglycones in the Herbal Laxatives Aloe barbadensis Mill., Cassia angustifolia Vahl Pods, Rhamnus frangula L. Bark, Rhamnus purshianus DC. Bark, and Rheum palmatum L. Roots
Abstract
:1. Introduction
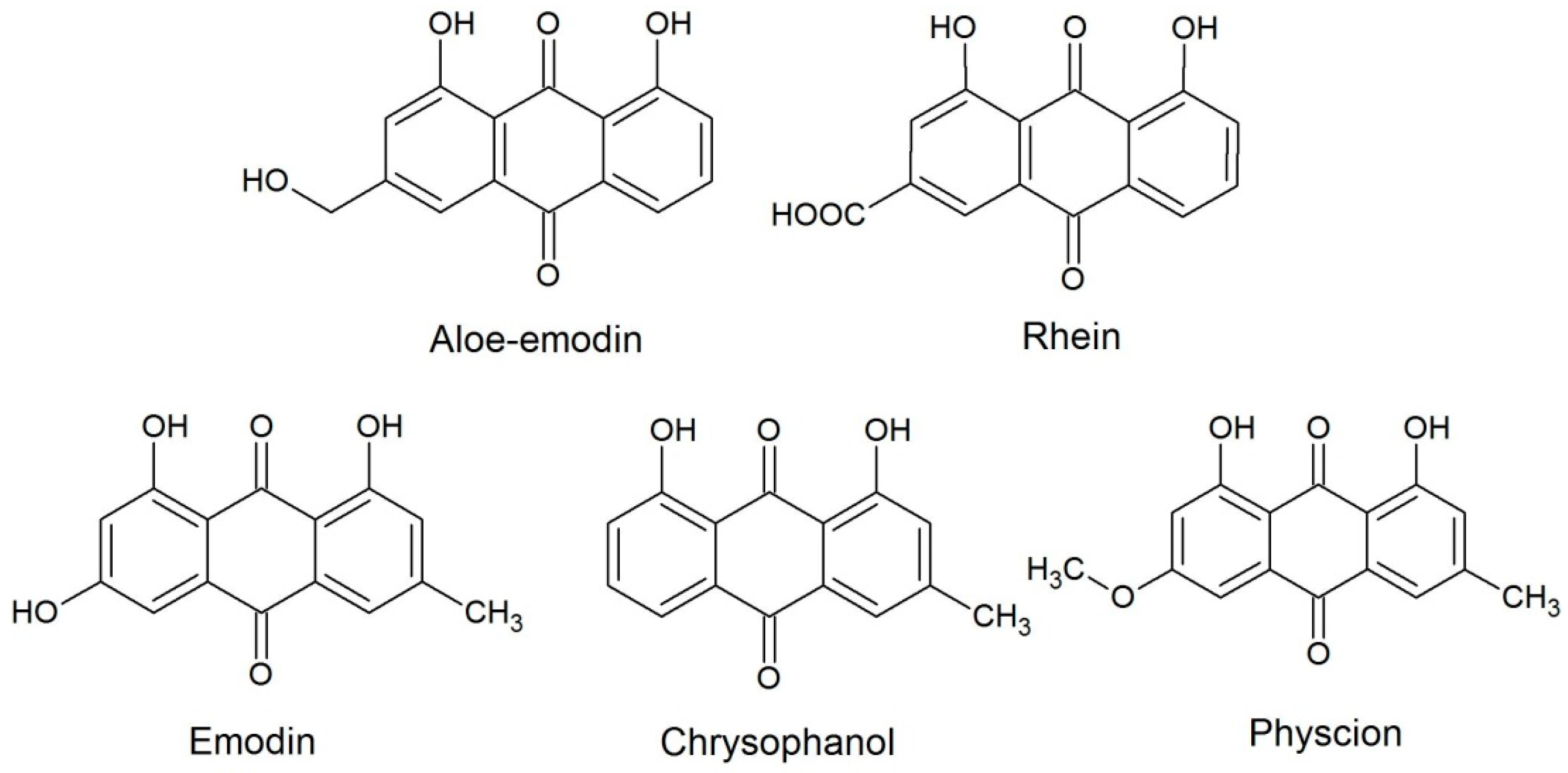
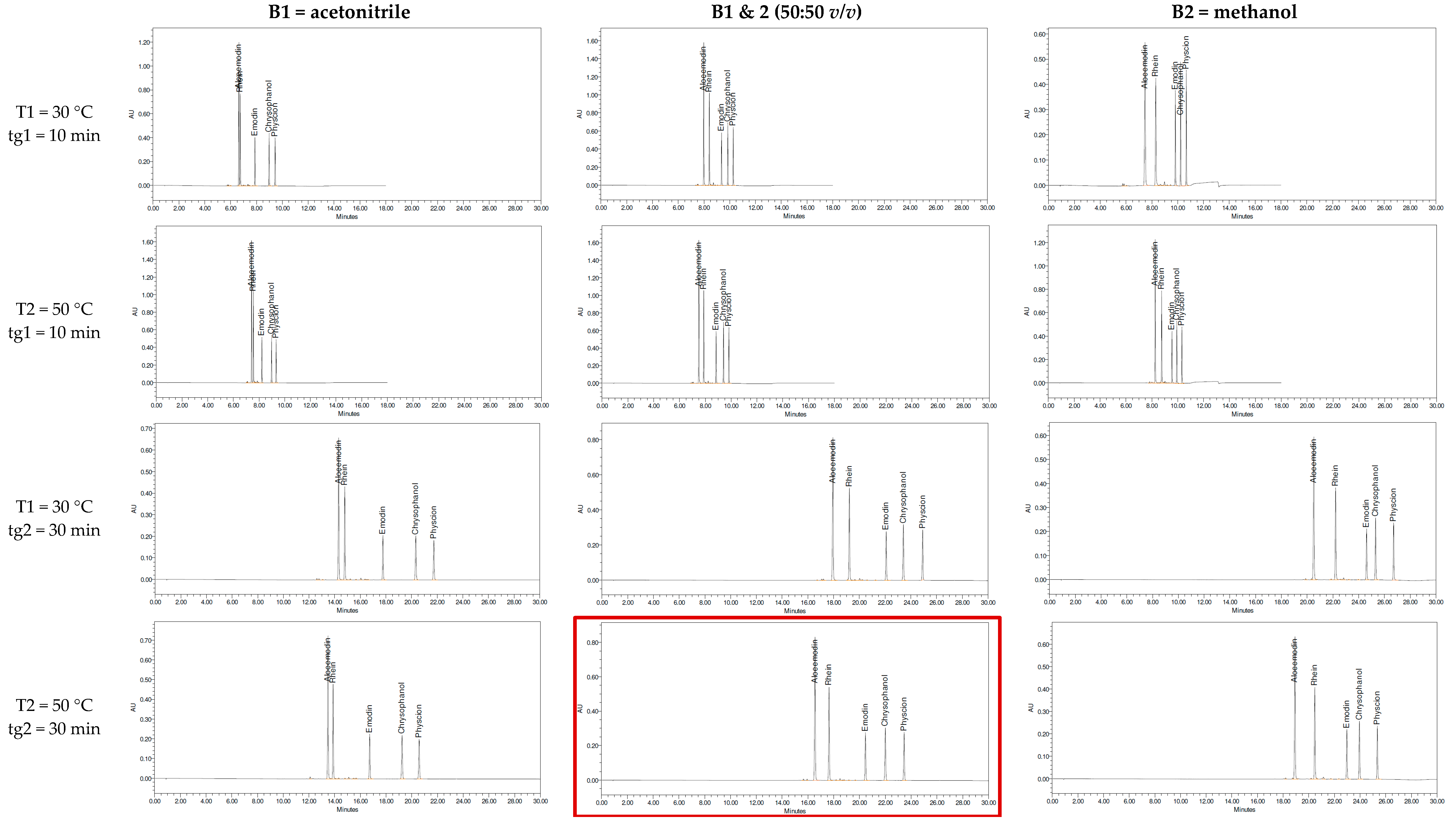
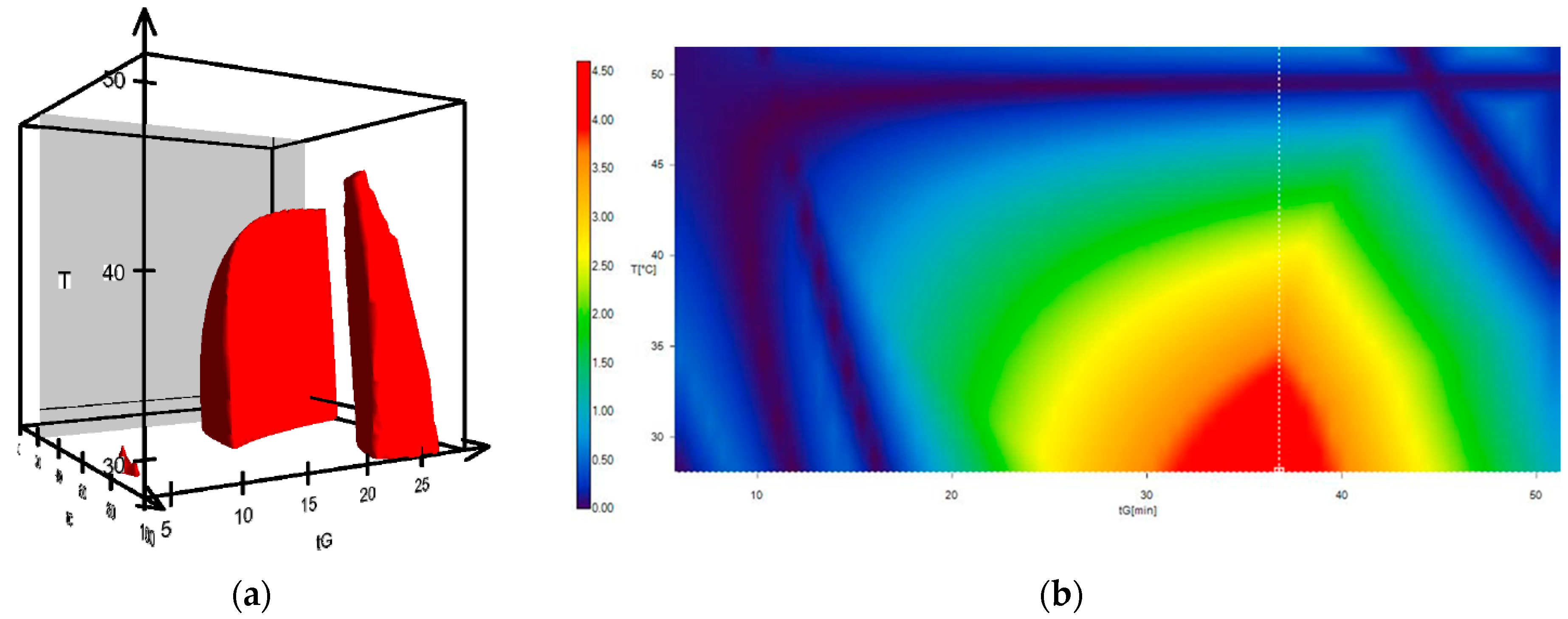
2. Results and Discussion
3. Materials and Methods
3.1. Chemicals
3.2. Standard Solutions
3.3. Plant Material and Sample Preparation
3.4. Equipment and Fixed Chromatographic Conditions
3.5. Model Parameters
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Melzig, M.F. Phytopharmaka als Laxanzien. Abführmittel auf pflanzlicher Basis. Pharm. Unserer Zeit 2008, 37, 136–141. [Google Scholar] [CrossRef] [PubMed]
- El-Najjar, N.; Gali-Muhtasib, H.; Vuorela, P.; Urtti, A.; Vuorela, H. Naphthoquinones and Anthraquinones: Chemical, Analytical and Biological Overview. In Handbook of Chemical and Biological Plant Analytical Methods, 1st ed.; Hostettmann, K., Stuppner, H., Marston, A., Chen, S., Eds.; JohnWiley & Sons, Ltd.: New York, NY, USA, 2014; pp. 595–610. ISBN 978-1-119-95275-6. [Google Scholar]
- Izhaki, I. Emodin—A secondary metabolite with multiple ecological functions in higher plants. New Phytol. 2002, 155, 205–217. [Google Scholar] [CrossRef]
- Abdissa, N.; Induli, M.; Fitzpatrick, P.; Alao, J.P.; Sunnerhagen, P.; Landberg, G.; Yenesew, A.; Erdélyi, M. Cytotoxic Quinones from the Roots of Aloe dawei. Molecules 2014, 19, 3264–3273. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Fu, J.; Yin, X.; Cao, S.; Li, X.; Lin, L.; Ni, J. Emodin: A Review of its Pharmacology, Toxicity and Pharmacokinetics. Phytother. Res. 2016, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- Heidemann, A.; Völkner, W.; Mengs, U. Genotoxicity of aloeemodin in vitro and in vivo. Mutat. Res. Genet. Toxicol. 1996, 367, 123–133. [Google Scholar] [CrossRef]
- Westendorf, J.; Marquardt, H.; Poginsky, B.; Dominiak, M.; Schmidt, J.; Marquardt, H. Genotoxicity of naturally occurring hydroxyanthraquinones. Mutat. Res. Genet. Toxicol. 1990, 240, 1–12. [Google Scholar] [CrossRef]
- Li, Y.; Luan, Y.; Qi, X.; Li, M.; Gong, L.; Xue, X.; Wu, X.; Wu, Y.; Chen, M.; Xing, G.; et al. Emodin triggers DNA double-strand breaks by stabilizing topoisomerase II-DNA cleavage complexes and by inhibiting ATP hydrolysis of topoisomerase II. Toxic Sci. 2010, 118, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Mueller, S.O.; Stopper, H. Characterization of the genotoxicity of anthraquinones in mammalian cells. Biochim. Biophys. Acta Gen. Subj. 1999, 1428, 406–414. [Google Scholar] [CrossRef]
- National Toxicology Program. Toxicology and Carcinogenesis Studies of Emodin (CAS No. 518-82-1) in F344/N Rats and B6C3F1 Mice. Natl. Toxicol. Program Tech. Rep. Ser. 2001, 493, 1–278. [Google Scholar]
- Commitee on Herbal Medicinal Products (HMPC). Assessment Report on Cassia senna L. and Cassia angustifolia Vahl, Folium and fructus, (Doc. Ref. EMEA/HMPC/51870/2006); European Medicines Agency: London, UK, 2007. [Google Scholar]
- Brusick, D.; Mengs, U. Assessment of the genotoxic risk from laxative senna products. Environ. Mol. Mutagen. 1997, 29, 1–9. [Google Scholar] [CrossRef]
- Morales, M.A.; Hernández, D.; Bustamante, S.; Bachiller, I.; Rojas, A. Is senna laxative use associated to cathartic colon, genotoxicity, or carcinogenicity? J. Toxicol. 2009, 2009, 287247. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.R.; Monteiro, M.R.; Rocha, H.M.; Ribeiro, A.F.; Caldeira-de-Araujo, A.; Leitão, A.C.; Bezerra, R.J.; Pádula, M. Assessment of antimutagenic and genotoxic potential of senna (Cassia angustifolia Vahl.) aqueous extract using in vitro assays. Toxicol. In Vitro 2008, 22, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Mueller, S.O.; Schmitt, M.; Dekant, W.; Stopper, H.; Schlatter, J.; Schreier, P.; Lutz, W.K. Occurrence of emodin, chrysophanol and physcion in vegetables, herbs and liquors. Genotoxicity and anti-genotoxicity of the anthraquinones and of the whole plants. Food Chem. Toxicol. 1999, 37, 481–491. [Google Scholar] [CrossRef]
- EDQM. Draft for the revision of the monographs Senna leaf (0206), Senna pods, Alexandrian (0207) and Tinnevelly (0208)—PA/PH/Exp. 13A/T (15) 19–21 ANP. Pharmeuropa 2015, 27, 169–186. [Google Scholar]
- Fernand, V.E.; Dinh, D.T.; Washington, S.J.; Fakayode, S.O.; Losso, J.N.; van Ravenswaay, R.O.; Warner, I.M. Determination of pharmacologically active compounds in root extracts of Cassia alata L. by use of high performance liquid chromatography. Talanta 2008, 74, 896–902. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Chan, C.O.; Lau, C.C.; Yu, Z.; Mok, D.K.W.; Chen, S. Simultaneous determination of eight anthraquinones in Semen Cassiae by HPLC-DAD. Phytochem. Anal. 2012, 23, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Yao, W.; Ji, W.; Wei, J.; Peng, S. Qualitative and quantitative analysis of anthraquinones in rhubarbs by high performance liquid chromatography with diode array detector and mass spectrometry. Food Chem. 2013, 141, 1710–1715. [Google Scholar] [CrossRef] [PubMed]
- Monschein, S. Entwicklung Einer HPLC-Gehaltsbestimmungsmethode für Rhei Radix. Master’s Thesis, Karl-Franzens-University Graz, Graz, Austria, 2005. [Google Scholar]
- Wang, J.; Li, H.; Jin, C.; Qu, Y.; Xiao, X. Development and validation of a UPLC method for quality control of rhubarb-based medicine: Fast simultaneous determination of five anthraquinone derivatives. J. Pharm. Biomed. Anal. 2008, 47, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.L.; Zhu, P.L.; Liu, M.C. Computer-aided development of a high-performance liquid chromatographic method for the determination of hydroxyanthraquinone derivatives in Chinese herb medicine rhubarb. J. Chromatogr. A 1999, 857, 167–174. [Google Scholar] [CrossRef]
- He, D.; Chen, B.; Tian, Q.; Yao, S. Simultaneous determination of five anthraquinones in medicinal plants and pharmaceutical preparations by HPLC with fluorescence detection. J. Pharm. Biomed. Anal. 2009, 49, 1123–1127. [Google Scholar] [CrossRef] [PubMed]
- Genovese, S.; Tammaro, F.; Menghini, L.; Carlucci, G.; Epifano, F.; Locatelli, M. Comparison of three different extraction methods and HPLC determination of the anthraquinones aloe-emodine, emodine, rheine, chrysophanol and physcione in the bark of Rhamnus alpinus L. (Rhamnaceae). Phytochem. Anal. 2010, 21, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Locatelli, M.; Genovese, S.; Carlucci, G.; Kremer, D.; Randic, M.; Epifano, F. Development and application of high-performance liquid chromatography for the study of two new oxyprenylated anthraquinones produced by Rhamnus species. J. Chromatogr. A 2012, 1225, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Kremer, D.; Kosalec, I.; Locatelli, M.; Epifano, F.; Genovese, S.; Carlucci, G.; Zovko Koncic, M. Anthraquinone profiles, antioxidant and antimicrobial properties of Frangula rupestris (Scop.) Schur and Frangula alnus Mill. bark. Food Chem. 2012, 131, 1174–1180. [Google Scholar] [CrossRef]
- Peter, S.; Meier, N.; Josic, G.; Meier, B.; Wolfram, E. Optimization of LC separation of anthraquinones in Sennae fructus and Sennae folium supported by an in-silico tool. Planta Med. 2016, 81, P1024. [Google Scholar] [CrossRef]
- Frey, O. Untersuchungen zum Anthranoid-Gehalt in Frangulae Cortex sowie zum Extraktionsverhalten der Anthranoide unter Berücksichtigung der Aglyka. Mather’s Thesis, Zuerich University of Applied Sciences, Winterthur, Switzerland, September 2014. [Google Scholar]
- Rosenthal, I.; Wolfram, E.; Peter, S.; Meier, B. A validated method for the analysis of frangulin A/B and glucofrangulin A/B with HPLC and UHPLC. J. Nat. Prod. 2014, 77, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Molnár, I.; Rieger, H.; Monks, K.E. Aspects of the ‘Design Space’ in high pressure liquid chromatography method development. J. Chromatogr. A 2010, 1217, 3193–3200. [Google Scholar] [CrossRef] [PubMed]
- Kormány, R.; Molnár, I.; Fekete, J. Renewal of an old European Pharmacopoeia method for Terazosin using modeling with mass spectrometric peak tracking. J. Pharm. Biomed. Anal. 2017, 135, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Fekete, S.; Fekete, J.; Ganzler, K. Validated UPLC method for the fast and sensitive determination of steroid residues in support of cleaning validation in formulation area. J. Pharm. Biomed. Anal. 2009, 49, 833–838. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |

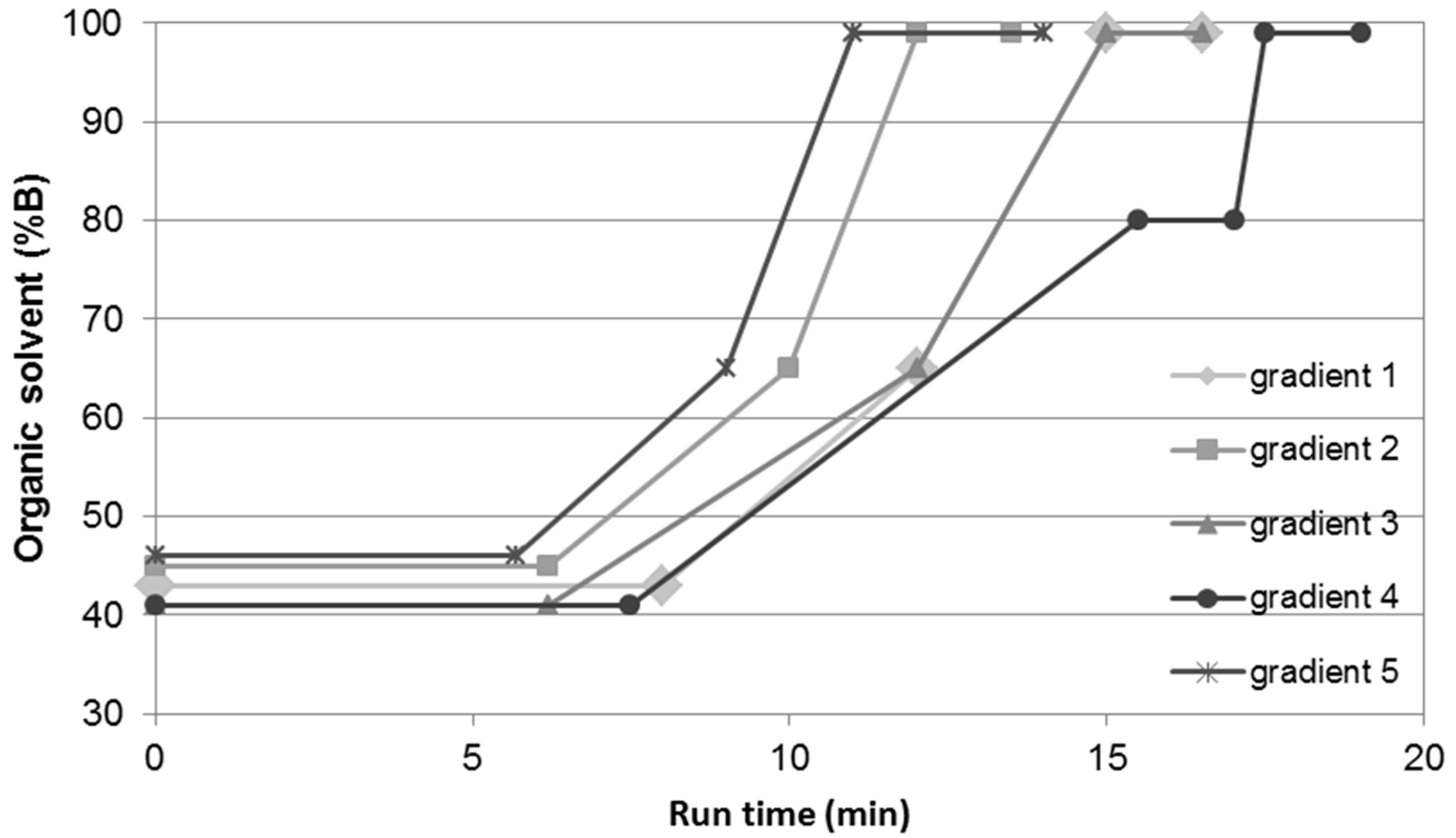
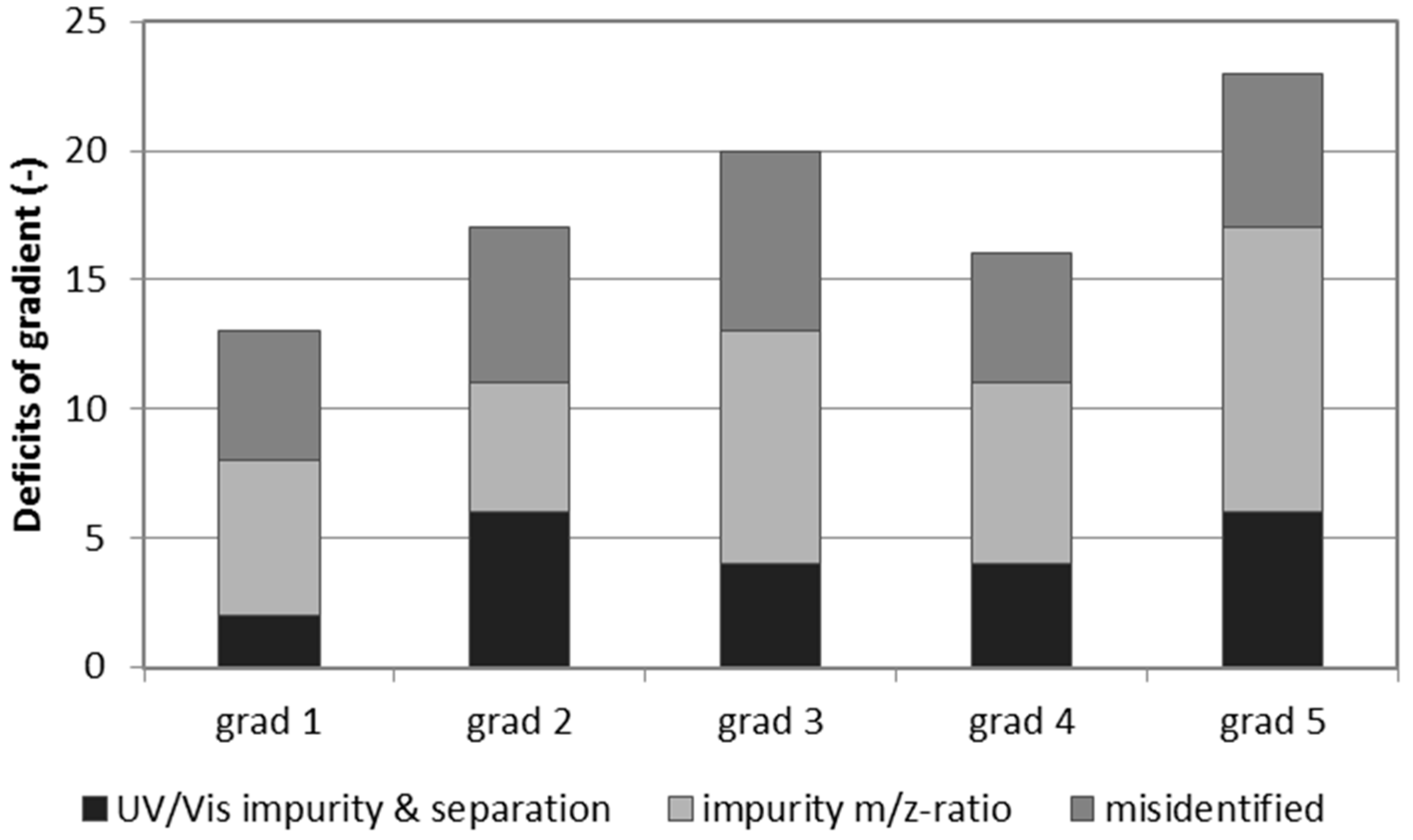
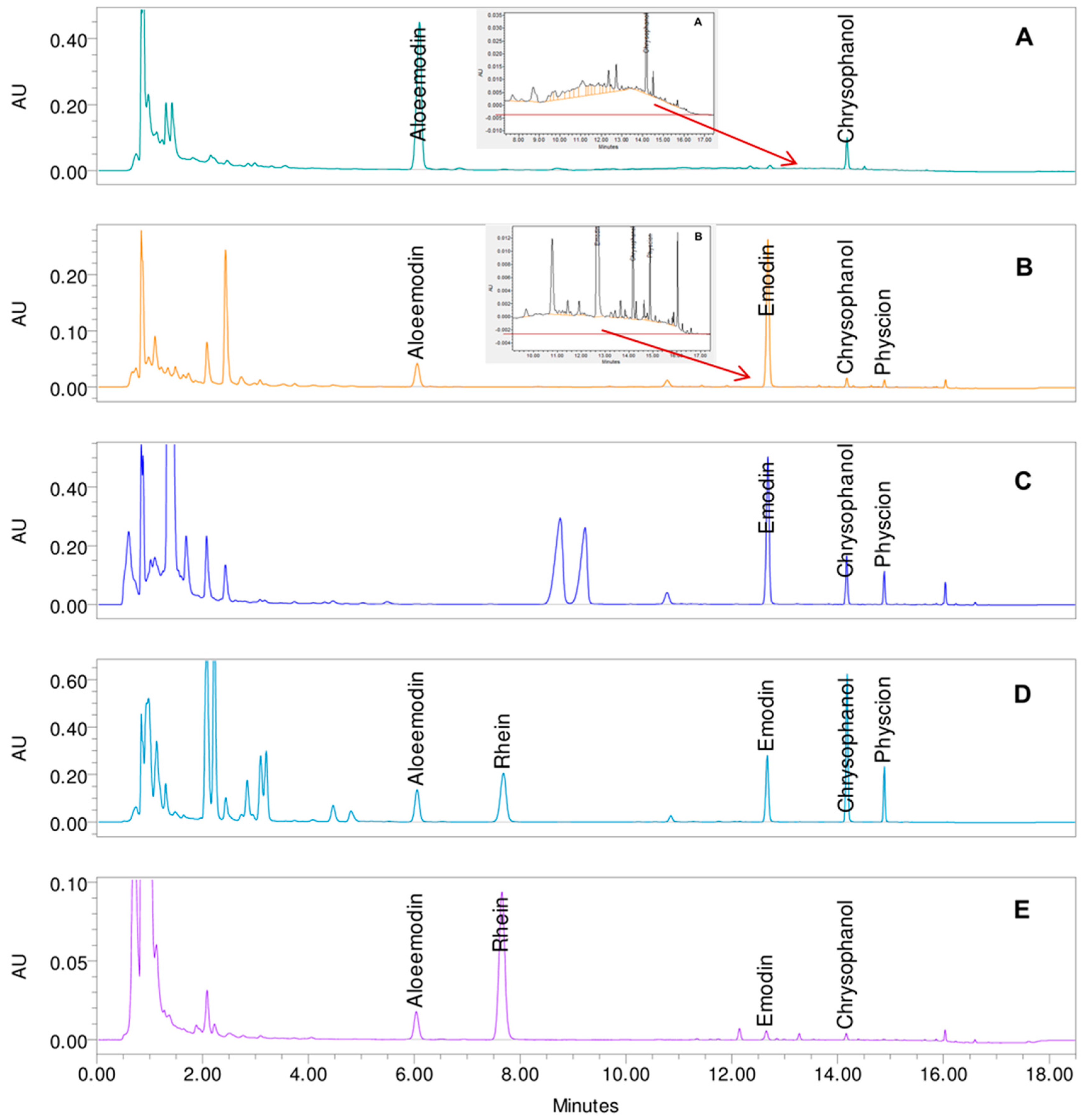
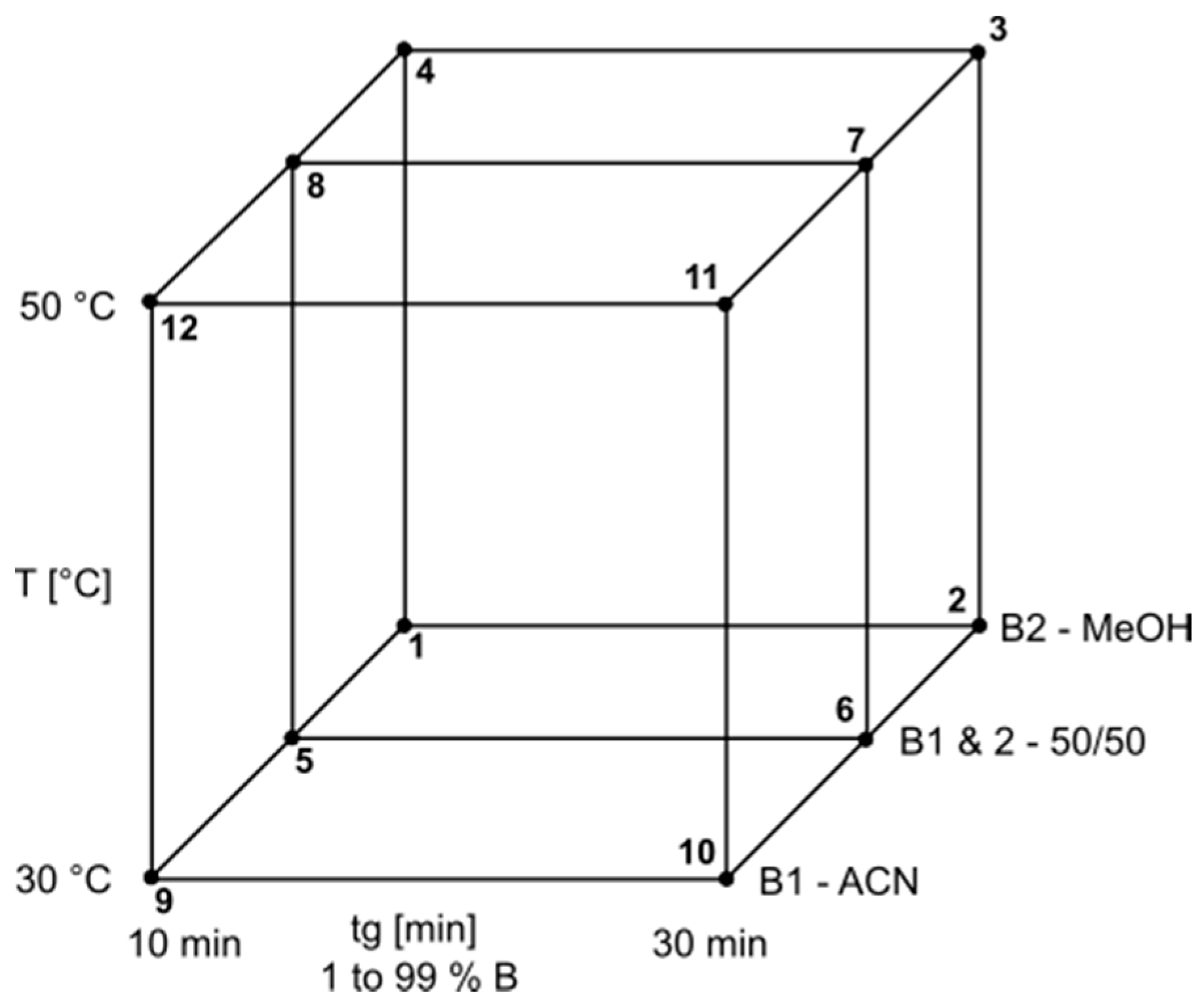
| Gradient 1 | Gradient 2 | Gradient 3 | Gradient 4 | Gradient 5 | ||
|---|---|---|---|---|---|---|
| Reference solutions | AE | |||||
| R | 7.19 | 6.48 | 7.88 | 8.73 | 6.06 | |
| E | 29.33 | 27.79 | 28.22 | 25.66 | 27.23 | |
| C | 17.66 | 17.18 | 18.85 | 18.31 | 16.39 | |
| P | 11.97 | 9.97 | 11.47 | 12.85 | 9.70 | |
| A. barbadensis | AE | 2.22 | impure | n/a | n/a | 1.74 |
| C | n/a | impure | impure | 2.22 | n/a | |
| R. purshianus | AE | 1.44 | n/a | 2.36 | 2.15 | 1.73 |
| E | 4.51 | 4.17 | 5.25 | 4.91 | n/a | |
| C | 1.97 | impure | 2.19 | 3.07 | impure | |
| P | 1.65 | n/a | 1.70 | 1.20 | n/a | |
| R. frangula | E | n/a | n/a | n/a | n/a | 5.52 |
| C | 4.76 | n/a | 4.97 | 5.05 | n/a | |
| P | 2.49 | n/a | impure | n/a | n/a | |
| R. palmatum | AE | 2.68 | 2.75 | 2.65 | impure | impure |
| R | 5.31 | 4.84 | n/a | n/a | 4.12 | |
| E | 4.13 | 4.65 | 5.59 | n/a | impure | |
| C | 3.26 | 3.52 | 3.17 | 13.08 | 2.68 | |
| P | impure | impure | impure | impure | impure | |
| C. angustifolia | AE | 2.77 | 1.41 | n/a | n/a | 2.76 |
| R | 5.65 | 5.12 | n/a | impure | 5.28 | |
| E | 2.10 | impure | 1.70 | 1.88 | impure | |
| C | impure | 2.27 | impure | impure | 1.73 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meier, N.; Meier, B.; Peter, S.; Wolfram, E. In-Silico UHPLC Method Optimization for Aglycones in the Herbal Laxatives Aloe barbadensis Mill., Cassia angustifolia Vahl Pods, Rhamnus frangula L. Bark, Rhamnus purshianus DC. Bark, and Rheum palmatum L. Roots. Molecules 2017, 22, 1838. https://doi.org/10.3390/molecules22111838
Meier N, Meier B, Peter S, Wolfram E. In-Silico UHPLC Method Optimization for Aglycones in the Herbal Laxatives Aloe barbadensis Mill., Cassia angustifolia Vahl Pods, Rhamnus frangula L. Bark, Rhamnus purshianus DC. Bark, and Rheum palmatum L. Roots. Molecules. 2017; 22(11):1838. https://doi.org/10.3390/molecules22111838
Chicago/Turabian StyleMeier, Nadja, Beat Meier, Samuel Peter, and Evelyn Wolfram. 2017. "In-Silico UHPLC Method Optimization for Aglycones in the Herbal Laxatives Aloe barbadensis Mill., Cassia angustifolia Vahl Pods, Rhamnus frangula L. Bark, Rhamnus purshianus DC. Bark, and Rheum palmatum L. Roots" Molecules 22, no. 11: 1838. https://doi.org/10.3390/molecules22111838




