Thymosin α1 Interacts with Hyaluronic Acid Electrostatically by Its Terminal Sequence LKEKK
Abstract
:1. Introduction
2. Results
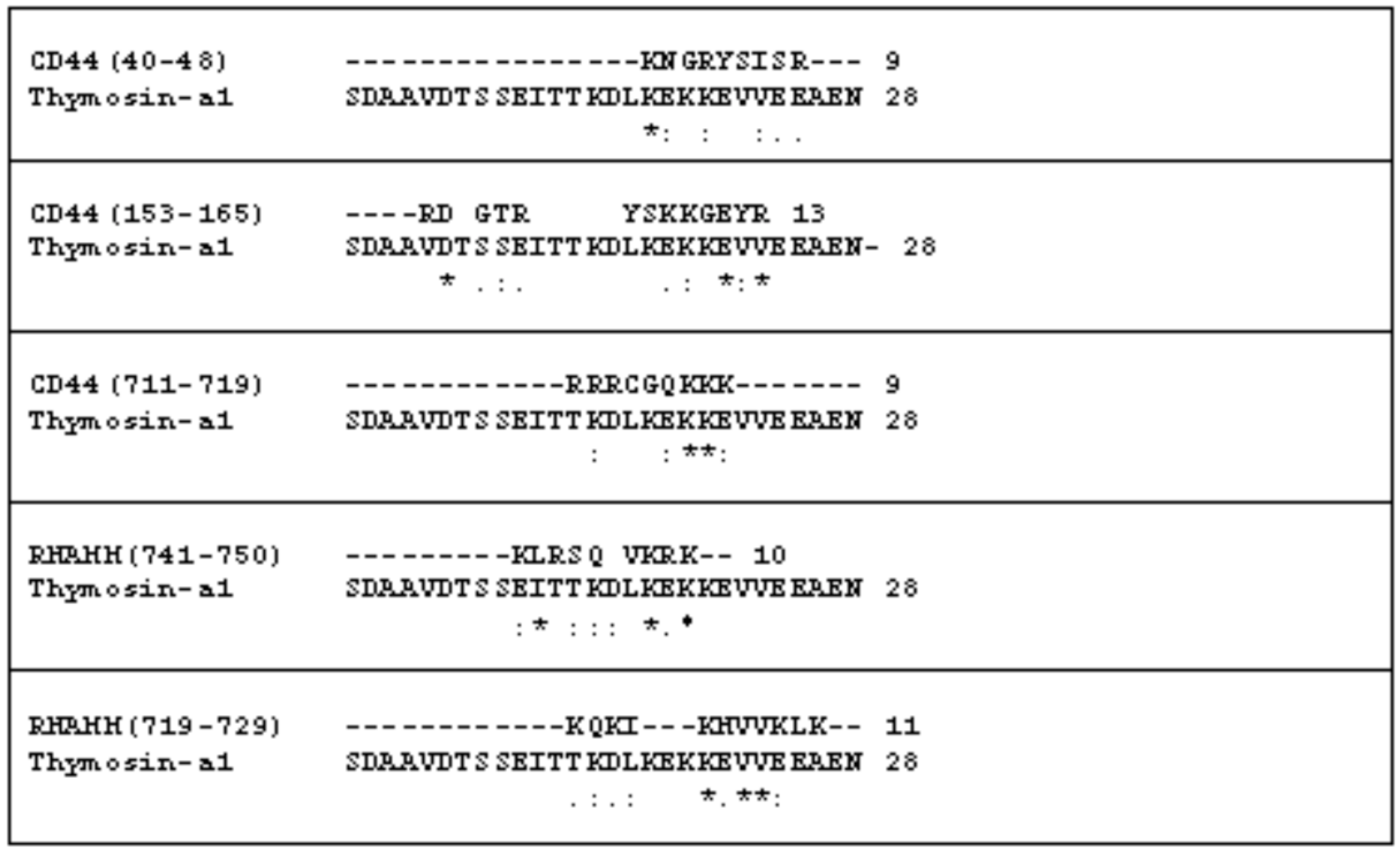
2.1. The Comparison of the Aminoacidic Sequence between Thymosin α1 and the Common HA Binding Sites of RHAMM and CD44
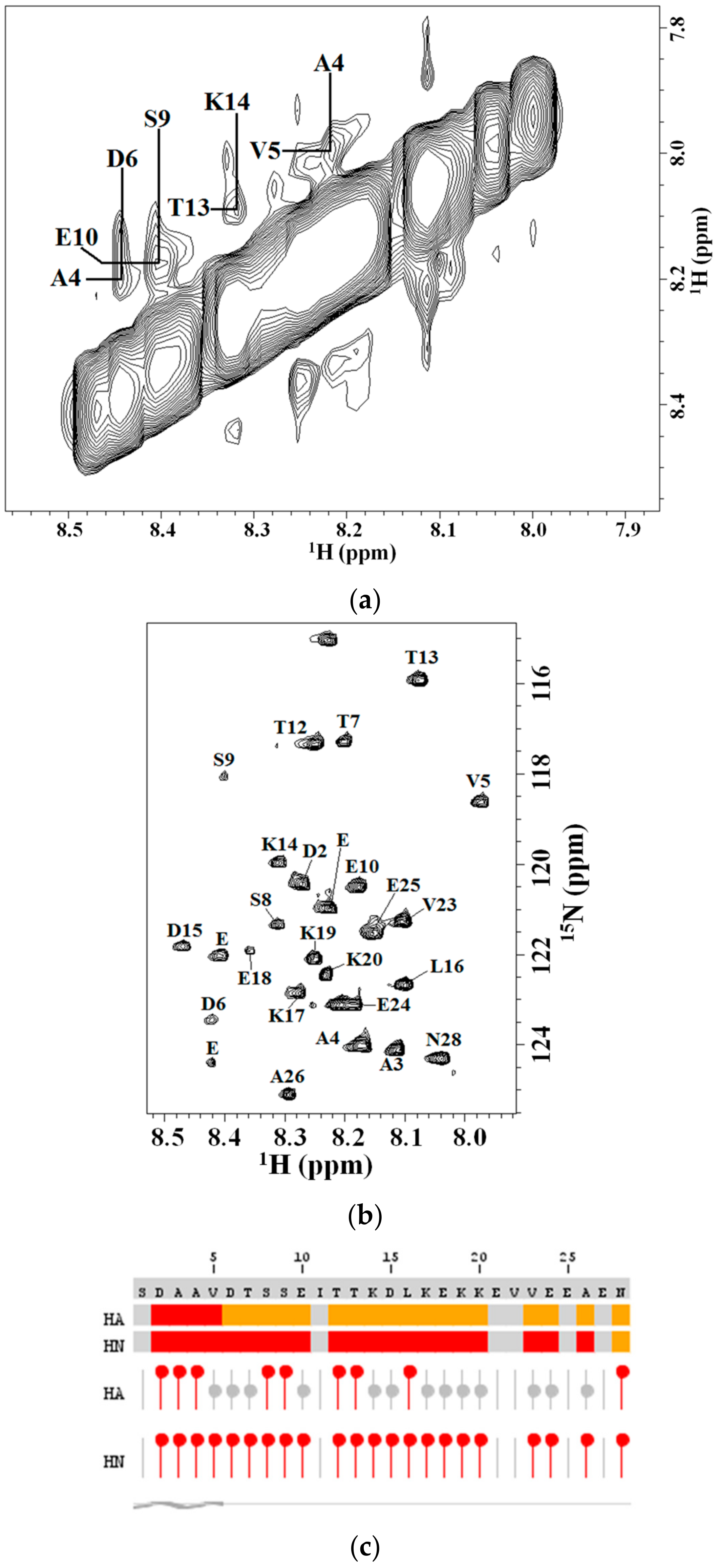
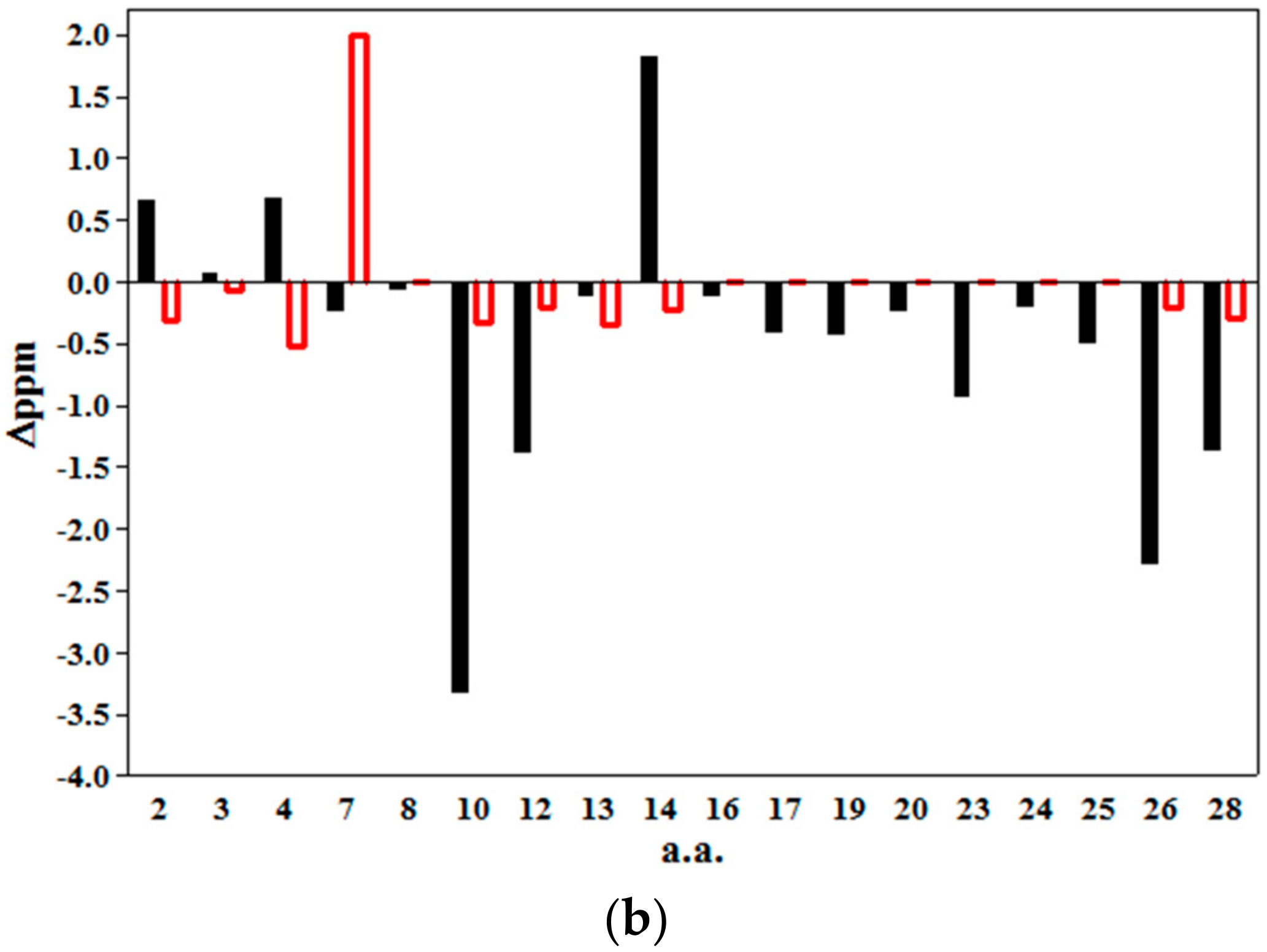
2.2. 2D NMR Studies: Structural Characterization of Tα1 HA Interaction
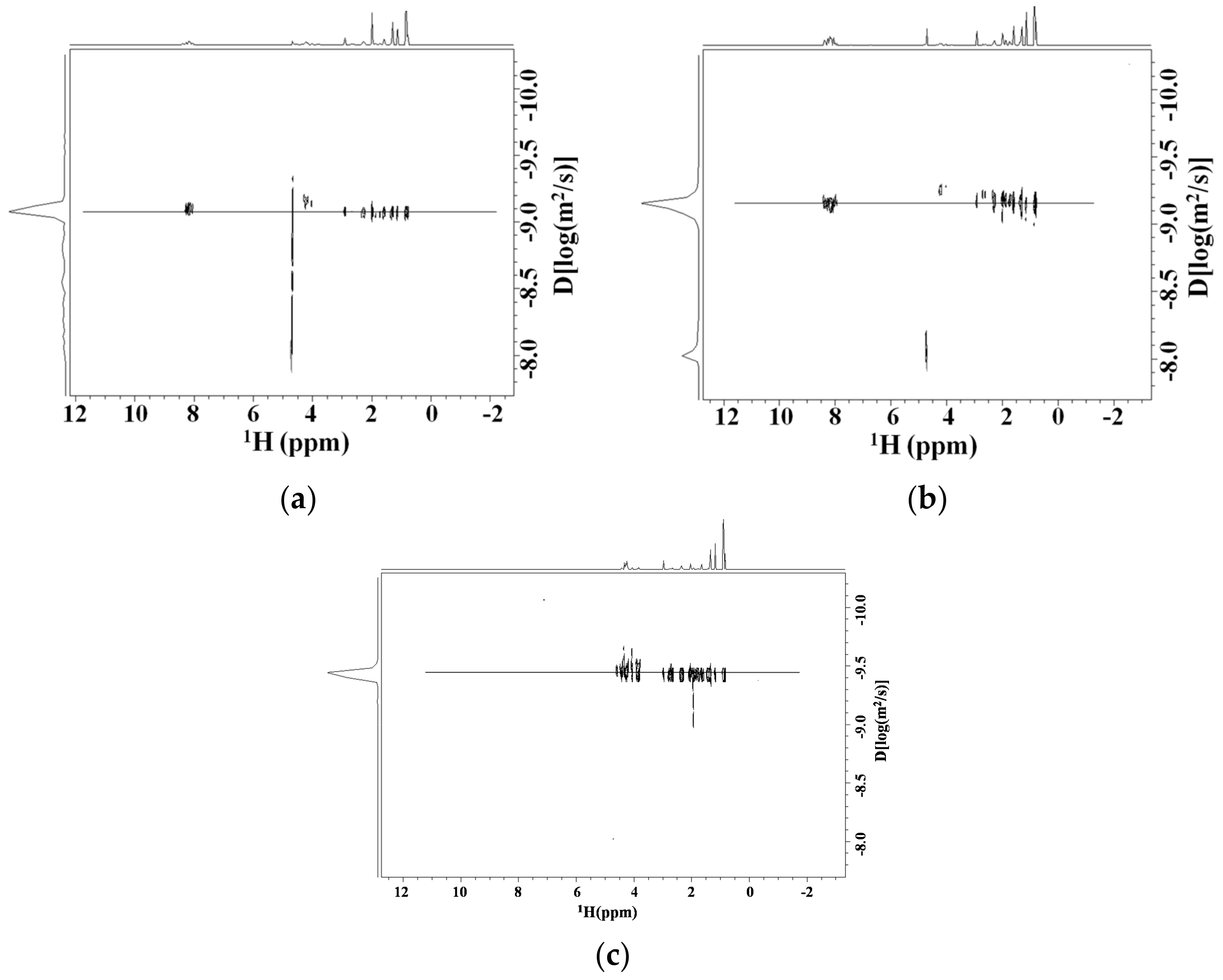
Diffusion NMR Studies
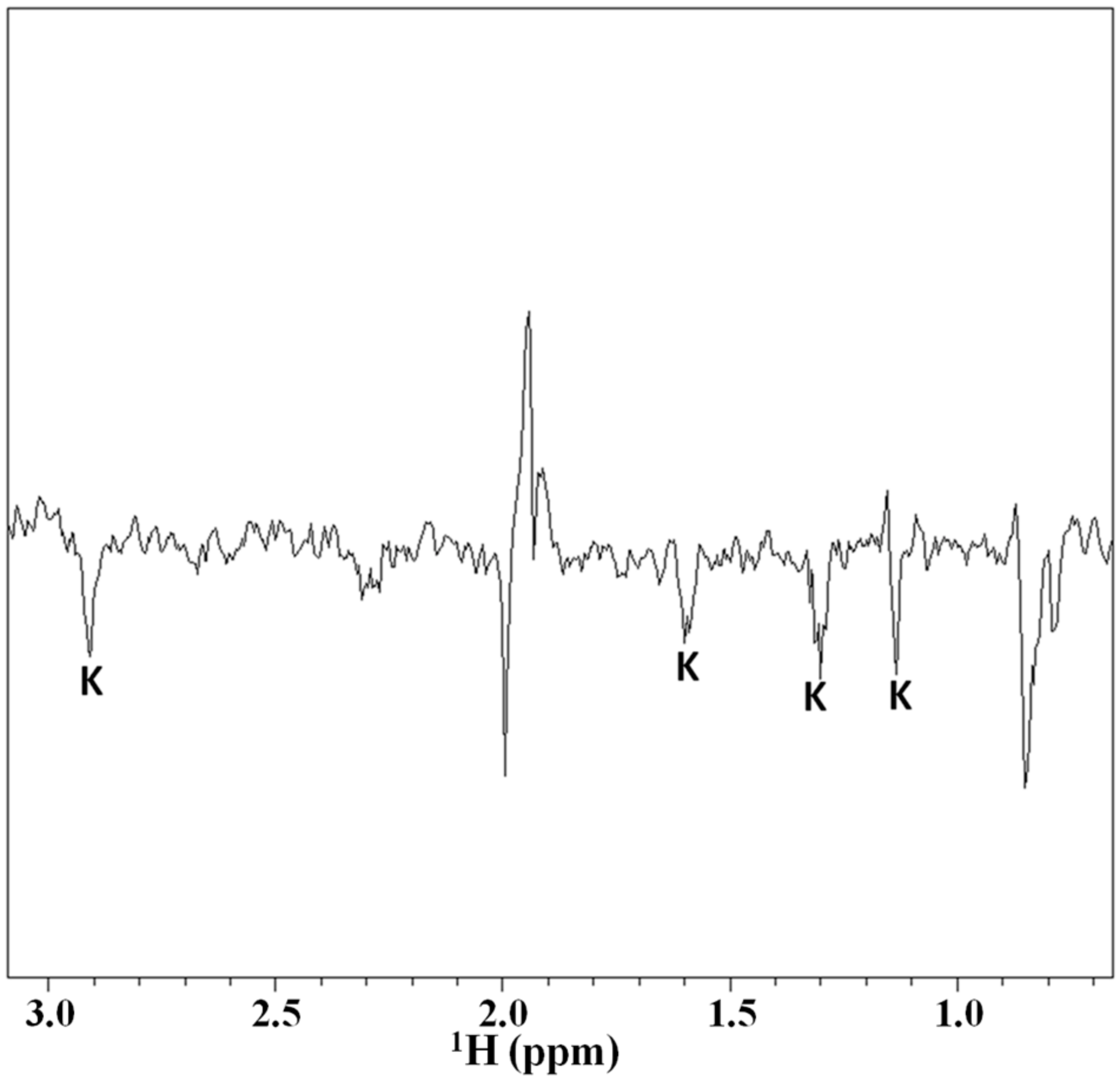
2.3. Magnetization Transfer by WaterLOGSY
2.4. Structuration Propensity of Thymosin α1: In Phospholipidic Vesicles and in Presence of Hyaluronic Acid
3. Discussion
4. Materials and Methods
4.1. Sequence Homology
4.2. Preparation of Samples for NMR Measurements
4.3. NMR Spectroscopy
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Goldstein, A.L.; Guha, A.; Zatz, M.M.; Hardy, M.A.; White, A. Purification and Biological Activity of Thymosin, a Hormone of the Thymus Gland. Proc. Natl. Acad. Sci. USA 1972, 69, 1800–1803. [Google Scholar] [CrossRef] [PubMed]
- Low, T.L.; Thurman, G.B.; McAdoo, M.; McClure, J.; Rossio, J.L.; Naylor, P.H.; Goldstein, A.L. The chemistry and biology of thymosin. I. Isolation, characterization, and biological activities of thymosin alpha1 and polypeptide beta1 from calf thymus. J. Biol. Chem. 1979, 254, 981–986. [Google Scholar]
- Low, T.L.; Goldstein, A.L. The chemistry and biology of thymosin. II. Amino acid sequence analysis of thymosin alpha1 and polypeptide beta1. J. Biol. Chem. 1979, 254, 987–995. [Google Scholar] [PubMed]
- Sarandeses, C.S.; Covelo, G.; Díaz-Jullien, C. Prothymosin alpha is processed to thymosin α1 and thymosin α11 by a lysosomal asparaginyl endopeptidase. J. Biol. Chem. 2003, 278, 13286–13293. [Google Scholar] [CrossRef] [PubMed]
- Franco, F.J.; Diaz, C.; Barcia, M.; Freire, M. Thymosin α1 is a native peptide in several tissues. Biochim. Biophys. Acta 1992, 1120, 43–48. [Google Scholar] [CrossRef]
- Chen, J.M.; Dando, P.M.; Rawlings, N.D.; Brown, M.A.; Young, N.E.; Stevens, R.A.; Hewitt, E.; Watts, C.; Barrett, A.J. Cloning, isolation, and characterization of mammalian legumain, an asparaginyl endopeptidase. J. Biol. Chem. 1997, 272, 8090–8098. [Google Scholar] [CrossRef] [PubMed]
- Romani, L.; Oikonomou, V.; Moretti, S.; Iannitti, R.G.; D’Adamo, M.C.; Villella, V.R.; Pariano, M.; Sforna, L.; Borghi, M.; Bellet, M.M.; et al. Thymosin α1 represents a potentialpotent single-molecule-basedtherapy for cystic fibrosis. Nat. Med. 2017. [Google Scholar] [CrossRef] [PubMed]
- Iino, S.; Toyota, J.; Kumada, H.; Kiyosawa, K.; Kakumu, S.; Sata, M.; Suzuki, H.; Martins, E.B. The efficacy and safety of thymosin alpha-1 in Japanese patients with chronic hepatitis B; results from a randomized clinical trial. J. Viral Hepat. 2005, 12, 300–306. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Zhuang, L.; Cheng, H.Y.; Yan, S.M.; Yu, L.; Huang, J.H.; Tang, B.Z.; Huang, M.L.; Ma, Y.L.; Chongsuvivatwong, V.; et al. Efficacy of thymosin alpha-1 and interferon alpha in treatment of chronic viral hepatitis B: A randomized controlled study. World J. Gastroenterol. 2006, 12, 6715–6721. [Google Scholar] [CrossRef] [PubMed]
- Andreone, P.; Cursaro, C.; Gramenzi, A.; Margotti, M.; Ferri, E.; Talarico, S.; Biselli, M.; Felline, F.; Tuthill, C.; Martins, E.; et al. In vitro effect of thymosin-alpha1 and interferon-alpha on Th1 and Th2 cytokine synthesis in patients with chronic hepatitis C. J. Viral Hepat. 2001, 8, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Kullavanuaya, P.; Treeprasertsuk, S.; Thong-Ngam, D.; Chaermthai, K.; Gonlachanvit, S.; Suwanagool, P. The combined treatment of interferon alpha-2a and thymosin alpha 1 for chronic epatitis C: The 48 weeks end of treatment results. J. Med. Assoc. Thail. 2001, 84 (Suppl. 1), S462–S468. [Google Scholar]
- Carraro, G.; Naso, A.; Montomoli, E.; Gasparini, R.; Camerini, R.; Panatto, D.; Tineo, M.C.; De Giorgi, L.; Piccirella, S.; Khadang, B.; et al. Thymosin-alpha 1 (Zadaxin) enhances the immunogenicity of an adjuvated pandemic H1N1v influenza vaccine (Focetria) in hemodialyzed patients: A pilot study. Vaccine 2011, 30, 1170–1180. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, A.L. From lab to bedside: Emerging clinical applications of thymosin α1. Expert Opin. Biol. Ther. 2009, 9, 593–608. [Google Scholar] [CrossRef] [PubMed]
- Maio, M.; Mackiewicz, A.; Testori, A.; Trefzer, U.; Ferraresi, V.; Jassem, J.; Garbe, C.; Lesimple, T.; Guillot, B.; Gascon, P.; et al. Large randomized study of thymosin α1, interferon alfa, or both in combination with dacarbazine in patients with metastatic melanoma. J. Clin. Oncol. 2010, 28, 1780–1787. [Google Scholar] [CrossRef] [PubMed]
- Billich, A. Thymosin alpha1 SciClone Pharmaceuticals. Curr. Opin. Investig. Drugs 2002, 3, 698–707. [Google Scholar] [PubMed]
- Garaci, E.; Pica, F.; Serafino, A.; Balestrieri, E.; Matteucci, C.; Moroni, G.; Sorrentino, R.; Zonfrillo, M.; Pierimarchi, P.; Sinibaldi Vallebona, P. Thymosin α1 and cancer: Action on immune effector and tumor target cells. Ann. N. Y. Acad. Sci. 2012, 1269, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Serafino, A.; Pica, F.; Andreola, F.; Gaziano, R.; Moroni, N.; Moroni, G.; Zonfrillo, M.; Pierimarchi, P.; Sinibaldi-Vallebona, P.; Garaci, E. Thymosin α1 activates complement receptor-mediated phagocytosis in human monocyte-derived macrophages. J. Innate Immun. 2014, 6, 72–88. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, C.; Napolitano, G.; Mastino, A.; Di Vincenzo, S.; D’Agostini, S.; Grelli, C.; Bucci, I.; Singer, D.S.; Kohn, L.D.; Monaco, F.; et al. Thymosin-α1 regulates MHC class I expression in FRTL-5 cells at transcriptional level. Eur. J. Immunol. 2000, 30, 778–786. [Google Scholar] [CrossRef]
- Knutsen, A.P.; Freeman, J.J.; Mueller, K.R.; Roodman, S.T.; Bouhasin, J.D. Thymosin-α1 stimulates maturation of CD34+ stemcellsinto CD3+4+ cells in an in vitro thymic epithelia organ coculture model. Int. J. Immunopharmacol. 1999, 21, 15–26. [Google Scholar] [CrossRef]
- Romani, L.; Bistoni, F.; Gaziano, R.; Bozza, S.; Montagnoli, C.; Perruccio, K.; Pitzurra, L.; Bellocchio, S.; Velardi, A.; Rasi, G.; et al. Thymosin α1 activates dendritic cells for antifungal Th1 resistance through toll-like receptor signaling. Blood 2004, 103, 4232–4239. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Chan, J.; Dragoi, A.M.; Gong, X.; Ivanov, S.; Li, Z.W.; Chuang, T.; Tuthill, C.; Wan, Y.; Karin, M.; et al. Activation of IKK by thymosin α1 requires the TRAF6 signaling pathway. EMBO Rep. 2005, 6, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Leichtling, K.D.; Serrate, S.A.; Sztein, M.B. Thymosin α1 modulates the expression of high-affinity IL-2 receptor on normal human lymphocytes. Int. J. Immunopharmacol. 1990, 12, 19–29. [Google Scholar] [CrossRef]
- Pica, F.; Chimenti, M.S.; Gaziano, R.; Buè, C.; Casalinuovo, I.A.; Triggianese, P.; Conigliaro, P.; Di Carlo, D.; Cordero, V.; Adorno, G.; et al. Serum thymosin α1 levels in patients with chronic inflammatory autoimmune diseases. Clin. Exp. Immunol. 2016, 186, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Romani, L.; Moretti, S.; Fallarino, F.; Bozza, S.; Ruggeri, L.; Casagrande, A.; Aversa, F.; Bistoni, F.; Velardi, A.; Garaci, E. Jack of all trades: Thymosin α1 and its pleiotropy. Ann. N. Y. Acad. Sci. 2012, 1269, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Grottesi, A.; Sette, M.; Palamara, T. The conformation of peptide thymosin α1 in solution and in a membrane like environment by circular dichroism and NMR spectroscopy. A possible model for its interaction with the lymphocyte membrane. Peptides 1998, 19, 1731–1738. [Google Scholar] [CrossRef]
- Elizondo-Riojas, M.A.; Chamow, S.M.; Tuthill, C.W.; Gorenstein, D.G.; Volk, D.E. NMR structure of human thymosin alpha-1. Biochem. Biophys. Res. Commun. 2011, 416, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Nepravishta, R.; Mandaliti, W.; Eliseo, T.; Vallebona, P.S.; Pica, F.; Garaci, E.; Paci, M. Thymosin α1 inserts N terminus into model membranes assuming a helical conformation. Expert Opin. Biol. Ther. 2015, 2, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mandaliti, W.; Nepravishta, R.; Vallebona, P.S.; Pica, F.; Garaci, E.; Paci, M. New studies about the insertion mechanism of Thymosin α1 in negative regions of model membranes as starting point of the bioactivity. Amino Acids 2016, 48, 1231–1239. [Google Scholar] [CrossRef] [PubMed]
- Mandaliti, W.; Nepravishta, R.; Sinibaldi Vallebona, P.; Pica, F.; Garaci, E.; Paci, M. Thymosin α1 Interacts with Exposed Phosphatidylserine in Membrane Models and in Cells and Uses Serum Albumin as a Carrier. Biochemistry 2016, 15, 1462–1472. [Google Scholar] [CrossRef] [PubMed]
- Nepravishta, R.; Mandaliti, W.; Sinibaldi Vallebona, P.; Pica, F.; Garaci, E.; Paci, M. Mechanism of action of Thymosin α1: Does it interact with membrane by recognition of exposed phosphatidylserine on cell surface? A structural approach. Vitam. Horm. 2016, 102, 101–119. [Google Scholar] [PubMed]
- Marino, G.; Kroemer, G. Mechanisms of apoptotic phosphatidylserine exposure. Cell Res. 2013, 23, 1247–1248. [Google Scholar] [CrossRef] [PubMed]
- Fraser, J.R.; Laurent, T.C.; Laurent, U.B. Hyaluronan: Its nature, distribution, functions and turnover. J. Intern. Med. 1997, 242, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Shirali, A.C.; Goldstein, D.R. Activation of the innate immune system by the endogenous ligand hyaluronan. Curr. Opin. Organ Transplant. 2008, 13, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Senbanjo, L.T.; Chellaiah, M.A. CD44: A Multifunctional Cell Surface Adhesion Receptor Is a Regulator ofProgression and Metastasis of Cancer Cells. Front. Cell Dev. Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Toole, B.P. Hyaluronan and its binding proteins, the hyaladherins. Curr. Opin. Cell Biol. 1990, 2, 839–844. [Google Scholar] [CrossRef]
- Lesley, J.; Hascall, V.C.; Tammii, M.; Hyman, R. Hyaluronan Binding by Cell Surface CD44. J. Biol. Chem. 2000, 275, 26967–26975. [Google Scholar] [CrossRef] [PubMed]
- Schwertfeger, K.L.; Cowman, M.K.; Telmer, P.G.; Turley, E.A.; McCarthy, J.B. Hyaluronan, Inflammation, and Breast Cancer Progression. Front. Immunol. 2015, 6, 236–238. [Google Scholar] [CrossRef] [PubMed]
- Ponta, H.; Sherman, L.; Herrlich, P.A. CD44: From Adhesion Molecules to Signalling Regulators. Nat. Rev. Mol. Cell Biol. 2003, 4, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Louderbough, J.M.; Schroeder, J.A. Understanding the dual nature of CD44 in breast cancer progression. Mol. Cancer Res. 2011, 12, 1573–1586. [Google Scholar] [CrossRef] [PubMed]
- Peterson, R.M.; Yu, Q.; Stamenkovic, I.; Toole, B.P. Perturbation of Hyaluronan Interactions by Soluble CD44 Inhibits Growth of Murine Mammary Carcinoma Cells in Ascites. Am. J. Pathol. 2000, 56, 2159–2167. [Google Scholar] [CrossRef]
- Peach, R.J.; Hollenbaugh, D.; Stamenkovic, I.; Aruffo, A. Identification of Hyaluronic Acid Binding Sites in the Extracellular Domain of CD44. J. Cell Biol. 1993, 122, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Bourguignon, L.Y.; Shiina, M.; Li, J.J. Hyaluronan-CD44 interaction promotes oncogenic signaling, microRNA functions, chemoresistance, and radiation resistance in cancer stem cells leading to tumor progression. Adv. Cancer Res. 2014, 123, 255–275. [Google Scholar] [PubMed]
- Perschl, A.; Lesley, J.; English, N.; Trowbridge, I.; Hyman, R. Role of CD44 cytoplasmic domain in hyaluronan binding. Eur. J. Immunol. 1995, 25, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Ghatak, S.; Hascall, V.C.; Markwald, R.R.; Misra, S. Stromal hyaluronan interaction with epithelial CD44 variants promotes prostate cancer invasiveness by augmenting expression and function of hepatocyte growth factor and androgen receptor. J. Biol. Chem. 2010, 285, 19821–19832. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Zhang, L.; Turley, E.A. Identification of Two Hyaluronan-binding Domains in the Hyaluronan Receptor RHAMM. J. Biol. Chem. 1993, 268, 8617–8623. [Google Scholar] [PubMed]
- Yang, B.; Yang, B.L.; Savani, R.C.; Turley, E.A. Identification of a common hyaluronan binding motif in the hyaluronan binding proteins RHAMM, CD44 and link protein. EMBO J. 1994, 13, 286–296. [Google Scholar] [PubMed]
- Misra, S.; Hascall, V.C.; Markwald, R.R.; Ghatak, S. Interactions between Hyaluronan and Its Receptors (CD44, RHAMM) Regulate the Activities of Inflammation and Cancer. Front. Immunol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Turley, E.A.; Naor, D. RHAMM and CD44 peptides-analytic tools and potential drugs. Front. Biosci. (Landmark Ed.) 2012, 17, 1775–1794. [Google Scholar] [CrossRef] [PubMed]
- Tolg, C.; Hamilton, S.R.; Zalinska, E.; McCulloch, L.; Amin, R.; Akentieva, N.; Winnik, F.; Savani, R.; Bagli, D.J.; Luyt, L.G.; et al. A RHAMM mimetic peptide blocks hyaluronan signaling and reduces inflammation and fibrogenesis in excisional skin wounds. Am. J. Pathol. 2012, 181, 1250–1270. [Google Scholar] [CrossRef] [PubMed]
- Esguerra, K.V.; Tolg, C.; Akentieva, N.; Price, M.; Cho, C.F.; Lewis, J.D.; McCarthy, J.B.; Turley, E.A.; Luyt, L.G. Identification, design and synthesis of tubulin-derived peptides as novel hyaluronan mimetic ligands for the receptor for hyaluronan-mediated motility (RHAMM/HMMR). Integr. Biol. 2015, 7, 1547–1560. [Google Scholar] [CrossRef] [PubMed]
- Mummert, M.E.; Mohamadzadeh, M.; Mummert, D.I.; Mizumoto, N.; Takashima, A. Development of a Peptide Inhibitor of Hyaluronan-Mediated Leukocyte Trafficking. J. Exp. Med. 2000, 192, 769–779. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Xu, X.M.; Chen, J.; Wang, L.; Yang, S.; Underhill, C.B.; Zhang, L. Hyaluronan-Binding Peptide Can Inhibit Tumor Growth by Interacting with Bcl-2. Int. J. Cancer 2004, 109, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.M.; Chen, Y.; Chen, J.; Yang, S.; Gao, F.; Underhill, C.B.; Creswell, K.; Zhang, L. A Peptide with Three Hyaluronan Binding Motifs Inhibits Tumor Growth and Induces Apoptosis. Cancer Res. 2003, 63, 5685–5690. [Google Scholar] [PubMed]
- Hamilton, S.R.; Fard, S.F.; Paiwand, F.F.; Tolg, C.; Veiseh, M.; Wang, C.; McCarthy, J.B.; Bissell, M.J.; Koropatnick, J.; Turley, E.A. The Hyaluronan Receptors CD44 and Rhamm (CD168) Form Complexes with ERK1,2 ThatSustain High BasalMotility in Breast Cancer Cells. J. Biol. Chem. 2007, 282, 16667–16680. [Google Scholar] [CrossRef] [PubMed]
- Teriete, P.; Banerji, S.; Noble, M.; Blundell, C.; Wright, A.; Pickford, A.; Lowe, E.; Mahoney, D.; Tammi, M.; Kahmann, J.; et al. Structure of the Regulatory Hyaluronan-Binding Domain in the Inflammatory Leukocyte Homing Receptor CD44. Mol. Cell 2004, 13, 483–489. [Google Scholar] [CrossRef]
- Wüthrich, K. NMR of Proteins and Nucleic Acids; John Wiley & Sons: New York, NY, USA, 1986. [Google Scholar]
- Wishart, D.S.; Sykes, B.D.; Richards, F.M. The Chemical Shift Index: A Fast and Simple Method for the Assignment of Protein Secondary Structure through NMR Spectroscopy. J. Mol. Biol. 1991, 222, 311–333. [Google Scholar] [CrossRef]
- Williamson, M.P. Using chemical shift perturbation to characterize ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 2013, 73, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Ziebell, M.R.; Prestwich, G.D. Interactions of peptide mimics of hyaluronic acid with the receptor for hyaluronan mediated motility (RHAMM). J. Comput. Aided Mol. Des. 2004, 18, 597–614. [Google Scholar] [CrossRef] [PubMed]
- Navolotskaya, E.V.; Zinchenko, D.V.; Zolotarev, Y.A.; Kolobov, A.A.; Lipkin, V.M. Binding of Synthetic LKEKK Peptide to Human T-Lymphocytes. Biochemistry (Mosc.) 2016, 81, 871–875. [Google Scholar] [CrossRef] [PubMed]
- Pilarski, L.M.; Pruski, E.; Wizniak, J.; Paine, D.; Seeberger, K.; Mant, M.J.; Brown, C.B.; Belch, A.R. Potential role for hyaluronan and the hyaluronan receptor RHAMM in mobilization and trafficking of hematopoietic progenitor cells. Blood 1999, 93, 2918–2927. [Google Scholar] [PubMed]
- Turley, E.A.; Austen, L.; Vandeligt, K.; Clary, C. Hyaluronan and a cell-associated hyaluronan binding protein regulate the locomotion of ras-transformed cells. J. Cell Biol. 1991, 112, 1041–1047. [Google Scholar] [CrossRef] [PubMed]
- Garaci, E.; Mastino, A.; Pica, F.; Favalli, C. Combination treatment using thymosin alpha 1 and interferon after cyclophosphamide is able to cure Lewis lung carcinoma in mice. Cancer Immunol. Immunother. 1990, 32, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Mastino, A.; Favalli, C.; Grelli, S.; Rasi, G.; Pica, F.; Goldstein, A.L.; Garaci, E. Combination therapy with thymosin alpha 1 potentiates the anti-tumor activity of interleukin-2 with cyclophosphamide in the treatment of the Lewis lung carcinoma in mice. Int. J. Cancer. 1992, 50, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Garaci, E.; Pica, F.; Mastino, A.; Palamara, A.T.; Belardelli, F.; Favalli, C. Antitumor effect of thymosin alpha 1/interleukin-2 or thymosin alpha 1/interferon alpha, beta following cyclophosphamide in mice injected with highly metastatic Friend erythroleukemia cells. J. Immunother. Emphas. Tumor Immunol. 1993, 13, 7–17. [Google Scholar] [CrossRef]
- Clemente, C.G.; Mihm, M.C., Jr.; Bufalino, R.; Zurrida, S.; Collini, P.; Cascinelli, N. Prognostic value of tumor infiltrating lymphocytes in the vertical growth phase of primary cutaneous melanoma. Cancer 1996, 77, 1303–1310. [Google Scholar] [CrossRef]
- Osheroff, P.L. The effect of thymosin on glucocorticoid receptors in lymphoid cells. Cell. Immunol. 1981, 60, 376–385. [Google Scholar] [CrossRef]
- Baumann, C.A.; Badamchian, M.; Goldstein, A.L. Thymosin alpha 1 antagonizes dexamethasone and CD3-induced apoptosis of CD4+ CD8+ thymocytes through the activation of cAMP and protein kinase C dependent second messenger pathways. Mech. Ageing Dev. 1997, 94, 85–101. [Google Scholar] [CrossRef]
- Ho, A.D.; Stehle, B.; Dietz, G.; Hunstein, W.; Hoffbrand, A.V. Terminal differentiation of cord blood lymphocytes induced by thymosin fraction 5 and thymosin alpha 1. Scand. J. Immunol. 1985, 21, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Ohta, Y.; Sueki, K.; Yoneyama, Y.; Tezuka, E.; Yagi, Y. Immunomodulating activity of thymosin fraction 5 and thymosin alpha 1 in immunosuppressed mice. Cancer Immunol. Immunother. 1983, 15, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Ohta, Y.; Tezuka, E.; Tamura, S.; Yagi, Y. Thymosin alpha 1 exerts protective effect against the 5-FU induced bone marrow toxicity. Int. J. Immunopharmacol. 1985, 7, 761–768. [Google Scholar] [CrossRef]
- Hu, S.K.; Low, T.L.; Goldstein, A.L. Modulation of terminal deoxynucleotidyl transferase activity by thymosin. Mol. Cell. Biochem. 1981, 41, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Garaci, E.; Pica, F.; Matteucci, C.; Gaziano, R.; D’Agostini, C.; Miele, M.T.; Camerini, R.; Palamara, A.T.; Favalli, C.; Mastino, A.; et al. Historicalreview on thymosin α1 in oncology: Preclinical and clinicalexperiences. Expert Opin. Biol. Ther. 2015, 15 (Suppl. 1), S31–S39. [Google Scholar] [CrossRef] [PubMed]
- Tuthill, C.W.; King, R.S. Thymosin Apha 1–A Peptide Immune Modulator with a Broad Range of Clinical Applications. Clin. Exp. Pharmacol. 2013, 3, 4. [Google Scholar]
- Malinda, K.M.; Sidhu, G.S.; Banaudha, K.K.; Gaddipati, J.P.; Maheshwari, R.K.; Goldstein, A.L.; Kleinman, H.K. Thymosin alpha 1 stimulates endothelial cell migration, angiogenesis, and wound healing. J. Immunol. 1998, 160, 1001–1006. [Google Scholar] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.G.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol. Syst. Biol. 2011. [Google Scholar] [CrossRef] [PubMed]
- Marion, D.; Wuthrich, K. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of (1)H-(1)H spin-spin coupling constants in proteins. Biochem. Biophys. Res. Commun. 1983, 113, 967–974. [Google Scholar] [CrossRef]
- Bax, A.; Davis, D.G. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 1985, 65, 355–360. [Google Scholar] [CrossRef]
- Braunschweiler, L.; Ernst, R.R. Coherence transfer by isotropic mixing: Application to proton correlation spectroscopy. J. Magn. Reson. 1983, 3, 521–528. [Google Scholar] [CrossRef]
- Jeener, J.; Meier, B.H.; Bachmann, P.; Ernst, R.R. Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys. 1979, 71, 4546–4553. [Google Scholar] [CrossRef]
- Bodenhausen, G.; Reuben, D.J. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 1980, 69, 185–189. [Google Scholar] [CrossRef] [Green Version]
- Morris, K.F.; Johnson, C.S., Jr. Diffusion-ordered two-dimensional nuclear magnetic resonance spectroscopy. J. Am. Chem. Soc. 1992, 114, 3139–3141. [Google Scholar] [CrossRef]
- Cohen, Y.; Avram, L.; Frish, L. DiffusionNMR Spectroscopy in Supramolecular and Combinatorial Chemistry: An OldParameter-New Insights. Angew. Chem. Int. Ed. 2005, 44, 520–554. [Google Scholar] [CrossRef] [PubMed]
- Dalvit, C.; Fogliatto, G.; Stewart, A.; Veronesi, M.; Stockman, B. WaterLOGSY as a method for primary NMR screening: Practicalaspects and range of applicability. J. Biomol. NMR 2001, 21, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Dalvit, C.; Pevarello, P.; Tatò, M.; Veronesi, M.; Vulpetti, A.; Sundström, M. Identification of compounds with bindingaffinity to proteins via magnetization transfer from bulk water. J. Biomol. NMR 2000, 18, 65–68. [Google Scholar] [CrossRef] [PubMed]
- Hwang, L.; Shaka, A.J. Water suppressionthatworks. Excitationsculptingusingarbitrarywaveforms and pulsedfieldgradients. J. Magn. Reson. Ser. A 1995, 112, 275–279. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mandaliti, W.; Nepravishta, R.; Pica, F.; Vallebona, P.S.; Garaci, E.; Paci, M. Thymosin α1 Interacts with Hyaluronic Acid Electrostatically by Its Terminal Sequence LKEKK. Molecules 2017, 22, 1843. https://doi.org/10.3390/molecules22111843
Mandaliti W, Nepravishta R, Pica F, Vallebona PS, Garaci E, Paci M. Thymosin α1 Interacts with Hyaluronic Acid Electrostatically by Its Terminal Sequence LKEKK. Molecules. 2017; 22(11):1843. https://doi.org/10.3390/molecules22111843
Chicago/Turabian StyleMandaliti, Walter, Ridvan Nepravishta, Francesca Pica, Paola Sinibaldi Vallebona, Enrico Garaci, and Maurizio Paci. 2017. "Thymosin α1 Interacts with Hyaluronic Acid Electrostatically by Its Terminal Sequence LKEKK" Molecules 22, no. 11: 1843. https://doi.org/10.3390/molecules22111843





