2-Substituted Aniline as a Simple Scaffold for LuxR-Regulated QS Modulation
Abstract
:1. Introduction
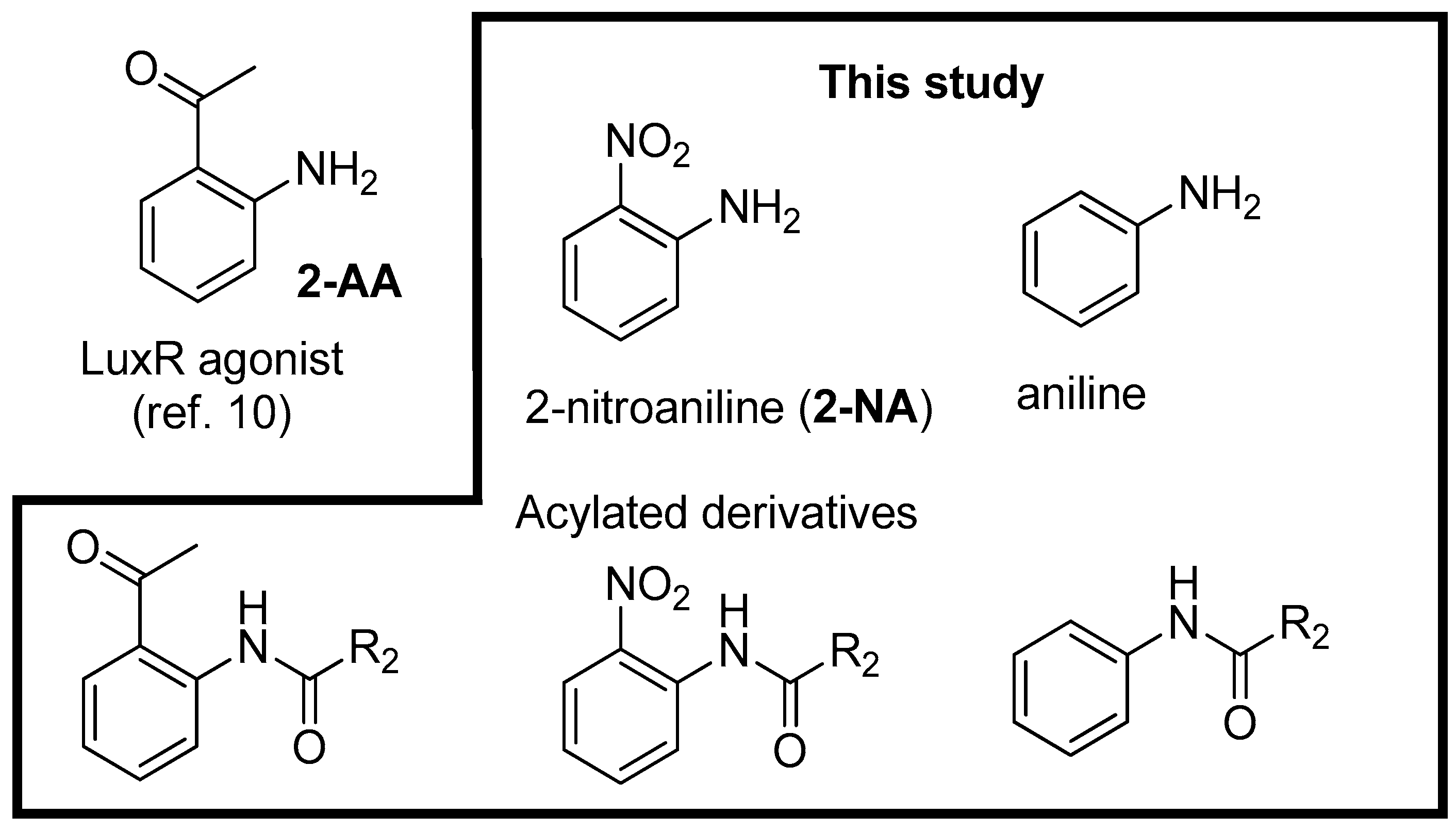
2. Results and Discussion
3. Materials and Methods
3.1. Binding Mode Studies
3.2. Biological Evaluation
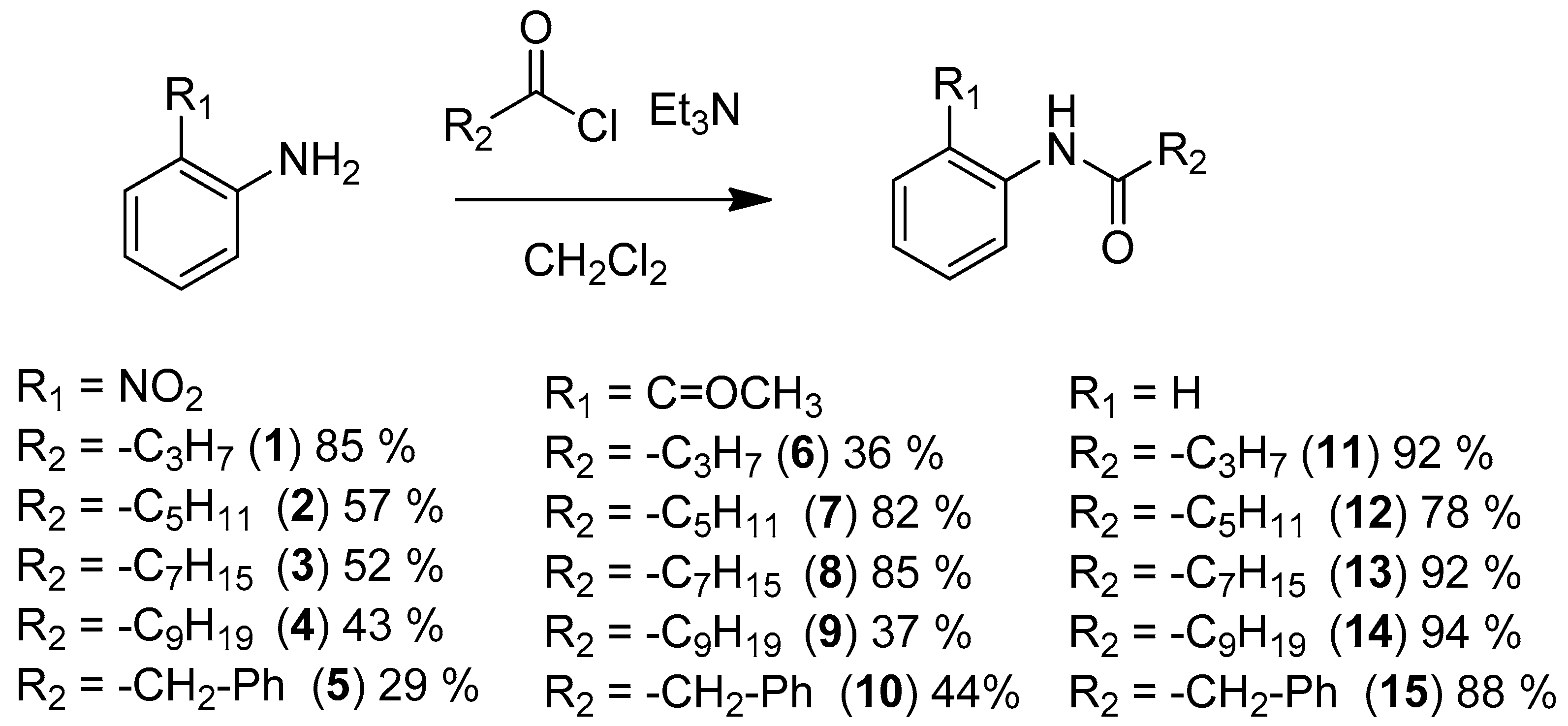
3.3. Synthesis
3.3.1. General Procedure for the Synthesis of N-(2-Nitrophenyl) Amide
3.3.2. General Procedure for the Synthesis of N-phenyl Amide Compounds and N-(2-acetylphenyl) Amide Compounds
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Garg, N.; Manchanda, G.; Kumar, A. Bacterial quorum sensing: Circuits and applications. Antonie Leeuwenhoek 2014, 105, 289–305. [Google Scholar] [CrossRef] [PubMed]
- Brackman, G.; Coenye, T. Quorum sensing inhibitors as anti-biofilm agents. Curr. Pharm. Des. 2015, 21, 5–11. [Google Scholar] [CrossRef] [PubMed]
- LaSarre, B.; Federle, M.J. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol. Mol. Biol. Rev. 2013, 77, 73–111. [Google Scholar] [CrossRef] [PubMed]
- Reuter, K.; Steinbach, A.; Helms, V. Interfering with bacterial quorum sensing. Perspect. Med. Chem. 2016, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Stevens, A.M.; Queneau, Y.; Soulere, L.; von Bodman, S.; Doutheau, A. Mechanisms and synthetic modulators of AHL-dependent gene regulation. Chem. Rev. 2011, 111, 4–27. [Google Scholar] [CrossRef] [PubMed]
- Tay, S.B.; Yew, W.S. Development of quorum-based anti-virulence therapeutics targeting Gram-negative bacterial pathogens. Int. J. Mol. Sci. 2013, 14, 16570–16599. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ma, S. Small molecules modulating AHL-based quorum sensing to attenuate bacteria virulence and biofilms as promising antimicrobial drugs. Curr. Med. Chem. 2014, 21, 296–311. [Google Scholar] [CrossRef] [PubMed]
- Dekimpe, V.; Deziel, E. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: The transcriptional regulator RhlR regulates LasR-specific factors. Microbiology 2009, 155, 712–723. [Google Scholar] [CrossRef] [PubMed]
- Drees, S.L.; Fetzner, S. PqsE of Pseudomonas aeruginosa acts as pathway-specific thioesterase in the biosynthesis of alkylquinolone signaling molecules. Chem. Biol. 2015, 22, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Kviatkovski, I.; Chernin, L.; Yarnitzky, T.; Frumin, I.; Sobel, N.; Helman, Y. Pseudomonas aeruginosa activates the quorum sensing LuxR response regulator through secretion of 2-aminoacetophenone. Chem. Commun. 2015, 51, 3258–3261. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Nair, S.K. Molecular basis for the recognition of structurally distinct autoinducer mimics by the Pseudomonas aeruginosa LasR quorum-sensing signaling receptor. Chem. Biol. 2009, 16, 961–970. [Google Scholar] [CrossRef] [PubMed]
- O'Reilly, M.C.; Blackwell, H.E. Structure-based design and biological evaluation of triphenyl scaffold-based hybrid compounds as hydrolytically stable modulators of a LuxR-Type quorum sensing receptor. ACS Infect. Dis. 2016, 2, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Thompson, M.A. ArgusLaB 4.0.1, Planetaria Software LLC.: Seattle, WA, USA, 2004.
- Estephane, J.; Dauvergne, J.; Soulere, L.; Reverchon, S.; Queneau, Y.; Doutheau, A. N-Acyl-3-amino-5H-furanone derivatives as new inhibitors of LuxR-dependent quorum sensing: Synthesis, biological evaluation and binding mode study. Bioorg. Med. Chem. Lett. 2008, 18, 4321–4324. [Google Scholar] [CrossRef] [PubMed]
- Soulere, L.; Frezza, M.; Queneau, Y.; Doutheau, A. Exploring the active site of acyl homoserine lactones-dependent transcriptional regulators with bacterial quorum sensing modulators using molecular mechanics and docking studies. J. Mol. Graph. Model. 2007, 26, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Darvesh, S.; McDonald, R.S.; Darvesh, K.V.; Mataija, D.; Mothana, S.; Cook, H.; Carneiro, K.M.; Richard, N.; Walsh, R.; Martin, E. On the active site for hydrolysis of aryl amides and choline esters by human cholinesterases. Bioorg. Med. Chem. 2006, 14, 4586–4599. [Google Scholar] [CrossRef] [PubMed]
- Reverchon, S.; Chantegrel, B.; Deshayes, C.; Doutheau, A.; Cotte-Pattat, N. New synthetic analogues of N-acyl homoserine lactones as agonists or antagonists of transcriptional regulators involved in bacterial quorum sensing. Bioorg. Med. Chem. Lett. 2002, 12, 1153–1157. [Google Scholar] [CrossRef]
- McInnis, C.E.; Blackwell, H.E. Design, synthesis, and biological evaluation of abiotic, non-lactone modulators of LuxR-type quorum sensing. Bioorg. Med. Chem. 2011, 19, 4812–4819. [Google Scholar] [CrossRef] [PubMed]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Frezza, M.; Castang, S.; Estephane, J.; Soulere, L.; Deshayes, C.; Chantegrel, B.; Nasser, W.; Queneau, Y.; Reverchon, S.; Doutheau, A. Synthesis and biological evaluation of homoserine lactone derived ureas as antagonists of bacterial quorum sensing. Bioorg. Med. Chem. 2006, 14, 4781–4791. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.P.; Anderson, K.W.; Buchwald, S.L. Sequential Cu-catalyzed amidation-base-mediated camps cyclization: A two-step synthesis of 2-Aryl-4-quinolones from o-halophenones. J. Org. Chem. 2007, 72, 7968–7973. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Kumar, S. Selective Oxidative Decarbonylative Cleavage of Unstrained C(sp(3))-C(sp(2)) Bond: Synthesis of Substituted Benzoxazinones. Org. Lett. 2016, 18, 4388–4391. [Google Scholar] [CrossRef] [PubMed]
- Correa, A.; Elmore, S.; Bolm, C. Iron-catalyzed N-Arylations of amides. Chem. Eur. J. 2008, 14, 3527–3529. [Google Scholar] [CrossRef] [PubMed]
- Jammi, S.; Sakthivel, S.; Rout, L.; Mukherjee, T.; Mandal, S.; Mitra, R.; Saha, P.; Punniyamurthy, T. CuO nanoparticles catalyzed C-N, C-O, and C-S cross-coupling reactions: Scope and mechanism. J. Org. Chem. 2009, 74, 1971–1976. [Google Scholar] [CrossRef] [PubMed]
- Hosseini-Sarvari, M.; Sodagar, E.; Doroodmand, M.M. Nano sulfated titania as solid acid catalyst in direct synthesis of fatty acid amides. J. Org. Chem. 2011, 76, 2853–2859. [Google Scholar] [CrossRef] [PubMed]
- Henry, C.; Bolien, D.; Ibanescu, B.; Bloodworth, S.; Harrowven, D.C.; Zhang, X.L.; Craven, A.; Sneddon, H.F.; Whitby, R.J. Generation and trapping of Ketenes in flow. Eur. J. Org. Chem. 2015, 2015, 1491–1499. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lim, K.C. Tin-free radical carbonylation of alkylsulfonyl derivatives into alkylcarbonyl derivatives. Chem. Asian J. 2008, 3, 1692–1701. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 1–15 are available from the authors. |
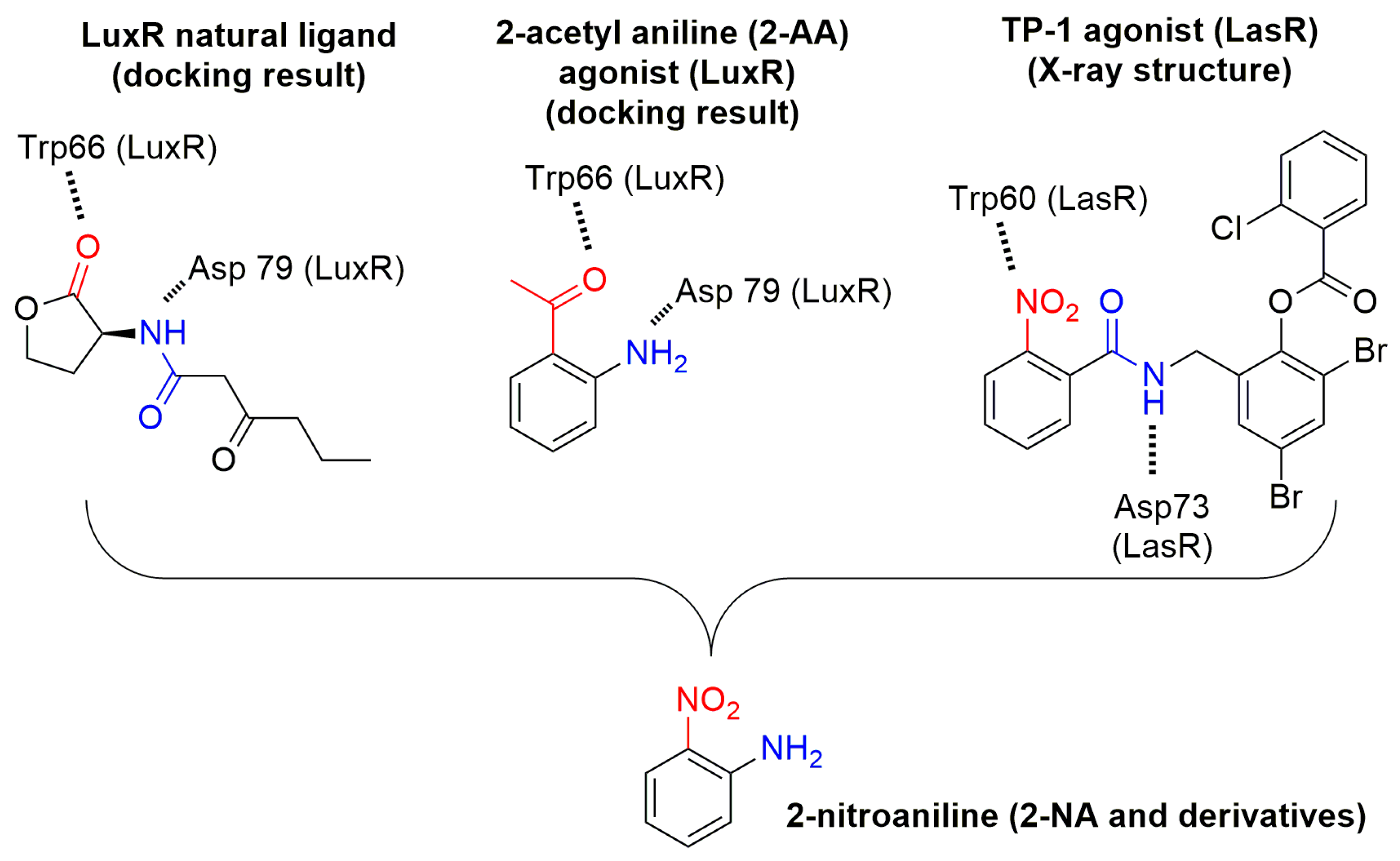
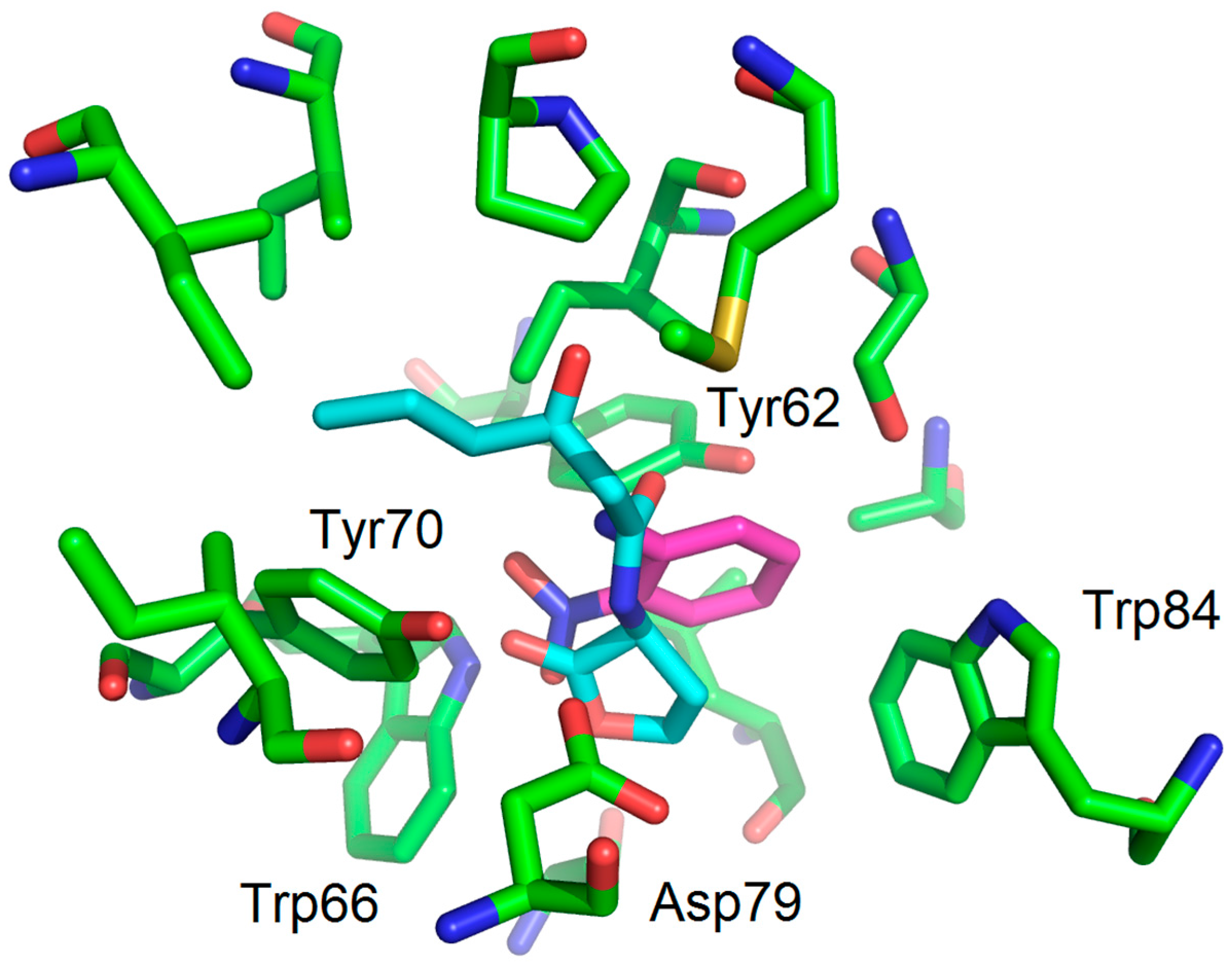
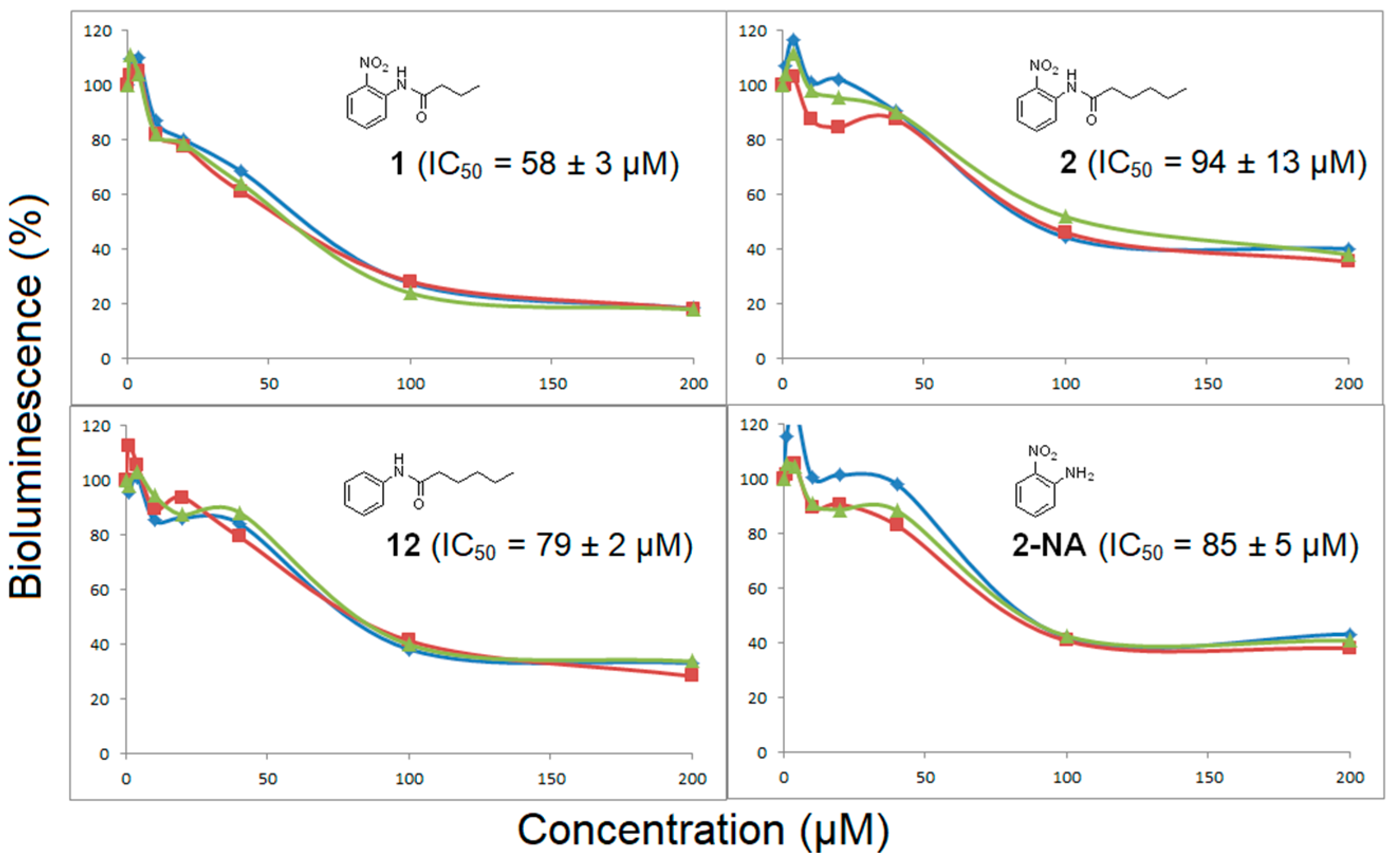
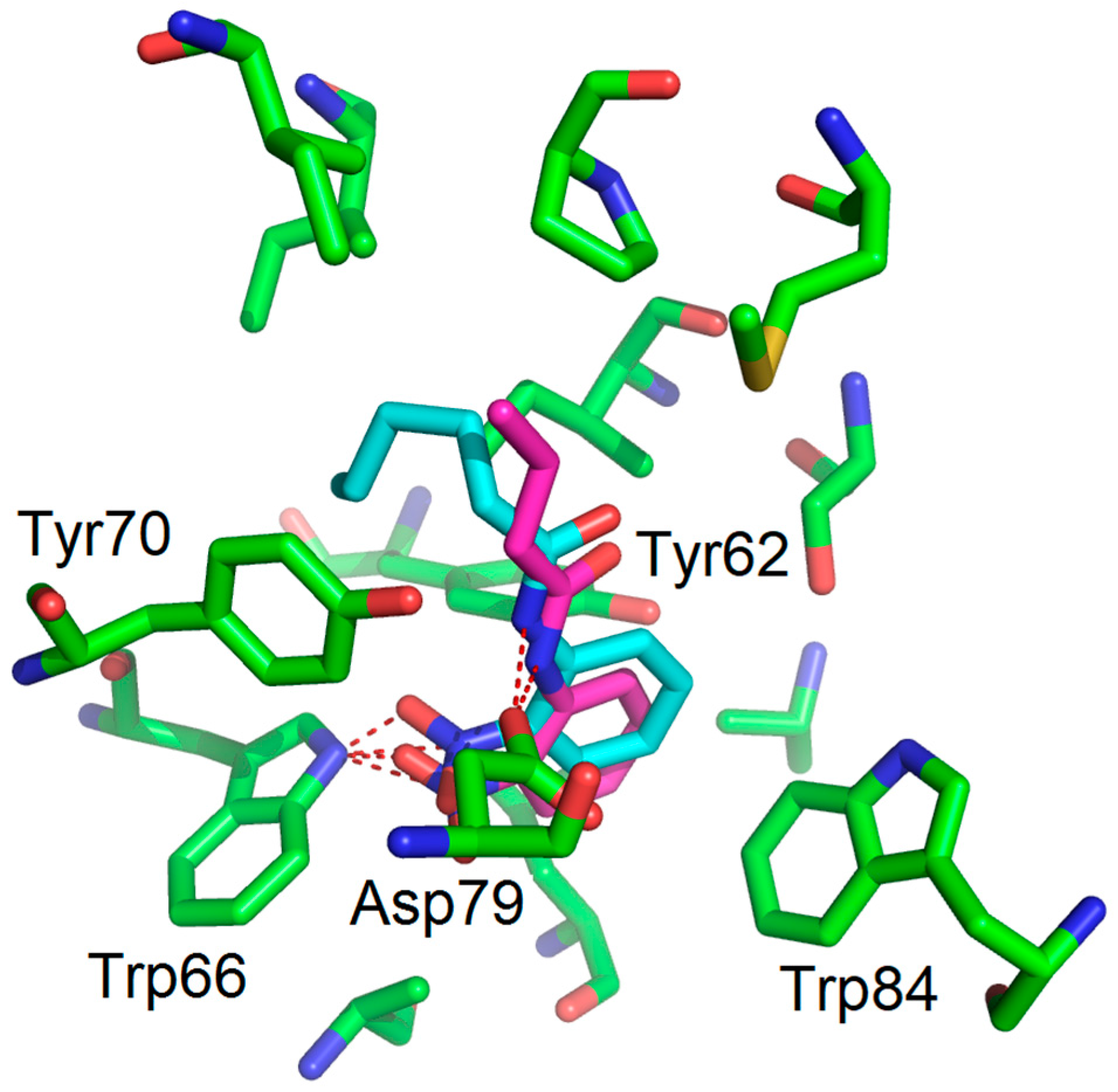
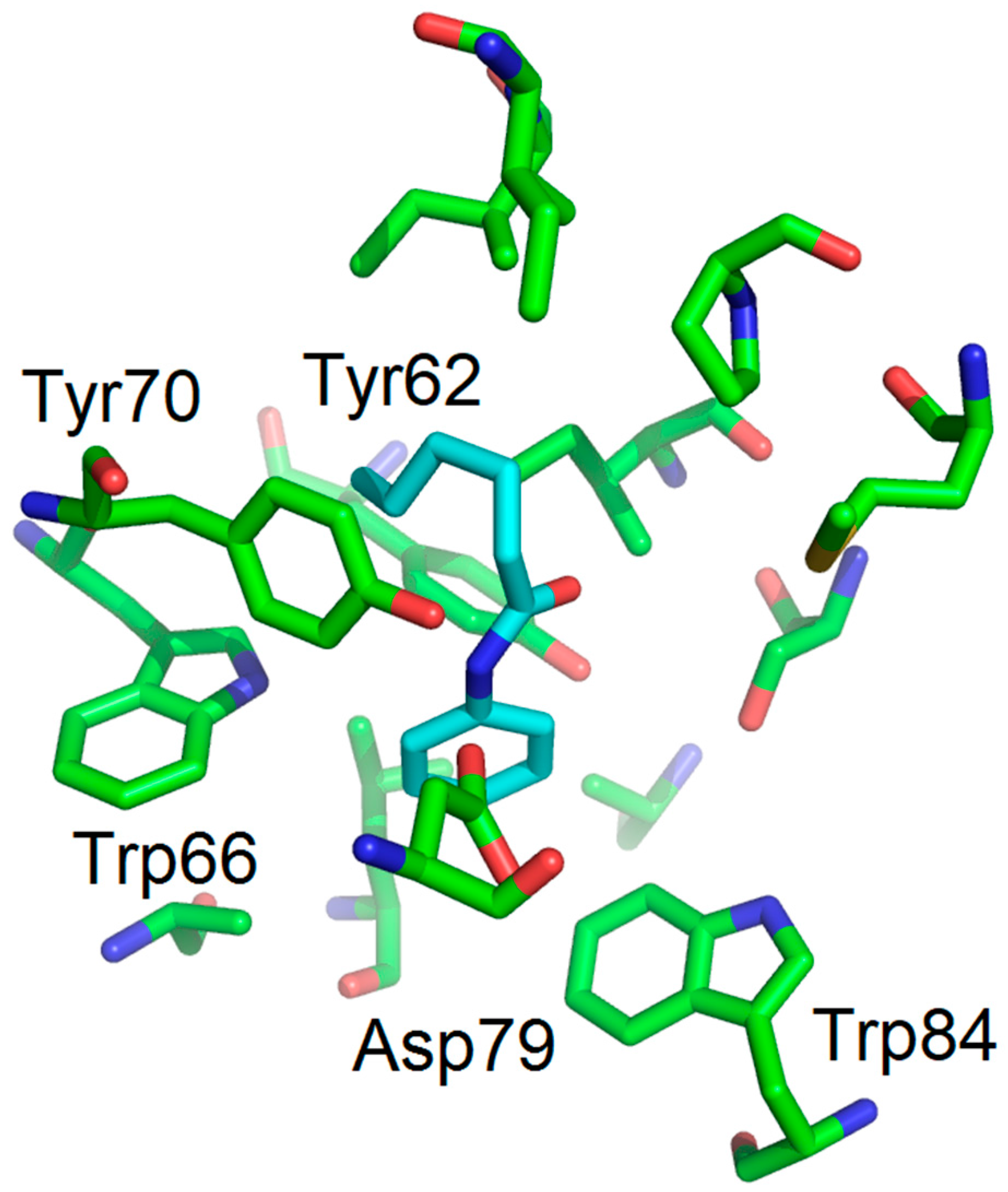

| Compounds | Trp66 | Asp79 | Tyr62 |
|---|---|---|---|
| OHHL (natural ligand) | +2.34 (C=O lactone) a | +3.01 (NH amide) | +3.01 (C=O amide) |
| 2-AA (agonist) | +2.20 (C=O) | +3.03 (NH2) | No function |
| 2-NA (antagonist) | +2.96 and 2.37 (NO2) | -NH2 too far | No function |
| 1 (antagonist) | +2.47 (NO2) | +3.00 (NH amide) | -C=O too far |
| 2 (antagonist) | +2.20 (NO2) | +3.01 (NH amide) | -C=O too far |
| 6 (No activity) | +1.99 (C=OCH3) | +3.10 (NH amide) | +2.97 (C=O amide) |
| 7 (No activity) | +2.01 (C=OCH3) | +2.98 (NH amide) | +2.94 (C=O amide) |
| 11 (No activity) | - | - | - |
| 12 (antagonist) | No function | +3.04 (NH amide) | -C=O too far |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Wawrzyniak, J.; Queneau, Y.; Soulère, L. 2-Substituted Aniline as a Simple Scaffold for LuxR-Regulated QS Modulation. Molecules 2017, 22, 2090. https://doi.org/10.3390/molecules22122090
Li S, Wawrzyniak J, Queneau Y, Soulère L. 2-Substituted Aniline as a Simple Scaffold for LuxR-Regulated QS Modulation. Molecules. 2017; 22(12):2090. https://doi.org/10.3390/molecules22122090
Chicago/Turabian StyleLi, Sizhe, Julien Wawrzyniak, Yves Queneau, and Laurent Soulère. 2017. "2-Substituted Aniline as a Simple Scaffold for LuxR-Regulated QS Modulation" Molecules 22, no. 12: 2090. https://doi.org/10.3390/molecules22122090




