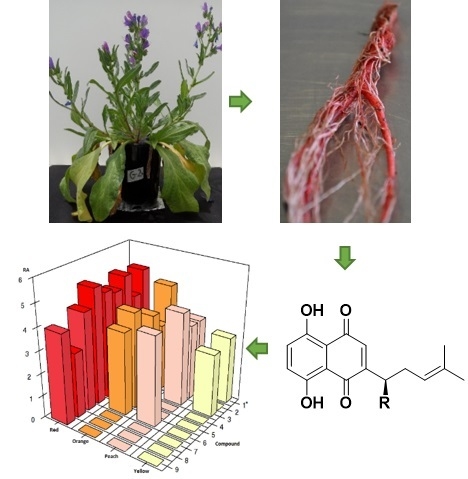
Metabolic Profiling and Identification of Shikonins in Root Periderm of Two Invasive Echium spp. Weeds in Australia
Abstract
:1. Introduction
2. Results and Discussion
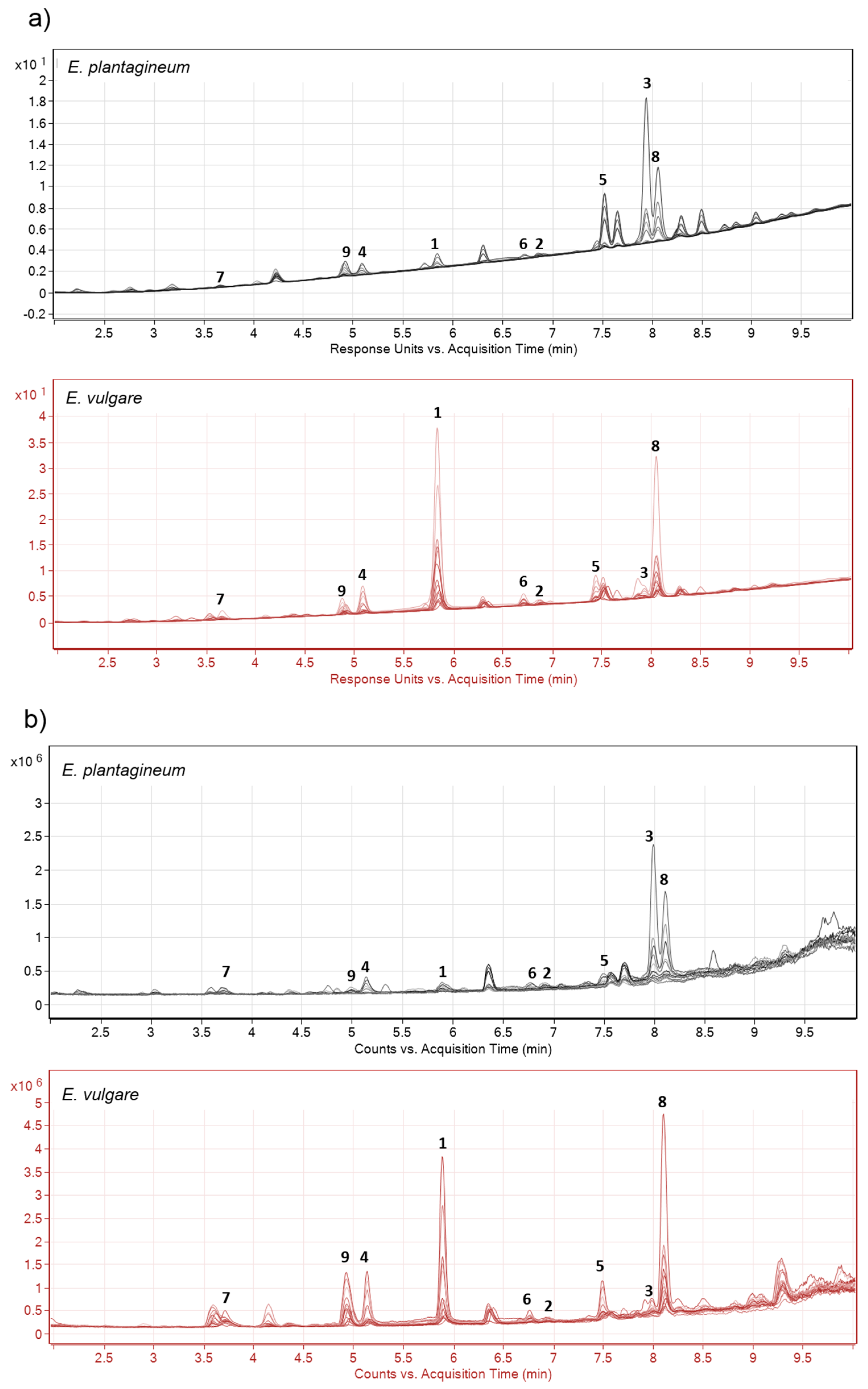
2.1. Identification of Isohexenylnaphthazarins in Echium plantagineum Root Periderm Extracts
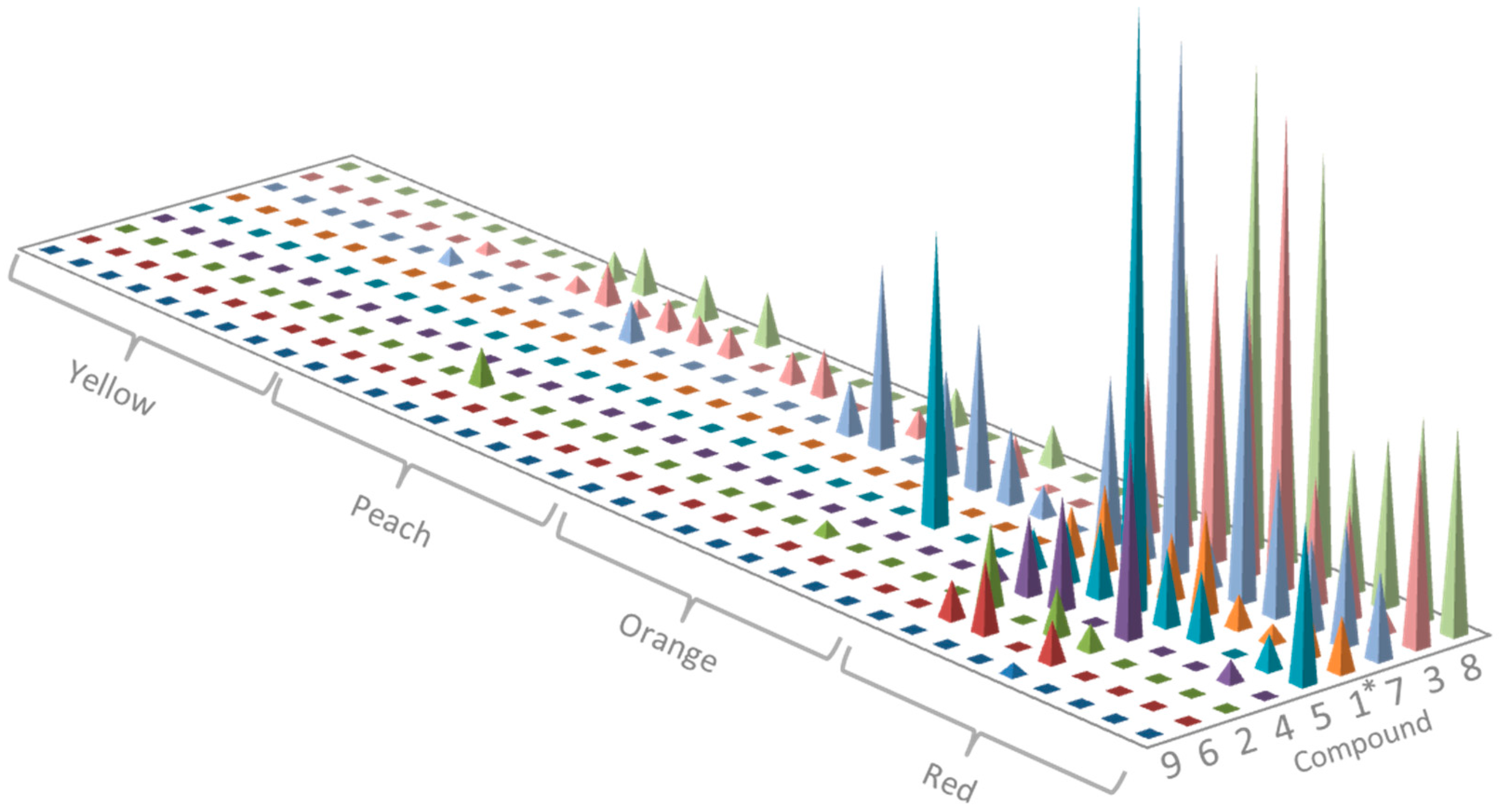
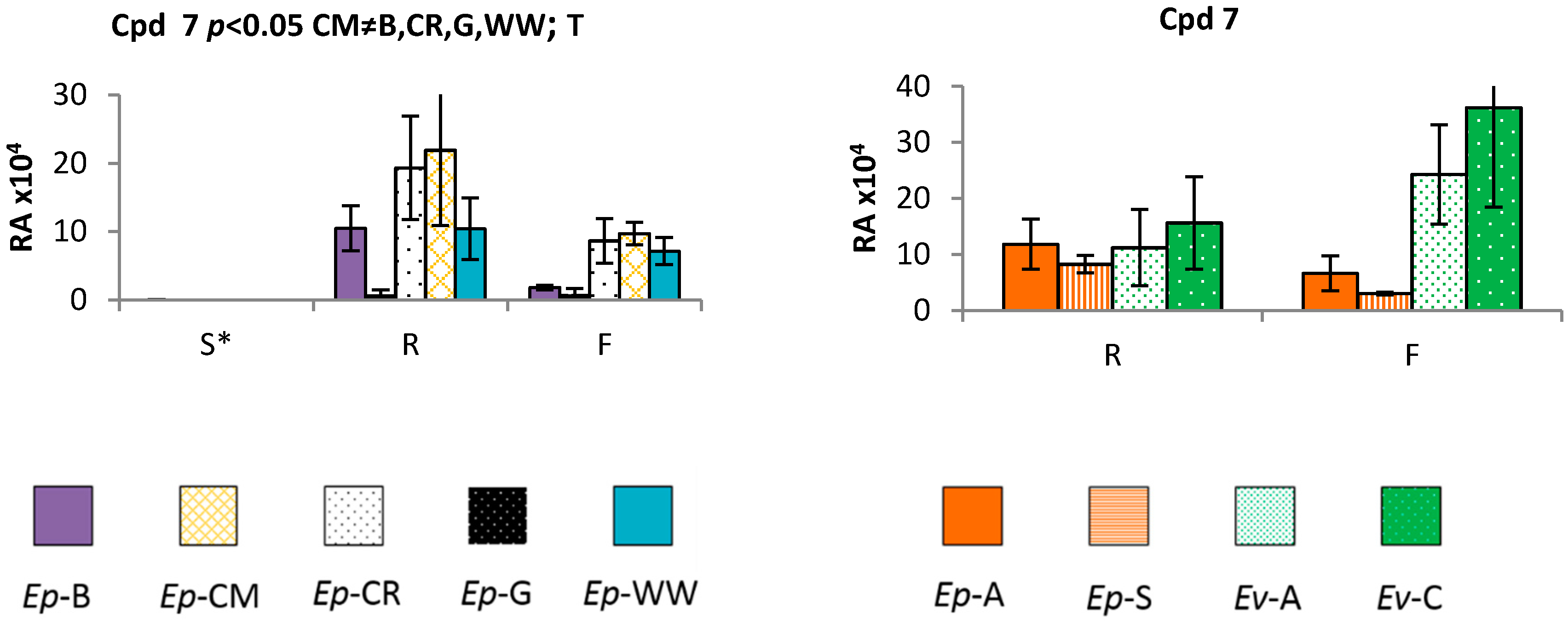
2.2. Profiling of Coloured Periderm Extracts
2.3. Time and Population Dependent Production of Isohexenylnaphthazarins
3. Materials and Methods
3.1. Chemicals and Standards
3.2. Source of Plant Material and Sample Preparation
3.3. Echium plantagineum Phenological Cycle Study
3.4. Comparative Profiling of Two Echium Species
3.5. UHPLC-MS Analysis
3.6. Spectrophotometry
3.7. Data Analysis
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgements
Author Contributions
Conflicts of Interest
References
- De Vos, R.C.; Moco, S.; Lommen, A.; Keurentjes, J.J.; Bino, R.J.; Hall, R.D. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2007, 2, 778–791. [Google Scholar] [CrossRef] [PubMed]
- Rochfort, S. Metabolomics reviewed: A new “omics” platform technology for systems biology and implications for natural products research. J. Nat. Prod. 2005, 68, 1813–1820. [Google Scholar] [CrossRef] [PubMed]
- Weston, L.A.; Skoneczny, D.; Weston, P.A.; Weidenhamer, J.D. Metabolic profiling: An overview—New approaches for the detection and functional analysis of biologically active secondary plant products. J. Allelochem. Interact 2015, 2, 15–27. [Google Scholar]
- Fiehn, O. Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks. Comp. Funct. Genom. 2001, 2, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Roessner, U.; Luedemann, A.; Brust, D.; Fiehn, O.; Linke, T.; Willmitzer, L.; Fernie, A.R. Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 2001, 13, 11–29. [Google Scholar] [CrossRef] [PubMed]
- Schauer, N.; Fernie, A.R. Plant metabolomics: Towards biological function and mechanism. Trends Plant Sci. 2006, 11, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Farag, M.A.; Huhman, D.V.; Dixon, R.A.; Sumner, L.W. Metabolomics reveals novel pathways and differential mechanistic and elicitor-specific responses in phenylpropanoid and isoflavonoid biosynthesis in Medicago truncatula cell cultures. Plant Physiol. 2008, 146, 387–402. [Google Scholar] [CrossRef] [PubMed]
- Skoneczny, D.; Garcia Duran, A.; Costas Gil, A.; Weston, P.A.; Torres, A.; Macias, F.A.; Gurr, G.M.; Weston, L.A. Metabolic profiling of secondary compounds in Echium plantagineum and Echium vulgare, two exotic invaders in Australia. In Proceedings of the 7th World Congress on Allelopathy, Vigo, Spain, 28 July–1 August 2014; Reigosa Roger, M., Sanchez-Moreiras, A., Eds.; International Allelopathy Society: Vigo, Spain, 2014. [Google Scholar]
- Skoneczny, D.; Weston, P.A.; Zhu, X.; Gurr, G.M.; Callaway, R.M.; Weston, L.A. Metabolic profiling of pyrrolizidine alkaloids in foliage of two Echium spp. Invaders in Australia—A case of novel weapons? IJMS 2015, 16, 26721–26737. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Yang, H.; Qi, L.W.; Liu, E.H.; Ren, M.T.; Yan, Y.T.; Chen, J.; Li, P. Unbiased metabolite profiling by liquid chromatography–quadrupole time-of-flight mass spectrometry and multivariate data analysis for herbal authentication: Classification of seven Lonicera species flower buds. J. Chromatogr. 2012, 1245, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.T.; Sparkman, O.D. Introduction to Mass Spectrometry: Instrumentation, Applications, and Strategies for Data Interpretation; John Wiley & Sons: Croydon, UK, 2007. [Google Scholar]
- Badri, D.V.; Weir, T.L.; van der Lelie, D.; Vivanco, J.M. Rhizosphere chemical dialogues: Plant–microbe interactions. Curr. Opin. Biotechnol. 2009, 20, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Çirak, C.; Radušienė, J.; Ivanauskas, L.; Janulis, V. Variation of bioactive secondary metabolites in Hypericum origanifolium during its phenological cycle. Acta Physiol. Plant. 2007, 29, 197–203. [Google Scholar] [CrossRef]
- Jones, O.H.; Maguire, M.; Griffin, J.; Jung, Y.H.; Shibato, J.; Rakwal, R.; Agrawal, G.; Jwa, N.S. Using metabolic profiling to assess plant-pathogen interactions: An example using rice (Oryza sativa) and the blast pathogen magnaporthe grisea. Eur. J. Plant Pathol. 2011, 129, 539–554. [Google Scholar] [CrossRef]
- Quinn, J.C.; Kessell, A.; Weston, L.A. Secondary plant products causing photosensitization in grazing herbivores: Their structure, activity and regulation. IJMS 2014, 15, 1441–1465. [Google Scholar] [CrossRef] [PubMed]
- Weston, P.A.; Weston, L.A.; Hildebrand, S. Environmental impact on biocontrol agents and secondary chemistry of Paterson’s curse (Echium plantagineum). In Proceedings of the Eighteenth Australasian Weeds Conference, Melbourne, Australia, 8–11 October 2012; pp. 203–207.
- Weston, L.A.; Ryan, P.R.; Watt, M. Mechanisms for cellular transport and release of allelochemicals from plant roots into the rhizosphere. J. Exp. Bot. 2012, 63, 3445–3454. [Google Scholar] [CrossRef] [PubMed]
- Bertin, C.; Yang, X.; Weston, L. The role of root exudates and allelochemicals in the rhizosphere. Plant Soil 2003, 256, 67–83. [Google Scholar] [CrossRef]
- Weston, L.A.; Duke, S.O. Weed and crop allelopathy. Crit. Rev. Plant Sci. 2003, 22, 367–389. [Google Scholar] [CrossRef]
- Mathesius, U.; Watt, M. Rhizosphere signals for plant–microbe interactions: Implications for field-grown plants. In Progress in Botany 72; Springer: Cham, Switzerland, 2011; pp. 125–161. [Google Scholar]
- Weston, L.A.; Mathesius, U. Flavonoids: Their structure, biosynthesis and role in the rhizosphere, including allelopathy. J. Chem. Ecol. 2013, 39, 283–297. [Google Scholar] [CrossRef] [PubMed]
- Hierro, J.L.; Callaway, R.M. Allelopathy and exotic plant invasion. Plant Soil 2003, 256, 29–39. [Google Scholar] [CrossRef]
- Brigham, L.A.; Michaels, P.J.; Flores, H.E. Cell-specific production and antimicrobial activity of naphthoquinones in roots of Lithospermum erythrorhizon. Plant Physiol. 1999, 119, 417–428. [Google Scholar] [CrossRef] [PubMed]
- Papageorgiou, V.P.; Assimopoulou, A.N.; Couladouros, E.A.; Hepworth, D.; Nicolaou, K.C. The chemistry and biology of alkannin, shikonin, and related naphthazarin natural products. Angew. Chem. Int. Ed. 1999, 38, 270–301. [Google Scholar] [CrossRef]
- Tappeiner, J.; Vasiliou, A.; Ganzera, M.; Fessas, D.; Stuppner, H.; Papageorgiou, V.P.; Assimopoulou, A.N. Quantitative determination of alkannins and shikonins in endemic Mediterranean Alkanna species. Biomed. Chromatogr. 2013, 28, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Weston, P.; Weston, L.; Hildebrand, S. Metabolic profiling in Echium plantagineum: Presence of bioactive pyrrolizidine alkaloids and napthoquinones from accessions across southeastern Australia. Phytochem. Rev. 2013, 12, 1–7. [Google Scholar] [CrossRef]
- Klemow, K.M.; Clements, D.R.; Threadgill, P.F.; Cavers, P.B. The biology of Canadian weeds. 116. Echium vulgare L. Can. J. Plant Sci. 2002, 82, 235–248. [Google Scholar] [CrossRef]
- Parsons, W.T.; Cuthbertson, E.G. Paterson’s curse, Salvation Jane. In Noxious Weeds of Australia, 2nd ed.; CSIRO Publishing: Melbourne, Australia, 2001; pp. 325–330. [Google Scholar]
- Grigulis, K.; Sheppard, A.W.; Ash, J.E.; Groves, R.H. The comparative demography of the pasture weed Echium plantagineum between its native and invaded ranges. J. Appl. Ecol. 2001, 38, 281–290. [Google Scholar] [CrossRef]
- Piggin, C.M. The biology of Australian weeds. 8. Echium plantagineum L. J. Aust. Inst. Agric. Sci. 1982, 48, 3–16. [Google Scholar]
- Australia’s Virtual Herbarium. Available online: http://avh.chah.org.au (accessed on 29 October 2013).
- Zhu, X.; Gopurenko, D.; Skoneczny, D.; Lepschi, B.J.; Spencer, M.A.; Gurr, G.M.; Callaway, R.M.; Serrano, M.; Weston, L.A. Introduction of Paterson’s Curse (Echium plantagineum) to Australia—Unravelling the Story by DNA Sequence Analysis. In Proceedings of the Twentieth Australasian Weeds Conference, Perth, Australia, 11–15 September 2016; pp. 157–161.
- Zhu, X.; Meyer, L.; Gopurenko, D.; Weston, P.A.; Gurr, G.M.; Callaway, R.M.; Lepschi, B.J.; Weston, L.A. Selection of DNA barcoding regions for identification and genetic analysis of two Echium invaders in Australia: E. plantagineum and E. vulgare. In Proceedings of the Nineteenth Australasian Weeds Conference, Hobart, Australia, 1–4 September 2014.
- Natural Resource Management. Available online: http://www.nrmsouth.org.au/wp-content/uploads/2014/10/patersons_curse.pdf (accessed on 29 October 2013).
- Colegate, S.M.; Edgar, J.A.; Knill, A.M.; Lee, S.T. Solid-phase extraction and HPLC-MS profiling of pyrrolizidine alkaloids and their N-oxides: A case study of Echium plantagineum. Phytochem. Anal. 2005, 16, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Skoneczny, D.; Weidenhamer, J.D.; Mwendwa, J.M.; Weston, P.A.; Gurr, G.M.; Callaway, R.M.; Weston, L.A. Identification and localization of bioactive naphthoquinones in the roots and rhizosphere of Paterson’s curse (Echium plantagineum), a noxious invader. J. Exp. Bot. 2016, 67, 3777–3788. [Google Scholar] [CrossRef] [PubMed]
- Albreht, A.; Vovk, I.; Simonovska, B.; Srbinoska, M. Identification of shikonin and its ester derivatives from the roots of Echium italicum L. J. Chromatogr. 2009, 1216, 3156–3162. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Jiang, Z.; Leung, K.S.-Y.; Zhao, Z. Simultaneous determination of naphthoquinone derivatives in boraginaceous herbs by high-performance liquid chromatography. Anal. Chim. Acta 2006, 577, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, Y.; Shinkai, Y.; Miura, T.; Cho, A.K. The chemical biology of naphthoquinones and its environmental implications. Annu. Rev. Pharmacol. Toxicol. 2012, 52, 221–247. [Google Scholar] [CrossRef] [PubMed]
- Nishibe, S.; Tamayama, Y.; Sasahara, M.; Andary, C. A phenylethanoid glycoside from plantago asiatica. Phytochemistry 1995, 38, 741–743. [Google Scholar] [CrossRef]
- Weston, L.A.; Weston, P.A.; McCully, M. Production of bioactive napthoquinones by roots of Paterson’s curse (Echium plantagineum)—Implications for invasion success? Pak. J. Weed Sci. Res. 2011, 18, 677–686. [Google Scholar]
- Gara, R.K.; Srivastava, V.K.; Duggal, S.; Bagga, J.K.; Bhatt, M.; Sanyal, S.; Mishra, D.P. Shikonin selectively induces apoptosis in human prostate cancer cells through the endoplasmic reticulum stress and mitochondrial apoptotic pathway. J. Biomed. Sci. 2015, 22, 26. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yang, L.; Oppenheim, J.; Howard, M. Cellular pharmacology studies of shikonin derivatives. Phytother. Res. 2002, 16, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Garcia Duran, A.; Chinchillan, N.; Skoneczny, D.; Torres Martinez, A.; Mollinillo, J.M.G.; Weston, L.A.; Macias, F.A. Isolation of bioactive naphthoquinones from the roots of Echium plantagineum L. (Paterson’s curse). In Proceedings of the 7th World Congress on Allelopathy, Vigo, Spain, 28 July–1 August 2014; Reigosa Roger, M., Sanchez-Moreiras, A., Eds.; International Allelopathy Society: Vigo, Spain, 2014. [Google Scholar]
- Cho, M.H.; Paik, Y.S.; Hahn, T.-R. Physical stability of shikonin derivatives from the roots of lithospermum erythrorhizon cultivated in Korea. J. Agric. Food Chem. 1999, 47, 4117–4120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xiao, S.; Li, X.; Zeng, X.; Cheng, Y. Ionization of shikonin derivatives using negative-ion electrospray mass spectrometry: [M − H]− versus [M + e]•−. J. Mass Spectrom. 2012, 47, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, Y.; Liang, Z.; Ho, A.; Wong, L.; Yong, P.; Chen, H.; Zhao, Z. Distribution of toxic alkaloids in tissues from three herbal medicine Aconitum species using laser micro-dissection, UHPLC–QTOF MS and LC-MS/MS techniques. Phytochemistry 2014, 107, 155–174. [Google Scholar] [CrossRef] [PubMed]
- Miyase, T.; Ishino, M.; Akahori, C.; Ueno, A.; Ohkawa, Y.; Tanizawa, H. Phenylethanoid glycosides from Plantago asiatica. Phytochemistry 1991, 30, 2015–2018. [Google Scholar] [CrossRef]
- Weidenhamer, J.D.; Xiaocheng, Z.; Skoneczny, D.; Mwendwa, J.; Weston, P.A.; Weston, L.A. Root exudation of lipophilic naphthoquionones by Paterson’s curse: A clue to their ecological role? In Proceedings of the 7th World Congress on Allelopathy, Vigo, Spain, 28 July–1 August 2014; Reigosa Roger, M., Sanchez-Moreiras, A., Eds.; International Allelopathy Society: Vigo, Spain, 2014. [Google Scholar]
- Callaway, R.M.; Aschehoug, E.T. Invasive plants versus their new and old neighbors: A mechanism for exotic invasion. Science 2000, 290, 521–523. [Google Scholar] [CrossRef] [PubMed]
- Wardle, D.A.; Karban, R.; Callaway, R.M. The ecosystem and evolutionary contexts of allelopathy. Trends Ecol. Evol. 2011, 26, 655–662. [Google Scholar]
- Pavarini, D.P.; Pavarini, S.P.; Niehues, M.; Lopes, N.P. Exogenous influences on plant secondary metabolite levels. Anim. Feed Sci. Technol. 2012, 176, 5–16. [Google Scholar] [CrossRef]
- Ben Farhat, M.; Jordán, M.J.; Chaouch-Hamada, R.; Landoulsi, A.; Sotomayor, J.A. Changes in phenolic profiling and antioxidant capacity of Salvia aegyptiaca L. By-products during three phenological stages. LWT-Food Sci. Technol. 2015, 63, 791–797. [Google Scholar] [CrossRef]
- Garcia Duran, A.; Skoneczny, D.; Torres, A.; Valdivia, M.M.; Molinillo, J.M.G.; Weston, L.A.; Macias, F.A. Bioactivity of isohexenylnapthazarins from root extracts of Echium spp. In Native and Invaded Ranges, Proceedings of the International Chemical Congress of Pacific Basin Societies; Honolulu, HI, USA, 15–20 December 2015, International Chemical Congress of Pacific Basin Societies: Honolulu, HI, USA.
- Boehm, R.; Sommer, S.; Shu-Ming, L.; Heide, L. Genetic engineering on shikonin biosynthesis: Expression of the bacterial ubiA gene in Lithospermum erythrorhizon. Plant Cell Physiol. 2000, 41, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.I. A phylogenetic analysis of morphological and molecular characters of boraginaceae: Evolutionary relationships, taxonomy, and patterns of character evolution. Cladistics 2013, 30, 139–169. [Google Scholar] [CrossRef]
- Ozgen, U.; Miloglu, F.D.; Bulut, G. Quantitative determination of shikonin derivatives with UV-vis spectrophotometric methods in the roots of Onosma nigricaule. Rev. Anal. Chem. 2011, 30, 59–63. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds 1, 2, 3, 7 are available from the authors.

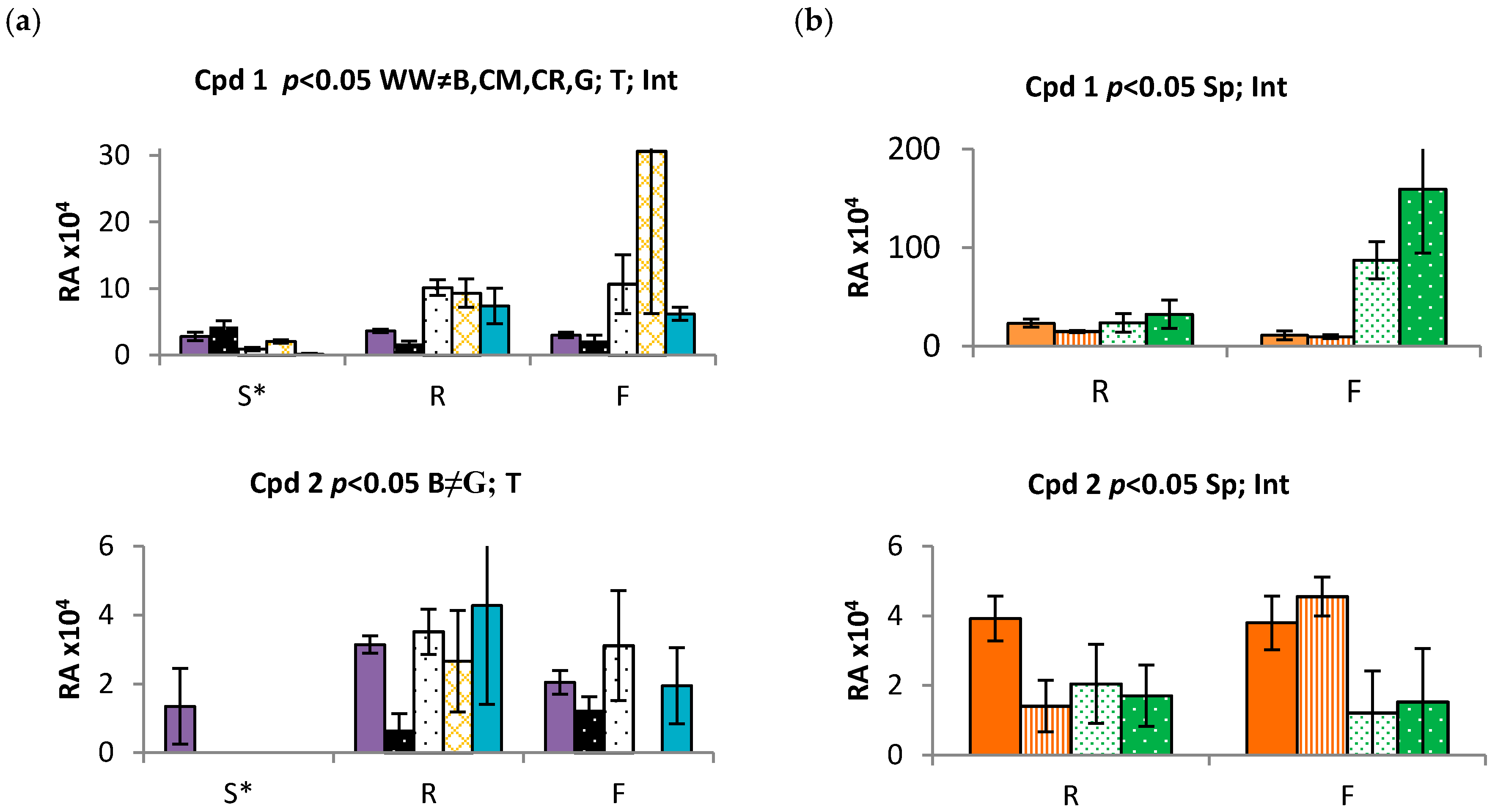
| Compound No. | Compound | Formula | Calculated [M − H] | Calculated [M−] | Measured (m/z) | Δppm | Production | Used Collision Energy (CE) | Approx. RT (min) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Acetylshikonin * | C18H18O6 | 329.1031 | 330.1109 | 330.1102 | 2.12 | 270.0915 | 10 | 5.9 |
| 2 | Deoxyshikonin * | C16H16O4 | 271.0976 | 272.1054 | 272.1048 | 2.21 | 203.0349 | 20 | 7.0 |
| 3 | Dimethylacrylshikonin * | C21H22O6 | 369.1344 | 370.1422 | 370.1407 | 4.05 | 270.0904 | 20 | 8.0 |
| 4 | β-hydroxyisovalerylshikonin | C21H24O7 | 387.1449 | 388.1528 | 387.1447 | 0.52 | 269.0814 | 10 | 5.2 |
| 5 | Isobutyrylshikonin | C20H22O6 | 357.1344 | 358.1422 | 358.1425 | 0.84 | 270.0902 | 10 | 7.6 |
| 6 | Propionylshikonin | C19H20O6 | 343.1187 | 344.1265 | 343.1169 | 5.25 | 269.0821 | 20 | 6.9 |
| 7 | Shikonin * | C16H16O5 | 287.0925 | 288.1003 | 287.0921 | 1.39 | 218.0221 | 20 | 3.8 |
| 8 | Compound 8 # | C21H24O6 | 371.1500 | 372.1578 | 372.1582 | 1.08 | 270.0896 | 10 | 8.2 |
| 9 | Compound 9 # | C21H24O6 | 371.1500 | 372.1578 | 371.1508 | 2.16 | 269.0820 | 20 | 4.9 |
| Species | Location | Collection Year | GPS Coordinates | |
|---|---|---|---|---|
| Longitude | Latitude | |||
| E. vulgare | Adaminaby | 2013 | −35.969875 | 148.712235 |
| Cooma | 2014 | −36.245188 | 149.028493 | |
| E. plantagineum | Adelong | 2013 | −35.296432 | 148.058302 |
| Bendigo | 2011 | −36.846232 | 144.181611 | |
| Grenfell | 2011 | −33.918941 | 148.160026 | |
| Coombah | 2011 | −32.980887 | 141.625002 | |
| Cobar | 2011 | −31.524130 | 145.589078 | |
| Silverton | 2013 | −31.879176 | 141.213968 | |
| Wagga Wagga | 2011 | −35.054791 | 147.350295 | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skoneczny, D.; Weston, P.A.; Zhu, X.; Gurr, G.M.; Callaway, R.M.; Barrow, R.A.; Weston, L.A. Metabolic Profiling and Identification of Shikonins in Root Periderm of Two Invasive Echium spp. Weeds in Australia. Molecules 2017, 22, 330. https://doi.org/10.3390/molecules22020330
Skoneczny D, Weston PA, Zhu X, Gurr GM, Callaway RM, Barrow RA, Weston LA. Metabolic Profiling and Identification of Shikonins in Root Periderm of Two Invasive Echium spp. Weeds in Australia. Molecules. 2017; 22(2):330. https://doi.org/10.3390/molecules22020330
Chicago/Turabian StyleSkoneczny, Dominik, Paul A. Weston, Xiaocheng Zhu, Geoff M. Gurr, Ragan M. Callaway, Russel A. Barrow, and Leslie A. Weston. 2017. "Metabolic Profiling and Identification of Shikonins in Root Periderm of Two Invasive Echium spp. Weeds in Australia" Molecules 22, no. 2: 330. https://doi.org/10.3390/molecules22020330