Functional Analysis of a Wheat AGPase Plastidial Small Subunit with a Truncated Transit Peptide
Abstract
:1. Introduction
2. Results and Discussion
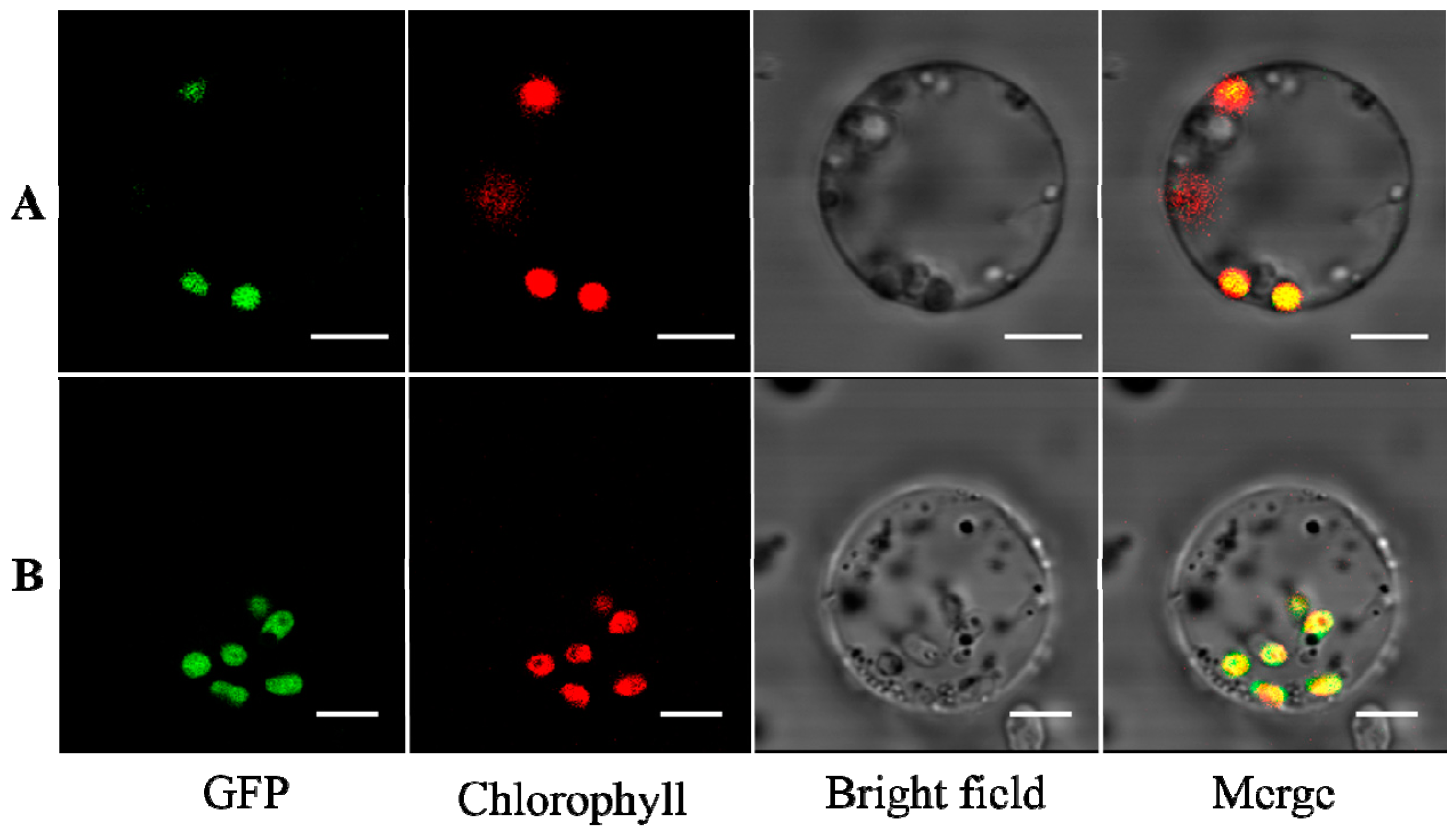
2.1. TaAGPS1b-EU586278 Subunit with the Truncated Transit Peptide Could Be Located in Plastid
2.2. Function of TaAGPS1b-EU586278 with the Truncated Transit Peptide Could Not Be Changed
3. Materials and Methods
3.1. Subcellular Localization of TaAGPS1b Subunit with the Truncated Transit Peptide
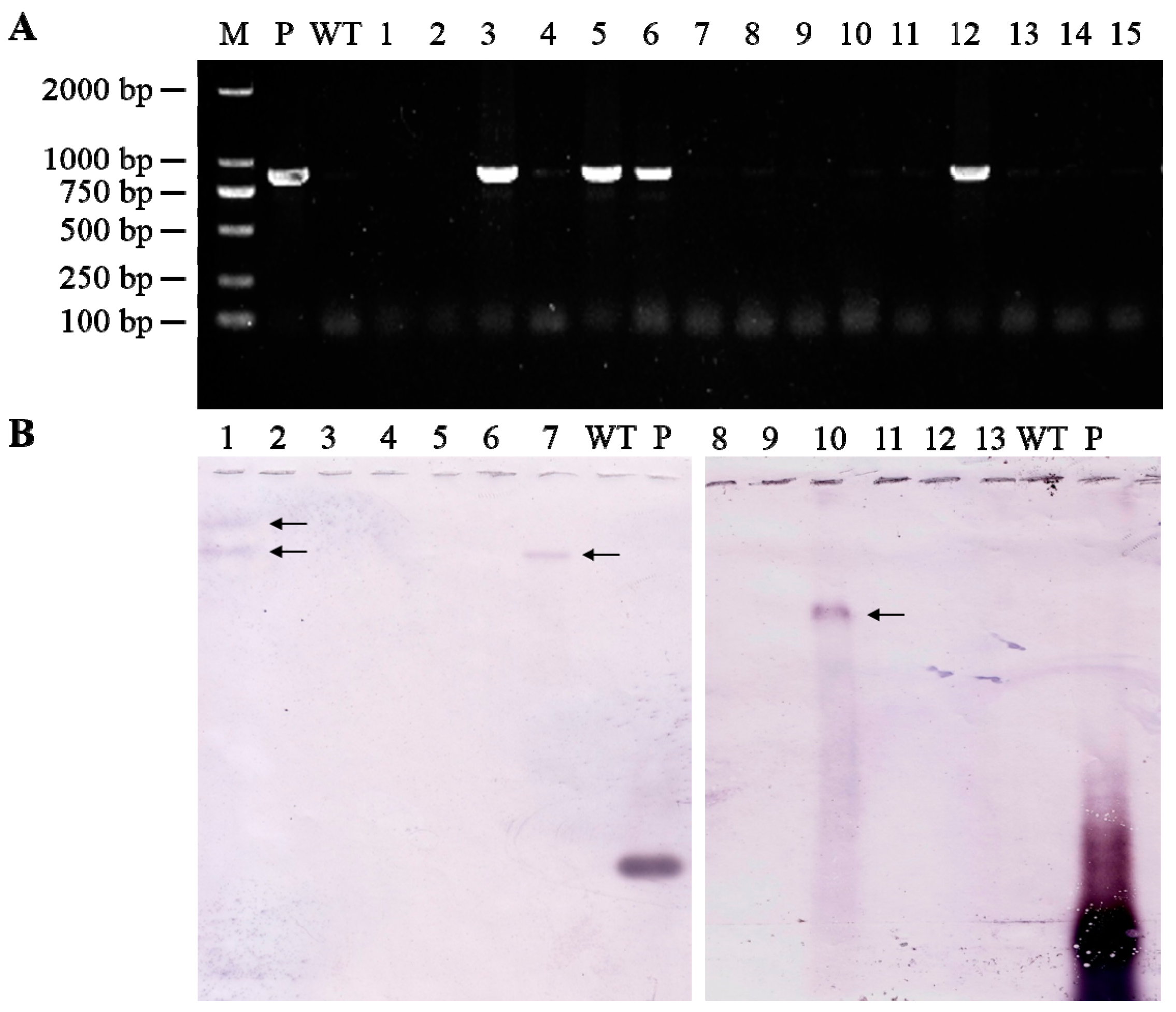
3.2. Identification of Transgenic Wheat Plants Expressing TaAGPS1b-EU586278 Transcript
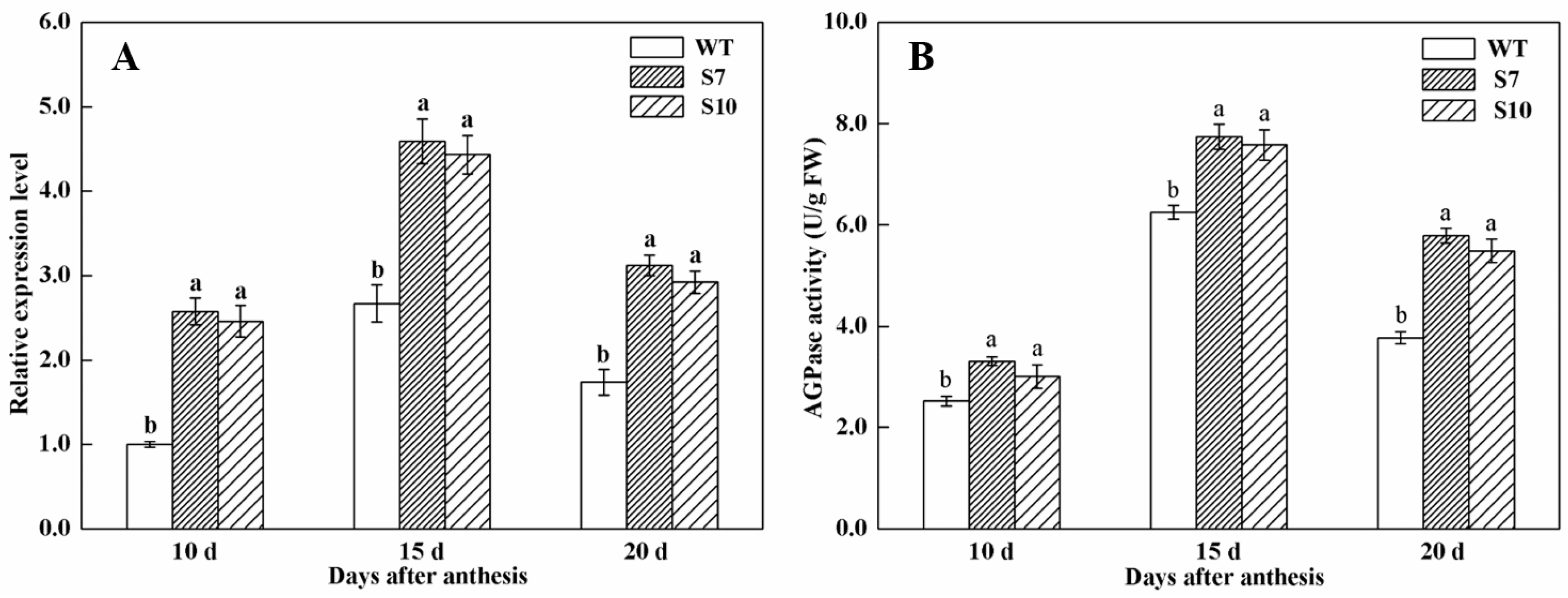
3.3. Transcript Levels of TaAGPS1b-EU586278 Gene in Transgenic Wheat Lines
3.4. Assays of AGPase Activity in Grains of Transgenic Wheat Lines
3.5. Analysis of Starch Content and Yield Characteristics between WT and Transgenic Wheat Lines
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgements
Author Contributions
Conflicts of Interest
References
- Zeeman, S.C.; Kossmann, J.; Smith, A.M. Starch: Its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant. Biol. 2010, 61, 209–234. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.S.; Ryoo, N.; Hahn, T.R.; Walia, H.; Nakamura, Y. Starch biosynthesis in cereal endosperm. Plant. Physiol. Biochem. 2010, 48, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wu, H.; Yuan, B.; Huang, M.; Yang, H.; Li, A.; Cheng, R. Synthesis of amphoteric starch-based grafting flocculants for flocculation of both positively and negatively charged colloidal contaminants from water. Chem. Eng. J. 2014, 244, 209–217. [Google Scholar] [CrossRef]
- Siedlecka, A.; Ciereszko, I.; Mellerowicz, E.; Martz, F.; Chen, J.; Kleczkowski, L.A. The small subunit ADP-glucose pyrophosphorylase (ApS) promoter mediates okadaic acid-sensitive uidA expression in starch-synthesizing tissues and cells in Arabidopsis. Planta 2003, 217, 184–192. [Google Scholar] [PubMed]
- Saripalli, G.; Gupta, P.K. AGPase: Its role in crop productivity with emphasis on heat tolerance in cereals. Theor. Appl. Genet. 2015, 128, 1893–1916. [Google Scholar] [CrossRef] [PubMed]
- Salamone, P.R.; Kavakli, I.H.; Slattery, C.J.; Okita, T.W. Directed molecular evolution of ADP-glucose pyrophosphorylase. Proc. Natl. Acad. Sci. USA 2002, 99, 1070–1075. [Google Scholar] [CrossRef] [PubMed]
- Burton, R.A.; Johnson, P.E.; Beckles, D.M.; Fincher, G.B.; Jenner, H.L.; Naldrett, M.J.; Denyer, K. Characterization of the genes encoding the cytosolic and plastidial forms of ADP-glucose pyrophosphorylase in wheat endosperm. Plant. Physiol. 2002, 130, 1464–1475. [Google Scholar] [CrossRef] [PubMed]
- Geigenberger, P. Regulation of starch biosynthesis in response to a fluctuating environment. Plant. Physiol. 2011, 155, 1566–1577. [Google Scholar] [CrossRef] [PubMed]
- Denyer, K.; Dunlap, F.; Thorbjørnsen, T.; Keeling, P.; Smith, A.M. The major form of ADP-glucose pyrophosphorylase in maize endosperm is extra-plastidial. Plant. Physiol. 1996, 112, 779–785. [Google Scholar] [CrossRef] [PubMed]
- Thorbjørnsen, T.; Villand, P.; Kleczkowski, L.A.; Olsen, O.A. A single gene encodes two different transcripts for the ADP-glucose pyrophosphorylase small subunit from barley (Hordeum vulgare). Biochem. J. 1996, 313, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Rösti, S.; Denyer, K. Two paralogous genes encoding small subunits of ADP-glucose pyrophosphorylase in maize, Bt2 and L2, replace the single alternatively spliced gene found in other cereal species. J. Mol. Evol. 2007, 65, 316–327. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.K.; Mir, R.R.; Mohan, A.; Kumar, J. Wheat genomics: Present status and future prospects. Int. J. Plant. Genomics. 2008, 2008, 896451. [Google Scholar] [CrossRef] [PubMed]
- Tetlow, I.J.; Davies, E.J.; Vardy, K.A.; Bowsher, C.G.; Burrell, M.M.; Emes, M.J. Subcellular localization of ADP-glucose pyrophosphorylase in developing wheat endosperm and analysis of the properties of a plastidial isoform. J. Exp. Bot. 2003, 54, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Beckles, D.M.; Smith, A.M.; ap Rees, T. A cytosolic ADP-glucose pyrophosphorylase is a feature of graminaceous endosperms, but not of other starch-storing organs. Plant. Physiol. 2001, 125, 818–827. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.Z.; Zheng, B.B.; Shen, B.Q.; Peng, H.F.; Guo, T.C. A novel Ta.AGP.S.1b transcript in Chinese common wheat (Triticum aestivum L.). C. R. Biol. 2010, 333, 716–724. [Google Scholar] [CrossRef] [PubMed]
- Bhushan, S.; Kuhn, C.; Berglund, A.K.; Roth, C.; Glaser, E. The role of the N-terminal domain of chloroplast targeting peptides in organellar protein import and miss-sorting. FEBS Lett. 2006, 580, 3966–3972. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, P.F.; Glaser, E. Processing peptidases in mitochondria and chloroplasts. BBA-Mol. Cell. Res. 2013, 1833, 360–370. [Google Scholar] [CrossRef] [PubMed]
- Bruce, B.D. Chloroplast transit peptides: Structure, function and evolution. Trends Cell. Biol. 2000, 10, 440–447. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef] [PubMed]
- Ohdan, T.; Francisco, P.B.; Sawada, J.T.; Hirose, T.; Nakamura, Y. Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J. Exp. Bot. 2005, 56, 3229–3244. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.C.; Zhang, X.K.; Yin, G.H.; Wang, L.N.; Han, Y.L.; Chen, L.; Huang, F.; Tang, J.W.; Xia, X.C.; He, Z.H. Genetic gains in grain yield, net photosynthesis and stomatal conductance achieved in Henan Province of China between 1981 and 2008. Field Crop. Res. 2011, 122, 225–233. [Google Scholar] [CrossRef]
- Giroux, M.J.; Shaw, J.; Barry, G.; Cobb, B.G.; Greene, T.; Okita, T.; Hannah, L.C. A single mutation that increases maize seed weight. Proc. Natl. Acad. Sci. USA 1996, 93, 5824–5829. [Google Scholar] [CrossRef]
- Smidansky, E.D.; Clancy, M.; Meyer, F.D.; Lanning, S.P.; Blake, N.K.; Talbert, L.E.; Giroux, M.J. Enhanced ADP-glucose pyrophosphorylase activity in wheat endosperm increase seed yield. Proc. Natl. Acad. Sci. USA 2002, 99, 1724–1729. [Google Scholar] [CrossRef] [PubMed]
- Smidansky, E.D.; Meyer, F.D.; Blakeslee, B.; Weglarz, T.E.; Greene, T.W.; Giroux, M.J. Expression of a modified ADP-glucose pyrophosphorylase large subunit in wheat seeds stimulates photosynthesis and carbon metabolism. Planta 2007, 225, 965–976. [Google Scholar] [CrossRef] [PubMed]
- Nagai, Y.S.; Sakulsingharoj, C.; Edwards, G.E.; Satoh, H.; Greene, T.W.; Blakeslee, B.; Okita, T.W. Control of starch synthesis in cereals: Metabolite analysis of transgenic rice expressing an up-regulated cytoplasmic ADP-glucose pyrophosphorylase in developing seeds. Plant. Cell. Physiol. 2009, 50, 635–643. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhang, S.; Zhao, Y.; Li, B.; Zhang, J. Over-expression of AGPase genes enhances seed weight and starch content in transgenic maize. Planta 2011, 233, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Yu, X.; Qi, X.; Yu, Q.; Deng, S.; Bai, B.; Li, N.; Zhang, A.; Zhu, C.; Liu, B.; Pang, J. Multigene engineering of starch biosynthesis in maize endosperm increases the total starch content and the proportion of amylase. Transgenic Res. 2013, 22, 1133–1142. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.; Liu, G.; Peng, X.; Wei, L.; Wang, C.; Zhu, Y.; Guo, T. Increasing the starch content and grain weight of common wheat by overexpression of the cytosolic AGPase large subunit gene. Plant. Physiol. Biochem. 2013, 73, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Huang, W.; Huang, Q.; Qin, X.; Dan, Z.; Yao, G.; Zhu, Y. The mechanism of ORFH79 suppression with the artificial restorer fertility gene Mt-GRP162. New Phytol. 2013, 199, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Su, J.; Duan, S.; Ao, Y.; Dai, J.; Liu, J.; Wang, J. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant. Methods 2011, 7, 1. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.H.; Liu, N.Y.; Tang, Z.S.; Liu, J.; Yang, W.C. Arabidopsis GLUTAMINE-RICH PROTEIN23 is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III. Plant. Cell. 2006, 18, 815–830. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.J.; Zhao, S.Y.; Chen, H.M.; Zhao, Q.Z.; Hu, Z.M.; Hou, B.K.; Xia, G.M. Transgenic wheat progeny resistant to powdery mildew generated by Agrobacterium inoculum to the basal portion of wheat seedling. Plant. Cell. Rep. 2006, 25, 1199–1204. [Google Scholar] [CrossRef] [PubMed]
- Saghai-Maroof, M.A.; Soliman, K.M.; Jorgensen, R.A.; Allard, R.W. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 1984, 81, 8014–8018. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Dai, T.; Jiang, D.; Cao, W. Effects of high temperature on key enzymes involved in starch and protein formation in grains of two wheat cultivars. J. Agron. Crop. Sci. 2008, 194, 47–54. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds are not available from the authors.
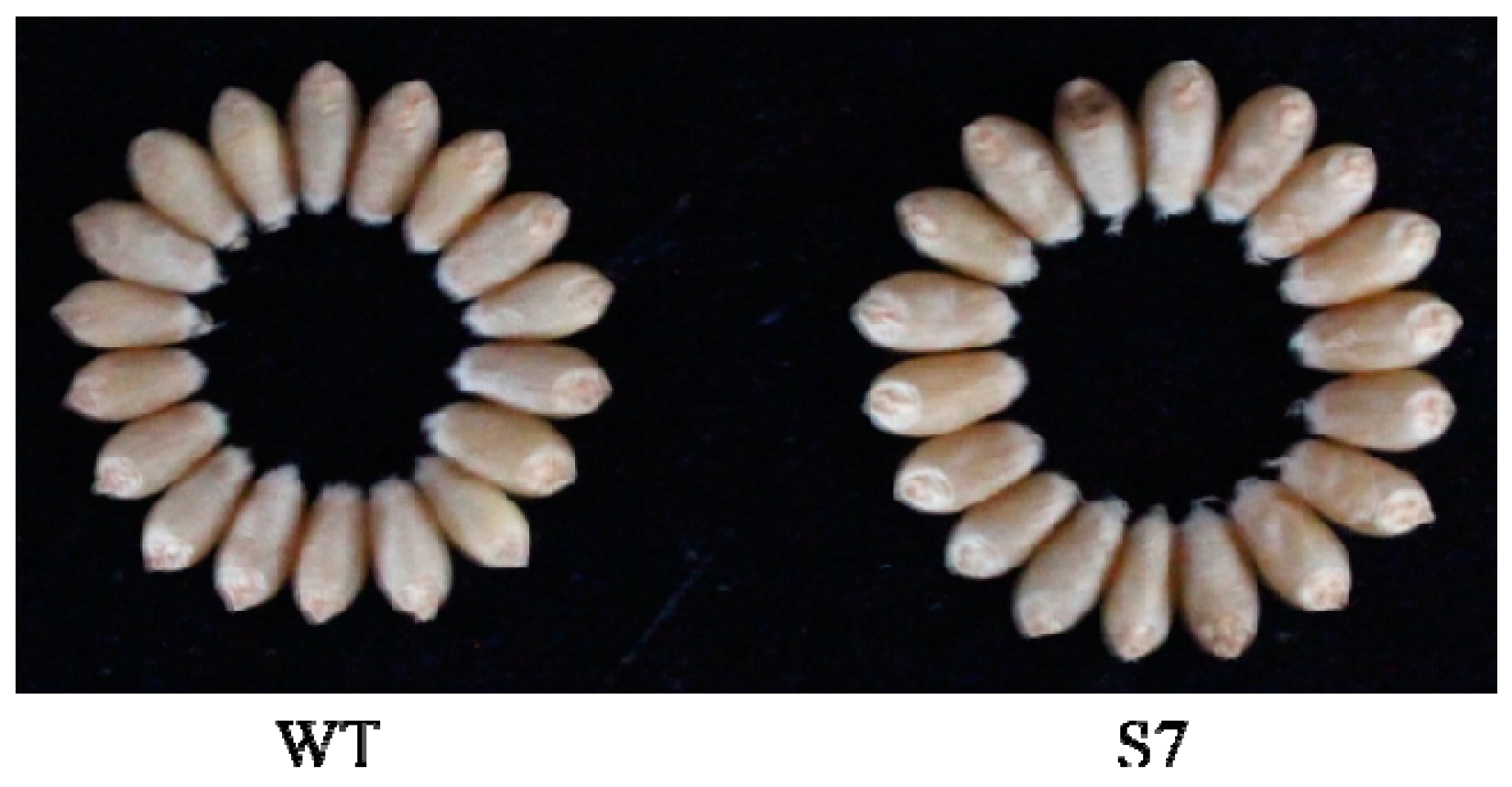
| Lines | Spike Number (per Plant) | Grain Number (per Spike) | Kernel Weight per Grain (mg) | Weight of Individual Spike (g) | Starch Content (mg/Grain) |
|---|---|---|---|---|---|
| WT | 6.3 ± 0.3a | 23.0 ± 0.6a | 53.28 ± 0.33b | 1.23 ± 0.03b | 34.77 ± 0.64b |
| S7 | 7.0 ± 0.6a | 23.3 ± 0.3a | 59.02 ± 0.61a | 1.38 ± 0.01a | 39.86 ± 0.52a |
| S10 | 6.7 ± 0.3a | 23.7 ± 0.3a | 57.12 ± 1.47a | 1.35 ± 0.16a | 38.77 ± 1.09a |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Gao, T.; Xu, M.; Dong, J.; Li, H.; Wang, P.; Li, G.; Guo, T.; Kang, G.; Wang, Y. Functional Analysis of a Wheat AGPase Plastidial Small Subunit with a Truncated Transit Peptide. Molecules 2017, 22, 386. https://doi.org/10.3390/molecules22030386
Yang Y, Gao T, Xu M, Dong J, Li H, Wang P, Li G, Guo T, Kang G, Wang Y. Functional Analysis of a Wheat AGPase Plastidial Small Subunit with a Truncated Transit Peptide. Molecules. 2017; 22(3):386. https://doi.org/10.3390/molecules22030386
Chicago/Turabian StyleYang, Yang, Tian Gao, Mengjun Xu, Jie Dong, Hanxiao Li, Pengfei Wang, Gezi Li, Tiancai Guo, Guozhang Kang, and Yonghua Wang. 2017. "Functional Analysis of a Wheat AGPase Plastidial Small Subunit with a Truncated Transit Peptide" Molecules 22, no. 3: 386. https://doi.org/10.3390/molecules22030386





