Role of Quinone Reductase 2 in the Antimalarial Properties of Indolone-Type Derivatives
Abstract
:1. Introduction
2. Results and Discussion
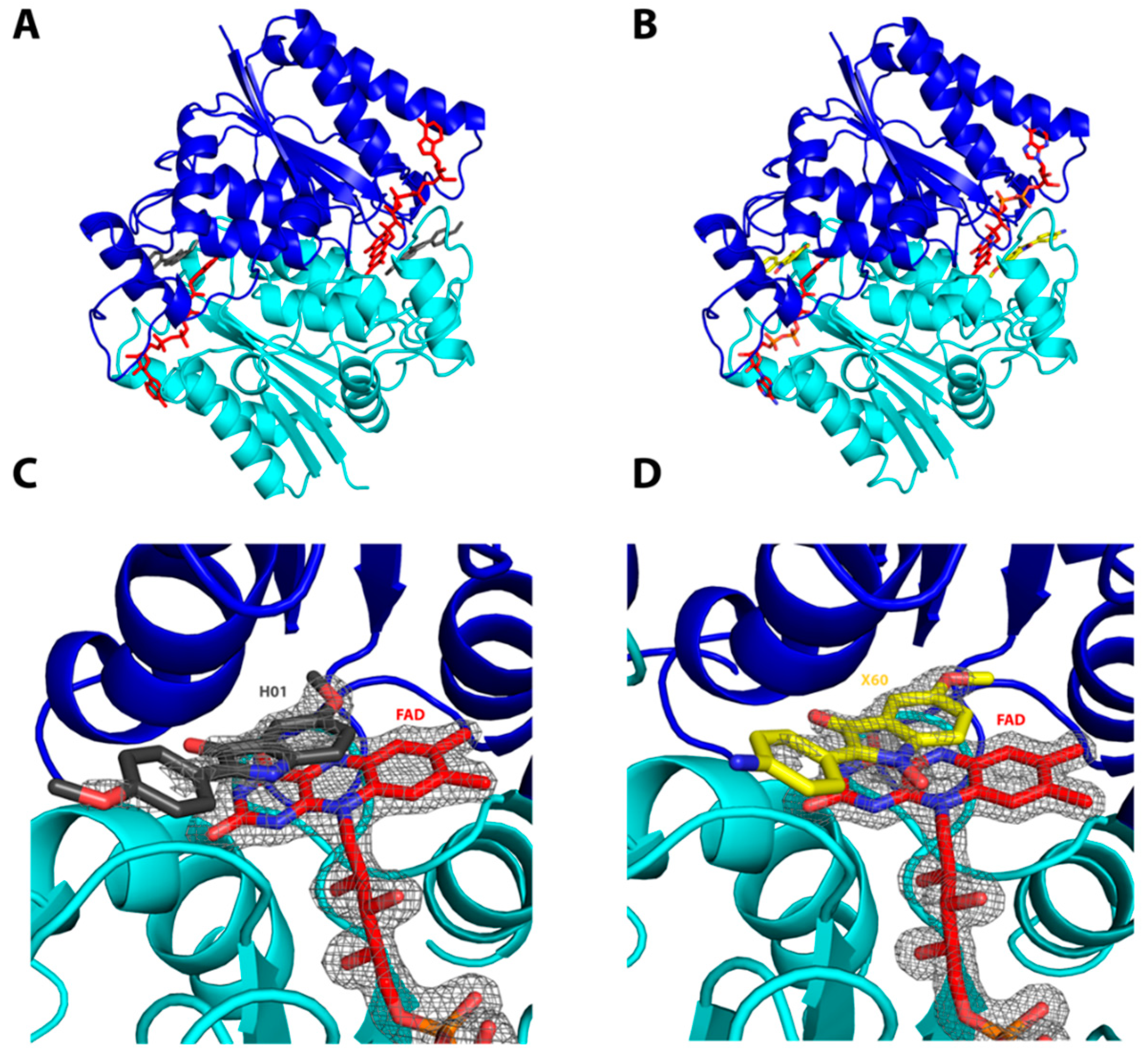
2.1. Interactions of the Compounds with the hQR2 Enzyme
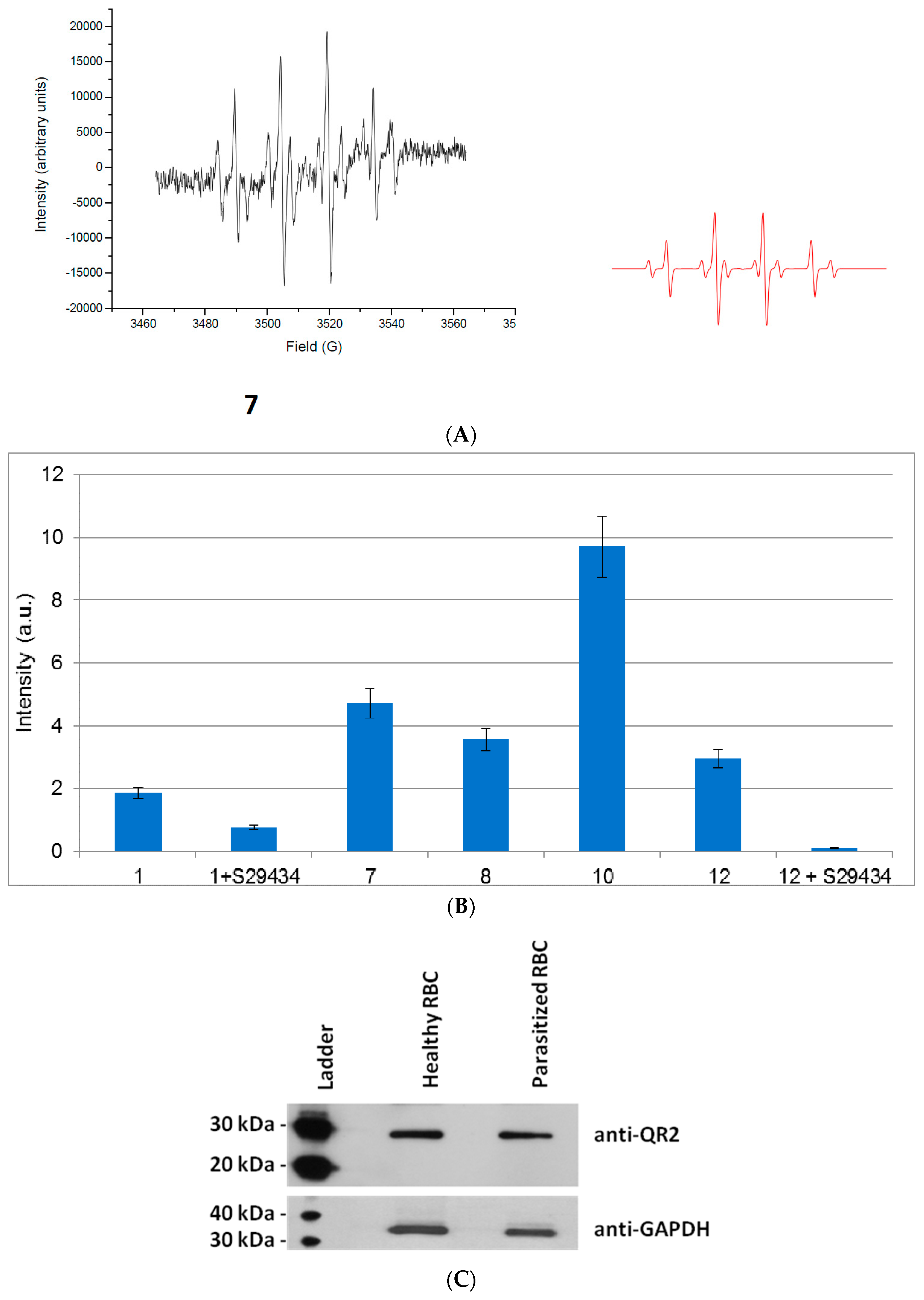
2.2. Free Radical Production during Metabolization of Indolone Derivatives by Purified hQR2
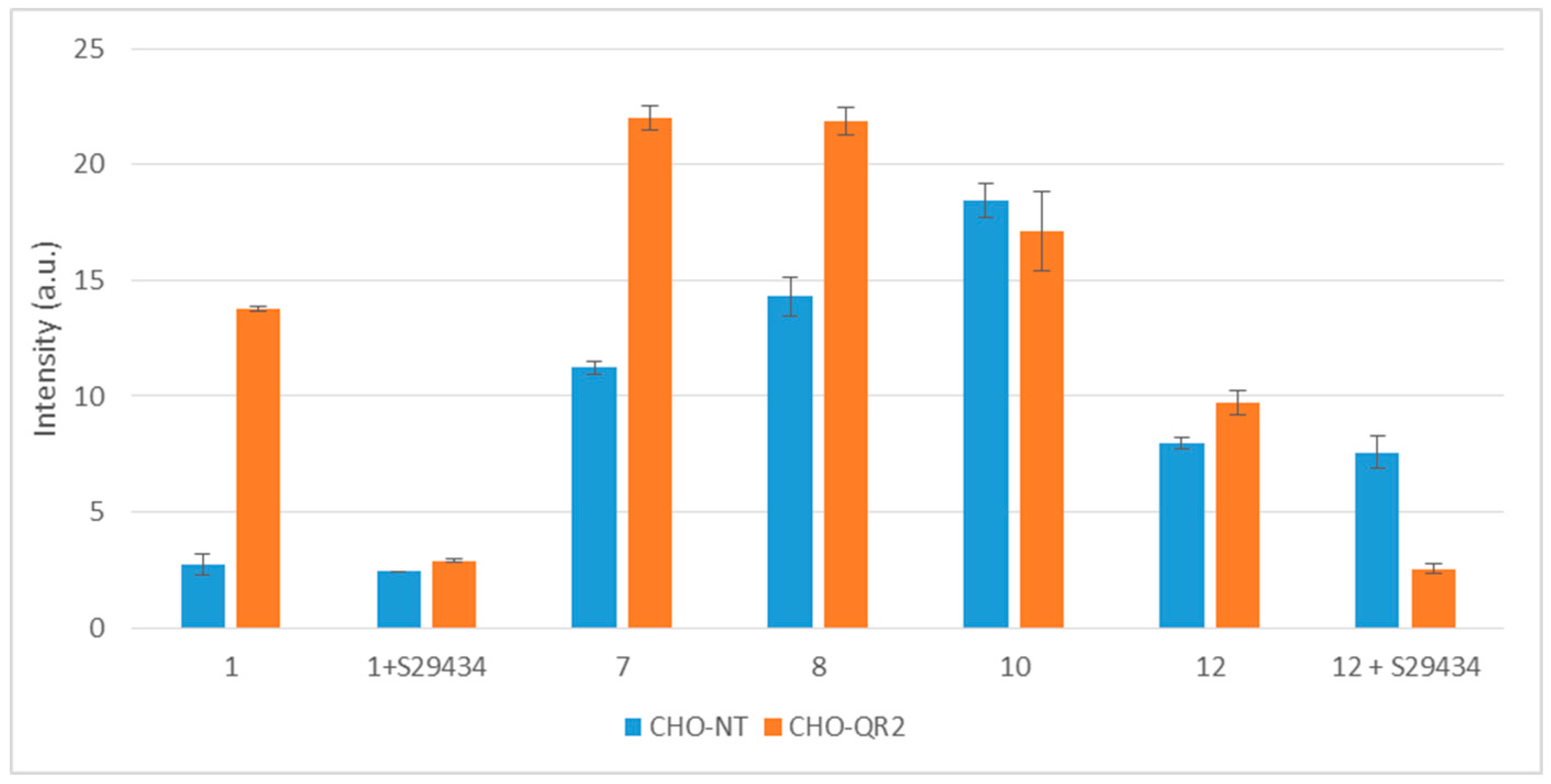
2.3. Free Radicals Produced by Cellular Metabolization of Indolone Derivatives
2.4. Intracellular Metabolization of the Compounds
3. Materials and Methods
3.1. Chemicals
3.2. hsQR2 Enzymatic Activity
3.3. Co-Crystallization of hQR2 with Bound Substrate—The RX Structure
3.4. In Vitro Cultures of P. falciparum
3.5. Isolation of Parasitized Red Blood Cells (pRBCs)
3.6. Cells Lysis and Protein Extraction
3.7. Western Blotting
3.8. Reactant and Substrate Preparations for the Free Radical Production Measurement
3.9. EPR Spectroscopy Experiments on hQR2
3.10. Reactant and Substrate Preparations for the Intracellular Metabolization Assay
3.11. Cellular Activity Assays
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- WHO World Malaria Report. Available online: http://www.who.int/malaria/publications/world-malaria-report-2015/report/en (accessed on 14 April 2016).
- Burrows, J.N.; van Huijsduijnen, R.H.; Mohrle, J.J.; Oeuvray, C.; Wells, T.N. Designing the next generation of medicines for malaria control and eradication. Malar J. 2013, 12, 187. [Google Scholar] [CrossRef] [PubMed]
- Medicines for Malaria Venture (MMV). Available online: http://www.mmv.org/research-development/interactive-rd-portfolio (accessed on 14 April 2016).
- Nepveu, F.; Kim, S.; Boyer, J.; Chatriant, O.; Ibrahim, H.; Reybier, K.; Monje, M.C.; Chevalley, S.; Perio, P.; Lajoie, B.H.; et al. Synthesis and antiplasmodial activity of new indolone N-oxide derivatives. J. Med. Chem. 2010, 53, 699–714. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, N.; Ibrahim, H.; Dormoi, J.; Briolant, S.; Pradines, B.; Moreno, A.; Mazier, D.; Legrand, P.; Nepveu, F. Albumin-bound nanoparticles of practically water-insoluble antimalarial lead greatly enhance its efficacy. Int. J. Pharm. 2014, 464, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, H.; Pantaleo, A.; Turrini, F.; Arese, P.; Nallet, J.P.; Nepveu, F. Pharmacological properties of indolone-N-oxides controlled by a bioreductive transformation in red blood cells? Med. Chem. Comm. 2011, 2, 860–869. [Google Scholar] [CrossRef]
- Nepveu, F.; Najahi, E.; Valentin, A. Antimalarial activities of indolones and derivatives. Curr. Top. Med. Chem. 2014, 14, 1643–1652. [Google Scholar] [CrossRef] [PubMed]
- Pantaleo, A.; Ferru, E.; Vono, R.; Giribaldi, G.; Lobina, O.; Nepveu, F.; Ibrahim, H.; Nallet, J.P.; Carta, F.; Mannu, F.; et al. New antimalarial indolone-N-oxides, generating radical species, destabilize the host cell membrane at early stages of Plasmodium falciparum growth: Role of band 3 tyrosine phosphorylation. Free Radic. Biol. Med. 2012, 52, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Nepveu, F.; Turrini, F. Targeting the redox metabolism of Plasmodium falciparum. Future Med. Chem. 2013, 5, 1993–2006. [Google Scholar] [CrossRef] [PubMed]
- Graves, P.R.; Kwiek, J.J.; Fadden, P.; Ray, R.; Hardeman, K.; Coley, A.M.; Foley, M.; Haystead, T.A. Discovery of novel targets of quinoline drugs in the human purine binding proteome. Mol. Pharmacol. 2002, 62, 1364–1372. [Google Scholar] [CrossRef]
- Kwiek, J.J.; Haystead, T.A.; Rudolph, J. Kinetic mechanism of quinone oxidoreductase 2 and its inhibition by the antimalarial quinolines. Biochemistry 2004, 43, 4538–4547. [Google Scholar] [CrossRef] [PubMed]
- Leung, K.K.; Shilton, B.H. Chloroquine binding reveals flavin redox switch function of quinone reductase 2. J. Biol. Chem. 2013, 288, 11242–11251. [Google Scholar] [CrossRef] [PubMed]
- Petri, W.A., Jr. Can a proteomics strategy be used to identify the anti-malarial activity of chloroquine? Trends Pharmacol. Sci. 2003, 24, 210–212. [Google Scholar] [CrossRef]
- Haystead, T.A. Response to Petri: Can a proteomics strategy be used to identify the anti-malarial activity of chloroquine? Trends Pharmacol. Sci. 2003, 24, 212–213. [Google Scholar] [CrossRef]
- Choi, Y.; Jermihov, K.; Nam, S.J.; Sturdy, M.; Maloney, K.; Qiu, X.; Chadwick, L.R.; Main, M.; Chen, S.N.; Mesecar, A.D.; et al. Screening natural products for inhibitors of quinone reductase-2 using ultrafiltration LC-MS. Anal. Chem. 2011, 83, 1048–1052. [Google Scholar] [CrossRef] [PubMed]
- Reddy, P.V.; Jensen, K.C.; Mesecar, A.D.; Fanwick, P.E.; Cushman, M. Design, synthesis, and biological evaluation of potent quinoline and pyrroloquinoline ammosamide analogues as inhibitors of quinone reductase 2. J. Med. Chem. 2012, 55, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Dufour, M.; Yan, C.; Siegel, D.; Colucci, M.A.; Jenner, M.; Oldham, N.J.; Gomez, J.; Reigan, P.; Li, Y.; De Matteis, C.I.; et al. Mechanism-based inhibition of quinone reductase 2 (NQO2): Selectivity for NQO2 over NQO1 and structural basis for flavoprotein inhibition. ChemBioChem 2011, 12, 1203–1208. [Google Scholar] [CrossRef] [PubMed]
- Nolan, K.A.; Humphries, M.P.; Bryce, R.A.; Stratford, I.J. Imidazoacridin-6-ones as novel inhibitors of the quinone oxidoreductase NQO2. Bioorg. Med. Chem. Lett. 2010, 20, 2832–2836. [Google Scholar] [CrossRef] [PubMed]
- Boussard, M.F.; Truche, S.; Rousseau-Rojas, A.; Briss, S.; Descamps, S.; Droual, M.; Wierzbicki, M.; Ferry, G.; Audinot, V.; Delagrange, P.; et al. New ligands at the melatonin binding site MT(3). Eur. J. Med. Chem. 2006, 41, 306–320. [Google Scholar] [CrossRef] [PubMed]
- Reybier, K.; Perio, P.; Ferry, G.; Bouajila, J.; Delagrange, P.; Boutin, J.A.; Nepveu, F. Insights into the redox cycle of human quinone reductase 2. Free Radic. Res. 2011, 45, 1184–1195. [Google Scholar] [CrossRef] [PubMed]
- Cassagnes, L.E.; Perio, P.; Ferry, G.; Moulharat, N.; Antoine, M.; Gayon, R.; Boutin, J.A.; Nepveu, F.; Reybier, K. In cellulo monitoring of quinone reductase activity and reactive oxygen species production during the redox cycling of 1,2 and 1,4 quinones. Free Radic. Biol. Med. 2015, 89, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Knox, R.J.; Jenkins, T.C.; Hobbs, S.M.; Chen, S.; Melton, R.G.; Burke, P.J. Bioactivation of 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB 1954) by human NAD(P)H quinone oxidoreductase 2: A novel co-substrate-mediated antitumor prodrug therapy. Cancer Res. 2000, 60, 4179–4186. [Google Scholar] [PubMed]
- Ferry, G.; Hecht, S.; Berger, S.; Moulharat, N.; Coge, F.; Guillaumet, G.; Leclerc, V.; Yous, S.; Delagrange, P.; Boutin, J.A. Old and new inhibitors of quinone reductase 2. Chem. Biol. Interact. 2010, 186, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Calamini, B.; Santarsiero, B.D.; Boutin, J.A.; Mesecar, A.D. Kinetic, thermodynamic and X-ray structural insights into the interaction of melatonin and analogues with quinone reductase 2. Biochem. J. 2008, 413, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Pegan, S.D.; Sturdy, M.; Ferry, G.; Delagrange, P.; Boutin, J.A.; Mesecar, A.D. X-ray structural studies of quinone reductase 2 nanomolar range inhibitors. Protein Sci. 2011, 20, 1182–1195. [Google Scholar] [CrossRef] [PubMed]
- Muller, T.; Johann, L.; Jannack, B.; Bruckner, M.; Lanfranchi, D.A.; Bauer, H.; Sanchez, C.; Yardley, V.; Deregnaucourt, C.; Schrevel, J.; et al. Glutathione reductase-catalyzed cascade of redox reactions to bioactivate potent antimalarial 1,4-naphthoquinones--a new strategy to combat malarial parasites. J. Am. Chem. Soc. 2011, 133, 11557–11571. [Google Scholar] [CrossRef] [PubMed]
- Belorgey, D.; Lanfranchi, D.A.; Davioud-Charvet, E. 1,4-Naphthoquinones and other NADPH-dependent glutathione reductase-catalyzed redox cyclers as antimalarial agents. Curr. Pharm. Des. 2013, 19, 2512–2528. [Google Scholar] [CrossRef] [PubMed]
- Bielitza, M.; Belorgey, D.; Ehrhardt, K.; Johann, L.; Lanfranchi, D.A.; Gallo, V.; Schwarzer, E.; Mohring, F.; Jortzik, E.; Williams, D.L.; et al. Antimalarial NADPH-Consuming Redox-Cyclers As Superior Glucose-6-Phosphate Dehydrogenase Deficiency Copycats. Antioxid. Redox Signal. 2015, 22, 1337–1351. [Google Scholar] [CrossRef] [PubMed]
- Bongard, R.; Lindemer, B.; Krenz, G.; Merker, M. Preferential utilization of NADPH as the endogenous electron donor for NAD(P)H:quinone oxidoreductase 1 (NQO1) in intact pulmonary arterial endothelial cells. Free Radic. Biol. Med. 2000, 46, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Najahi, E.; Valentin, A.; Fabre, P.L.; Reybier, K.; Nepveu, F. 2-Aryl-3H-indol-3-ones: Synthesis, electrochemical behaviour and antiplasmodial activities. Eur. J. Med. Chem. 2014, 78, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Mailliet, F.; Ferry, G.; Vella, F.; Berger, S.; Coge, F.; Chomarat, P.; Mallet, C.; Guenin, S.P.; Guillaumet, G.; Viaud-Massuard, M.C.; et al. Characterization of the melatoninergic MT3 binding site on the NRH:quinone oxidoreductase 2 enzyme. Biochem. Pharmacol. 2005, 71, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Nosjean, O.; Ferro, M.; Coge, F.; Beauverger, P.; Henlin, J.M.; Lefoulon, F.; Fauchere, J.L.; Delagrange, P.; Canet, E.; Boutin, J.A. Identification of the melatonin-binding site MT3 as the quinone reductase 2. J. Biol. Chem. 2000, 275, 31311–31317. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Collaborative Computational project N.4. The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994, 50, 760–763. [Google Scholar]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Bricogne, G.; Blanc, E.; Brandl, M.; Flensburg, C.; Keller, P.; Paciorek, W.; Roversi, P.; Sharff, A.; Smart, O.; Vonrhein, C.; et al. BUSTER, version X.Y.Z; Global Phasing Ltd.: Cambridge, UK, 2011. [Google Scholar]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta. Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Lambros, C.; Vanderberg, J.P. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 1979, 65, 418–420. [Google Scholar] [CrossRef] [PubMed]
- Ribaut, C.; Berry, A.; Chevalley, S.; Reybier, K.; Morlais, I.; Parzy, D.; Nepveu, F.; Benoit-Vical, F.; Valentin, A. Concentration and purification by magnetic separation of the erythrocytic stages of all human Plasmodium species. Malar J. 2008, 7, 45. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds 1–18 are available from the authors.
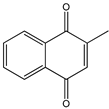 Menadione Menadione |  S29434 S29434 | |||
| Indolone-N-oxide (INODs) |  1 1 | 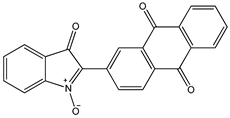 2 2 | 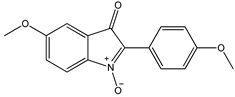 3 3 | |
 4 4 | 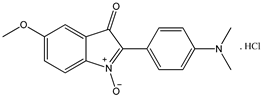 5 5 |  6 6 | ||
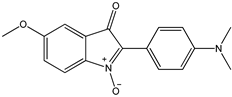 7 7 | 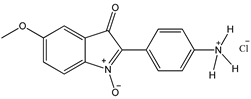 8 8 | 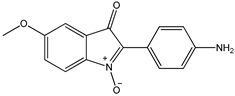 8’ 8’ | ||
| 2-Aryl-3H-indol-3-ones |  9 9 |  10 10 | 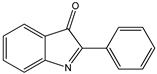 11 11 | |
 12 12 | ||||
| 2-aryl-3H-indol-3-ol |  13 13 | |||
| N-hydroxyindoles | ||||
 14 14 | 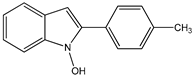 15 15 | |||
| 1H-indoles |  16 16 |  17 17 |  18 18 | |


| Substrate a | Vmax (µmol·mL−1·min−1) | KM (µM) |
|---|---|---|
| Menadione | 4264 ± 134 | 10 ± 1 |
| 1 | 789 ± 39 | 47 ± 5 |
| 8 | 1471 ± 98 | 12 ± 2 |
| 10 | 730 ± 59 | 21 ± 4 |
| 12 | 1255 ± 93 | 30 ± 6 |
| Nature of the Interaction | Monomer | Compound Atom | Protein Atom | H20 | Distance (Å) |
|---|---|---|---|---|---|
| hQR2-FAD Bound to Compound 10, PDB Entry: 4XDH | |||||
| Hydrogen bond | A | O12/233 | / | W199 | 2.73 Å |
| O4/233 | / | W142 | 2.84 Å | ||
| B | O12/233 | / | W180 | 2.72 Å | |
| π–π stacking | Interaction between the isoalloxazine ring and compound 10 | ||||
| hQR2-FAD Bound to Compound 8’, PDB Entry: 4XDG | |||||
| Hydrogen bond | A | O10/233 | / | W459 | 3.33 Å |
| O13/233 | / | W175 | 2.76 Å | ||
| B | O13/233 | / | W558 | 2.66 Å | |
| π–π stacking | Interaction between the isoalloxazine ring and compound 8′ | ||||
| Data Collection and Refinement Statistics | ||
|---|---|---|
| hQR2-FAD Bound to Compound 10, PDB Entry: 4XDH | hQR2-FAD Bound to Compound 8′, PDB Entry: 4XDG | |
| X-ray source | SOLEIL synchrotron, Proxima1 | SOLEIL synchrotron, Proxima1 |
| Oscillation range (°) | 0.1° | 0.1° |
| Number of frames | 2000 | 2000 |
| Exposure (s) | 0.1 | 0.1 |
| Detector distance (mm) | 296.299 | 270.864 |
| Wavelength (Å) | 0.97857 | 0.97857 |
| Space group | P212121 | P212121 |
| Unit cell parameters a, b, c (Å) | 56.514, 84.030, 106.420, 90.000, 90.000, 90.000 | 56.487, 83.634, 106.375, 90.000, 90.000, 90.000 |
| Resolution (Å) | 50.0–1.9 (2.01–1.90) | 50.0–1.5 (1.59–1.50) |
| Rsym | 20.2 (109.1) | 19 (61.9) |
| I/sI | 6.83 (1.61) | 12.58 (3.11) |
| Completeness (%) | 99.4 (97.6) | 99.9 (99.5) |
| Multiplicity | 7.32 (7.34) | 7.31 (7.14) |
| Number of reflections | 40641 | 81302 |
| (XX) statistics for high resolution range | ||
| Refinement Statistics | ||
| Resolution (Å) | 27.82–1.90 (1.95–1.90) | 46.81–1.50 (1.54–1.50) |
| Rwork/Rfree (%) | 17.09/19.88 (22.41/23.13) | 16.63/18.66 (23.98/28.35) |
| Number of atoms (protein/waters) | 3635/255 | 3727/524 |
| Mean B-factor (Å2) | 32.17 | 26.25 |
| R.m.s deviations: Bond lengths (Å)/Bond angles (°) | 0.010/0.99 | 0.010/1.02 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cassagnes, L.-E.; Rakotoarivelo, N.; Sirigu, S.; Pério, P.; Najahi, E.; Chavas, L.M.G.; Thompson, A.; Gayon, R.; Ferry, G.; Boutin, J.A.; et al. Role of Quinone Reductase 2 in the Antimalarial Properties of Indolone-Type Derivatives. Molecules 2017, 22, 210. https://doi.org/10.3390/molecules22020210
Cassagnes L-E, Rakotoarivelo N, Sirigu S, Pério P, Najahi E, Chavas LMG, Thompson A, Gayon R, Ferry G, Boutin JA, et al. Role of Quinone Reductase 2 in the Antimalarial Properties of Indolone-Type Derivatives. Molecules. 2017; 22(2):210. https://doi.org/10.3390/molecules22020210
Chicago/Turabian StyleCassagnes, Laure-Estelle, Nambinina Rakotoarivelo, Serena Sirigu, Pierre Pério, Ennaji Najahi, Léonard M. G. Chavas, Andrew Thompson, Régis Gayon, Gilles Ferry, Jean A. Boutin, and et al. 2017. "Role of Quinone Reductase 2 in the Antimalarial Properties of Indolone-Type Derivatives" Molecules 22, no. 2: 210. https://doi.org/10.3390/molecules22020210





