Sulforaphane Alters β-Naphthoflavone-Induced Changes in Activity and Expression of Drug-Metabolizing Enzymes in Rat Hepatocytes
Abstract
:1. Introduction
2. Results
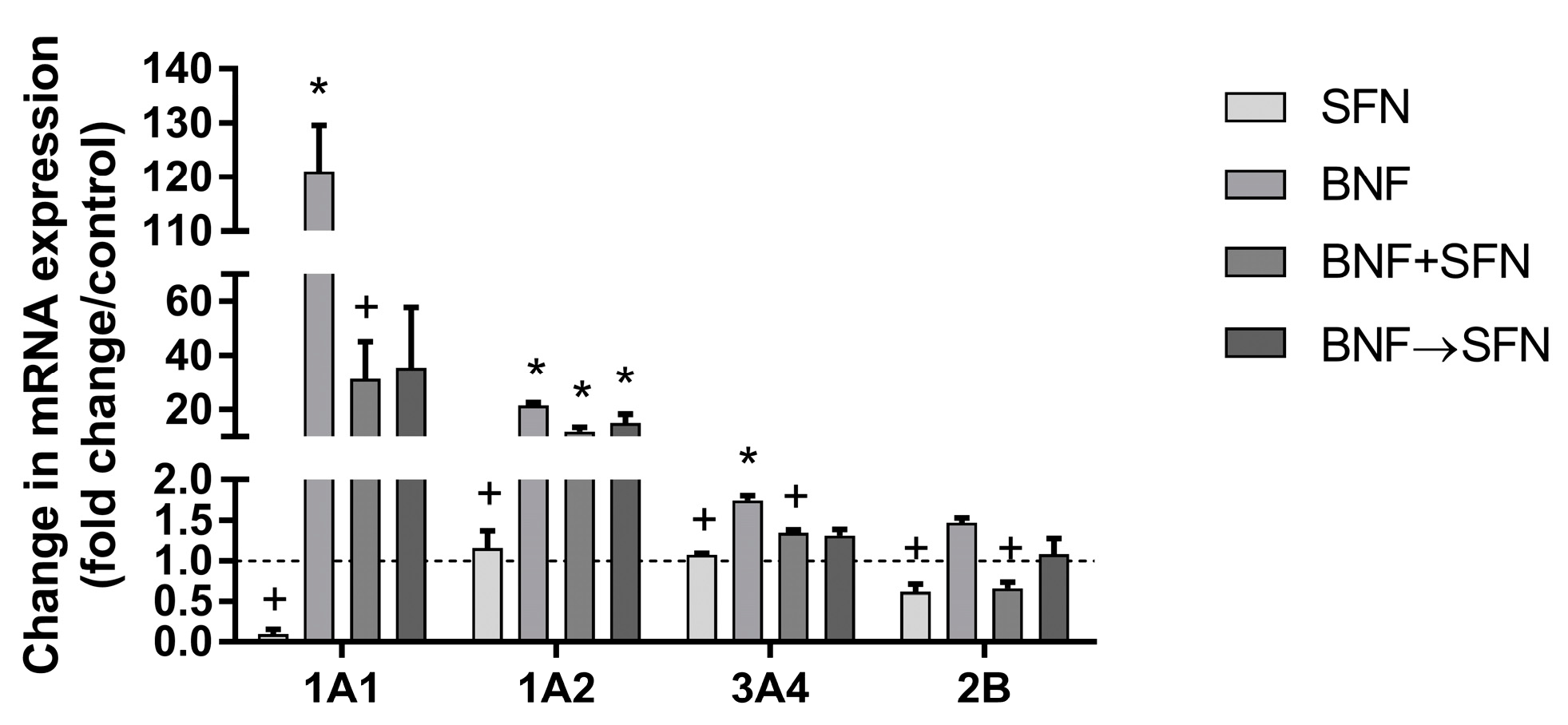
2.1. Effects of SFN on Cytochrome P450 Enzymes
2.2. Effects of SFN on Carbonyl-Reducing Enzymes
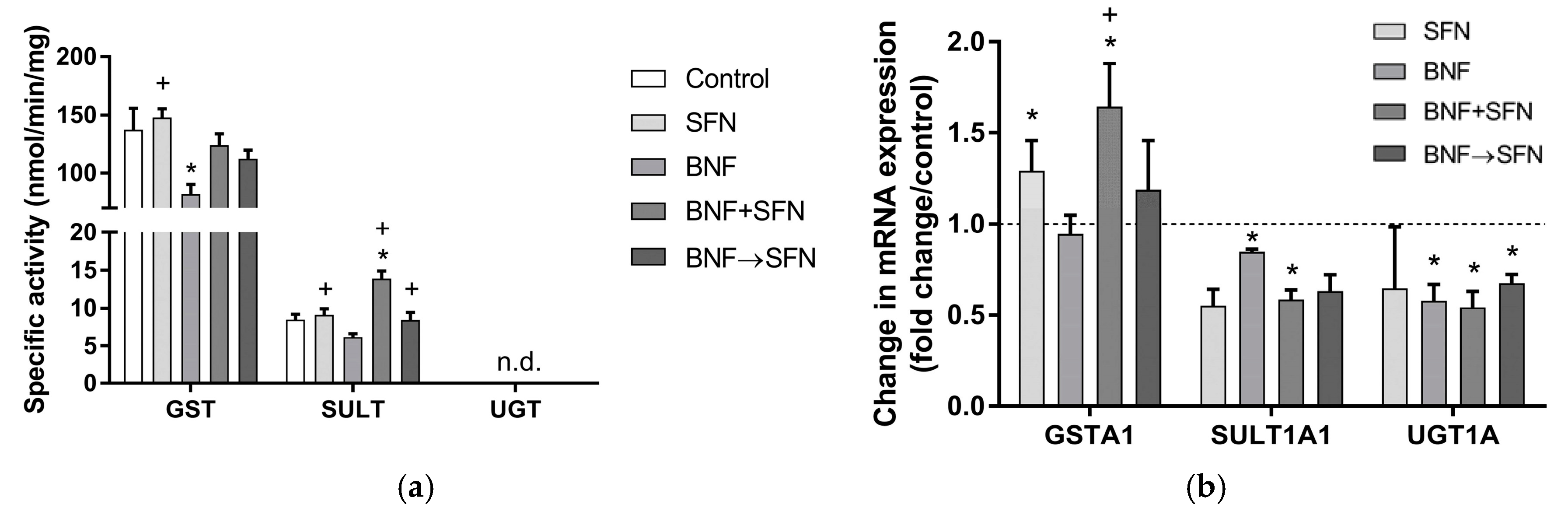
2.3. Effects of SFN on Conjugation Enzymes
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Isolation and Culture of Rat Hepatocytes
4.3. Preparation of Subcellular Fractions
4.4. RNA Isolation and Quantitative Real-Time PCR (qPCR)
4.5. Enzyme Assays
4.6. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Royston, K.J.; Tollefsbol, T.O. The Epigenetic Impact of Cruciferous Vegetables on Cancer Prevention. Curr. Pharmacol. Rep. 2015, 1, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Tarozzi, A.; Angeloni, C.; Malaguti, M.; Morroni, F.; Hrelia, S.; Hrelia, P. Sulforaphane as a Potential Protective Phytochemical against Neurodegenerative Diseases. Oxid. Med. Cell. Longev. 2013, 2013, 415078. [Google Scholar] [CrossRef] [PubMed]
- Angelino, D.; Jeffery, E. Glucosinolate hydrolysis and bioavailability of resulting isothiocyanates: Focus on glucoraphanin. J. Funct. Foods 2014, 7, 67–76. [Google Scholar] [CrossRef]
- Atwell, L.L.; Hsu, A.; Wong, C.P.; Stevens, J.F.; Bella, D.; Yu, T.W.; Pereira, C.B.; Lohr, C.V.; Christensen, J.M.; Dashwood, R.H.; et al. Absorption and chemopreventive targets of sulforaphane in humans following consumption of broccoli sprouts or a myrosinase-treated broccoli sprout extract. Mol. Nutr. Food Res. 2015, 59, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Gasper, A.V.; Al-Janobi, A.; Smith, J.A.; Bacon, J.R.; Fortun, P.; Atherton, C.; Taylor, M.A.; Hawkey, C.J.; Barrett, D.A.; Mithen, R.F. Glutathione S-transferase M1 polymorphism and metabolism of sulforaphane from standard and high-glucosinolate broccoli. Am. J. Clin. Nutr. 2005, 82, 1283–1291. [Google Scholar] [PubMed]
- La Marca, M.; Beffy, P.; Della Croce, C.; Gervasi, P.G.; Iori, R.; Puccinelli, E.; Longo, V. Structural influence of isothiocyanates on expression of cytochrome P450, phase II enzymes, and activation of Nrf2 in primary rat hepatocytes. Food Chem. Toxicol. 2012, 50, 2822–2830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheung, K.L.; Kong, A.N. Molecular targets of dietary phenethyl isothiocyanate and sulforaphane for cancer chemoprevention. AAPS J. 2010, 12, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Petri, N.; Tannergren, C.; Holst, B.; Mellon, F.A.; Bao, Y.; Plumb, G.W.; Bacon, J.; O’Leary, K.A.; Kroon, P.A.; Knutson, L.; et al. Absorption/metabolism of sulforaphane and quercetin, and regulation of phase II enzymes, in human jejunum in vivo. Drug. Metab. Dispos. 2003, 31, 805–813. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.B.; Brooks, J.D. Modest induction of phase 2 enzyme activity in the F-344 rat prostate. BMC Cancer 2006, 6, 62. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.D.; Kelleher, M.O.; Eggleston, I.M. The cancer chemopreventive actions of phytochemicals derived from glucosinolates. Eur. J. Nutr. 2008, 47, 73–88. [Google Scholar] [CrossRef] [PubMed]
- Higgins, L.G.; Hayes, J.D. Mechanisms of induction of cytosolic and microsomal glutathione transferase (GST) genes by xenobiotics and pro-inflammatory agents. Drug Metab. Rev. 2011, 43, 92–137. [Google Scholar] [CrossRef] [PubMed]
- McMahon, M.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 2003, 278, 21592–21600. [Google Scholar] [CrossRef] [PubMed]
- Sahi, J.; Grepper, S.; Smith, C. Hepatocytes as a tool in drug metabolism, transport and safety evaluations in drug discovery. Curr. Drug Discov. Technol. 2010, 7, 188–198. [Google Scholar] [PubMed]
- Maheo, K.; Morel, F.; Langouet, S.; Kramer, H.; Le Ferrec, E.; Ketterer, B.; Guillouzo, A. Inhibition of cytochromes P-450 and induction of glutathione S-transferases by sulforaphane in primary human and rat hepatocytes. Cancer Res. 1997, 57, 3649–3652. [Google Scholar] [PubMed]
- Srovnalova, A.; Vanduchova, A.; Svecarova, M.; Anzenbacherova, E.; Tomankova, V.; Anzenbacher, P.; Dvorak, Z. Effects of sulforaphane and its S- and R-enantiomers on the expression and activities of human drug-metabolizing cytochromes P450. J. Funct. Foods 2015, 14, 487–501. [Google Scholar] [CrossRef]
- Yoxall, V.; Kentish, P.; Coldham, N.; Kuhnert, N.; Sauer, M.J.; Ioannides, C. Modulation of hepatic cytochromes P450 and phase II enzymes by dietary doses of sulforaphane in rats: Implications for its chemopreventive activity. Int. J. Cancer 2005, 117, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Matusheski, N.V.; Jeffery, E.H. Comparison of the bioactivity of two glucoraphanin hydrolysis products found in broccoli, sulforaphane and sulforaphane nitrile. J. Agric. Food Chem. 2001, 49, 5743–5749. [Google Scholar] [CrossRef] [PubMed]
- Gross-Steinmeyer, K.; Stapleton, P.L.; Tracy, J.H.; Bammler, T.K.; Strom, S.C.; Eaton, D.L. Sulforaphane- and Phenethyl Isothiocyanate-Induced Inhibition of Aflatoxin B1-Mediated Genotoxicity in Human Hepatocytes: Role of GSTM1 Genotype and CYP3A4 Gene Expression. Toxicol. Sci. 2010, 116, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Abdull Razis, A.F.; Hanlon, N.; Soltys, E.; Krizova, V.; Iori, R.; Plant, K.E.; Plant, N.; Ioannides, C. The naturally occurring aliphatic isothiocyanates sulforaphane and erucin are weak agonists but potent non-competitive antagonists of the aryl hydrocarbon receptor. Arch. Toxicol. 2012, 86, 1505–1514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, C.; Poulton, E.J.; Grun, F.; Bammler, T.K.; Blumberg, B.; Thummel, K.E.; Eaton, D.L. The dietary isothiocyanate sulforaphane is an antagonist of the human steroid and xenobiotic nuclear receptor. Mol. Pharmacol. 2007, 71, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Li, L.; Fuhrman, J.; Ferguson, S.; Wang, H. The role of constitutive androstane receptor in oxazaphosphorine-mediated induction of drug-metabolizing enzymes in human hepatocytes. Pharm. Res. 2011, 28, 2034–2044. [Google Scholar] [CrossRef] [PubMed]
- Angeloni, C.; Leoncini, E.; Malaguti, M.; Angelini, S.; Hrelia, P.; Hrelia, S. Modulation of phase II enzymes by sulforaphane: Implications for its cardioprotective potential. J. Agric. Food Chem. 2009, 57, 5615–5622. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.Q.; Chen, C.; Yang, B.; Hebbar, V.; Kong, A.N. Differential responses from seven mammalian cell lines to the treatments of detoxifying enzyme inducers. Life Sci. 2003, 72, 2243–2253. [Google Scholar] [CrossRef]
- Singh, P.; Sharma, R.; McElhanon, K.; Allen, C.D.; Megyesi, J.K.; Benes, H.; Singh, S.P. Sulforaphane protects the heart from doxorubicin-induced toxicity. Free Radic. Biol. Med. 2015, 86, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Bonnesen, C.; Eggleston, I.M.; Hayes, J.D. Dietary indoles and isothiocyanates that are generated from cruciferous vegetables can both stimulate apoptosis and confer protection against DNA damage in human colon cell lines. Cancer Res. 2001, 61, 6120–6130. [Google Scholar] [PubMed]
- Lakhman, S.S.; Chen, X.; Gonzalez-Covarrubias, V.; Schuetz, E.G.; Blanco, J.G. Functional characterization of the promoter of human carbonyl reductase 1 (CBR1). Role of XRE elements in mediating the induction of CBR1 by ligands of the aryl hydrocarbon receptor. Mol. Pharmacol. 2007, 72, 734–743. [Google Scholar] [CrossRef] [PubMed]
- Nannelli, A.; Rossignolo, F.; Tolando, R.; Rossato, P.; Longo, V.; Gervasi, P.G. Effect of beta-naphthoflavone on AhR-regulated genes (CYP1A1, 1A2, 1B1, 2S1, Nrf2, and GST) and antioxidant enzymes in various brain regions of pig. Toxicology 2009, 265, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Payen, L.; Courtois, A.; Loewert, M.; Guillouzo, A.; Fardel, O. Reactive oxygen species-related induction of multidrug resistance-associated protein 2 expression in primary hepatocytes exposed to sulforaphane. Biochem. Biophys. Res. Commun. 2001, 282, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Buckley, D.B.; Klaassen, C.D. Induction of Mouse UDP-Glucuronosyltransferase mRNA Expression in Liver and Intestine by Activators of Aryl-Hydrocarbon Receptor, Constitutive Androstane Receptor, Pregnane X Receptor, Peroxisome Proliferator-Activated Receptor alpha, and Nuclear Factor Erythroid 2-Related Factor 2. Drug Metab. Dispos. 2009, 37, 847–856. [Google Scholar] [PubMed]
- Falany, C.N.; Strom, P.; Swedmark, S. Sulphation of o-desmethylnaproxen and related compounds by human cytosolic sulfotransferases. Br. J. Clin. Pharmacol. 2005, 60, 632–640. [Google Scholar] [CrossRef] [PubMed]
- Maier, T.; Guell, M.; Serrano, L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009, 583, 3966–3973. [Google Scholar] [CrossRef] [PubMed]
- Bartels, C.L.; Tsongalis, G.J. MicroRNAs: Novel biomarkers for human cancer. Clin. Chem. 2009, 55, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Richert, L.; Tuschl, G.; Abadie, C.; Blanchard, N.; Pekthong, D.; Mantion, G.; Weber, J.C.; Mueller, S.O. Use of mRNA expression to detect the induction of drug metabolising enzymes in rat and human hepatocytes. Toxicol. Appl. Pharmacol. 2009, 235, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Lnenickova, K.; Skalova, L.; Stuchlikova Raisova, L.; Szotakova, B.; Matouskova, P. The induction of xenobiotic-metabolizing enzymes in hepatocytes by beta-naphthoflavone: Time-dependent changes in activities, protein and mRNA levels. Acta Pharm. 2017, in press. [Google Scholar]
- Berry, M.; Edwards, A.; Barritt, G. Isolated Hepatocytes: Preparation, Properties and Applications; Elsevier Science Publishers: Amsterdam, The Netherlands, 1991; Volume 21, p. 460. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Burke, M.D.; Mayer, R.T. Ethoxyresorufin: Direct fluorimetric assay of a microsomal O-dealkylation which is preferentially inducible by 3-methylcholanthrene. Drug Metab. Dispos. 1974, 2, 583–588. [Google Scholar] [PubMed]
- Burke, M.D.; Thompson, S.; Elcombe, C.R.; Halpert, J.; Haaparanta, T.; Mayer, R.T. Ethoxy-, pentoxy- and benzyloxyphenoxazones and homologues: A series of substrates to distinguish between different induced cytochromes P-450. Biochem. Pharmacol. 1985, 34, 3337–3345. [Google Scholar] [CrossRef]
- Kobayashi, K.; Urashima, K.; Shimada, N.; Chiba, K. Substrate specificity for rat cytochrome P450 (CYP) isoforms: Screening with cDNA-expressed systems of the rat. Biochem. Pharmacol. 2002, 63, 889–896. [Google Scholar] [CrossRef]
- Teel, R.W.; Huynh, H. Modulation by phytochemicals of cytochrome P450-linked enzyme activity. Cancer Lett. 1998, 133, 135–141. [Google Scholar] [CrossRef]
- Elbarbry, F.; Attia, A.; Shoker, A. Validation of a new HPLC method for determination of midazolam and its metabolites: Application to determine its pharmacokinetics in human and measure hepatic CYP3A activity in rabbits. J. Pharm. Biomed. Anal. 2009, 50, 987–993. [Google Scholar] [CrossRef] [PubMed]
- Maté, L.; Virkel, G.; Lifschitz, A.; Ballent, M.; Lanusse, C. Hepatic and extra-hepatic metabolic pathways involved in flubendazole biotransformation in sheep. Biochem. Pharmacol. 2008, 76, 773–783. [Google Scholar] [CrossRef] [PubMed]
- Cullen, J.J.; Hinkhouse, M.M.; Grady, M.; Gaut, A.W.; Liu, J.; Zhang, Y.P.; Weydert, C.J.; Domann, F.E.; Oberley, L.W. Dicumarol inhibition of NADPH:quinone oxidoreductase induces growth inhibition of pancreatic cancer via a superoxide-mediated mechanism. Cancer Res. 2003, 63, 5513–5520. [Google Scholar] [PubMed]
- Mizuma, T.; Machida, M.; Hayashi, M.; Awazu, S. Correlation of drug conjugative metabolism rates between in vivo and in vitro: Glucuronidation and sulfation of p-nitrophenol as a model compound in rat. J. Pharmacobiodyn. 1982, 5, 811–817. [Google Scholar] [CrossRef] [PubMed]
- Frame, L.T.; Ozawa, S.; Nowell, S.A.; Chou, H.C.; DeLongchamp, R.R.; Doerge, D.R.; Lang, N.P.; Kadlubar, F.F. A simple colorimetric assay for phenotyping the major human thermostable phenol sulfotransferase (SULT1A1) using platelet cytosols. Drug Metab. Dispos. 2000, 28, 1063–1068. [Google Scholar] [PubMed]
- Habig, W.H.; Pabst, M.J.; Jakoby, W.B. Glutathione S-Transferases: The first enzymatic step in mercapturic acid formation. J. Biol. Chem. 1974, 249, 7130–7139. [Google Scholar] [PubMed]
Sample Availability: Not available. |

| Gene | NCBI Accession Number | Forward Primer | Reverse Primer | Amplicon Size (bp) |
|---|---|---|---|---|
| CYP1A1 | NM_012540.2 | GGGTGGCCTTGAACTCCTTA | TGGTGTAGCGGTTCATGACT | 83 |
| CYP1A2 | NM_012541.3 | CCAACCCAGCCCTCAAGAG | GGATGAGACCACCGTTGTCT | 168 |
| CYP3A | NM_013105 | GCCCTTTGGAAATGGACCCA | TGCAGAACTTTAGTGAGAGCGA | 84 |
| CYP2B | NM_012940.2 | CACCAAAGACACCATGTTCCG | TGGTCAAAGTACTGTGGGTCA | 99 |
| NQO1 | NM_017000.3 | TTCCAGCCGACAACCAGATC | AGCCTCCTCCTTTTCCTATCCT | 141 |
| CBR1 | NM_019170.2 | ACCCAAGATGTCTGCAAGGAG | CTGAGACTCACGCTGCTTGAT | 83 |
| AKR1C14 | NM_138547.3 | GGGTTGAAGAGTGTTGCAGG | AAGACCTAGGTTTGGCTCCC | 89 |
| GSTA1 | NM_031509 | CGAAAGCTTTGCAACAATCGC | GCATTAGAAAACGTGTTGGCCT | 77 |
| SULT1A1 | NM_031834.1 | CCTGTCCTTGCTCCCTCAGA | GGAGACAACCACATCCTTTGC | 85 |
| UGT1A | NM_201425.2 | CCTGGAAATGACTGCCGATG | GCGCATGATGTTCTCCTTGT | 82 |
| GAPDH | NM_017008.4 | GCAACTCCCATTCTTCCACC | CCACCACCCTGTTGCTGTAG | 114 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lněničková, K.; Dymáková, A.; Szotáková, B.; Boušová, I. Sulforaphane Alters β-Naphthoflavone-Induced Changes in Activity and Expression of Drug-Metabolizing Enzymes in Rat Hepatocytes. Molecules 2017, 22, 1983. https://doi.org/10.3390/molecules22111983
Lněničková K, Dymáková A, Szotáková B, Boušová I. Sulforaphane Alters β-Naphthoflavone-Induced Changes in Activity and Expression of Drug-Metabolizing Enzymes in Rat Hepatocytes. Molecules. 2017; 22(11):1983. https://doi.org/10.3390/molecules22111983
Chicago/Turabian StyleLněničková, Kateřina, Andrea Dymáková, Barbora Szotáková, and Iva Boušová. 2017. "Sulforaphane Alters β-Naphthoflavone-Induced Changes in Activity and Expression of Drug-Metabolizing Enzymes in Rat Hepatocytes" Molecules 22, no. 11: 1983. https://doi.org/10.3390/molecules22111983





