Distal Proton Shuttle Mechanism of Ribosome Catalysed Peptide Bond Formation—A Theoretical Study
Abstract
:1. Introduction
2. Results and Discussions
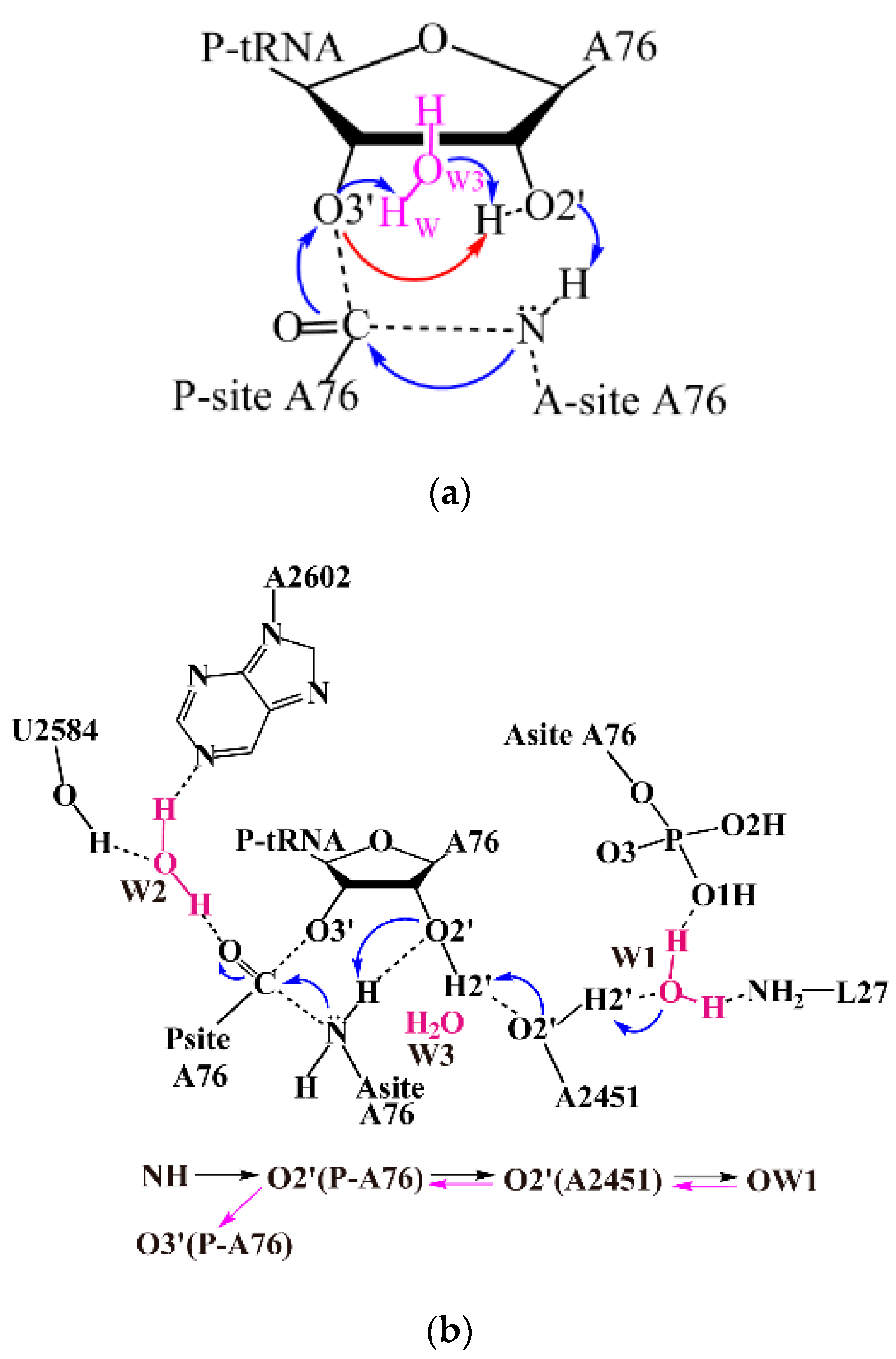
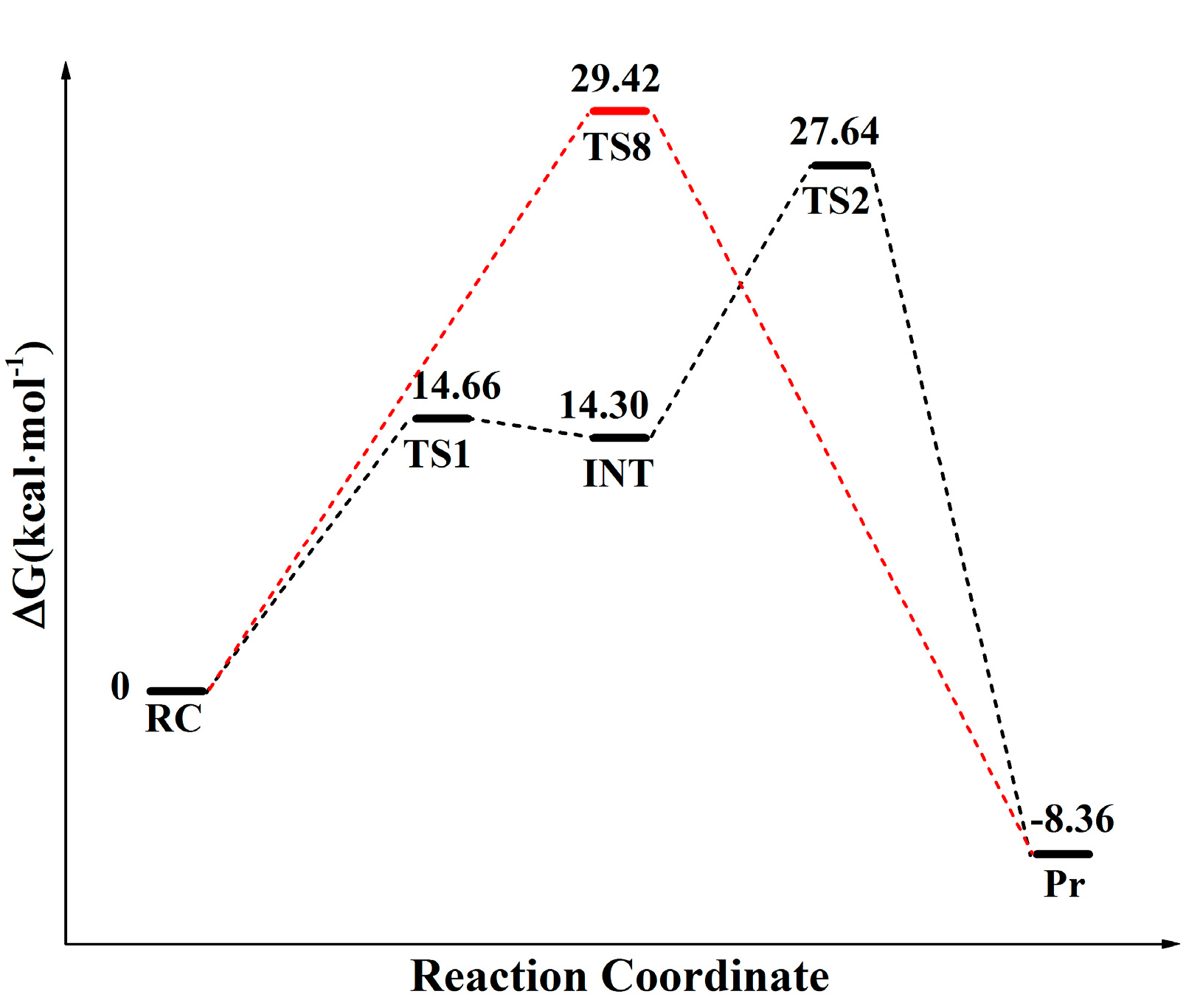
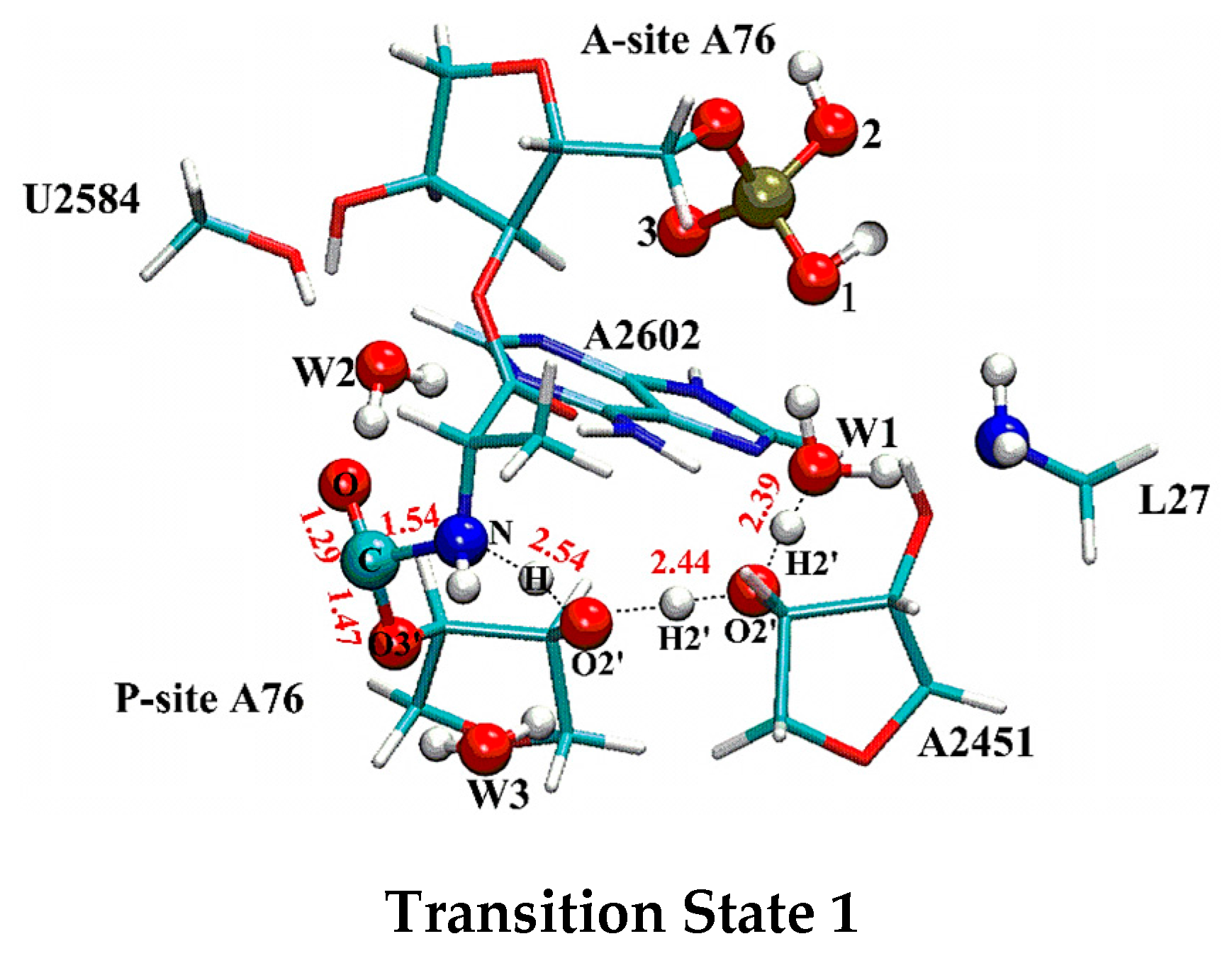
2.1. Comparison of Distal Proton Transfer Mechanism and Eight-Membered Ring Mechanism
2.2. The Important Role of Water Molecules in the Peptide Formation Reaction
2.3. The Function of L27 and 5′-Phosphate of A-Site A76
3. Materials and Methods
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yonath, A. Polar bears, antibiotics, and the evolving ribosome (Nobel Lecture). Angew. Chem. Int. Ed. 2010, 49, 4341–4354. [Google Scholar] [CrossRef] [PubMed]
- Schmeing, T.M.; Huang, K.S.; Kitchen, D.E.; Strobel, S.A.; Steitz, T.A. Structural insights into the roles of water and the 2′ hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol. Cell 2005, 20, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Kuhlenkoetter, S.; Wintermeyer, W.; Rodnina, M.V. Different substrate-dependent transition states in the active site of the ribosome. Nature 2011, 476, 351–354. [Google Scholar] [CrossRef] [PubMed]
- Ball, P. Water as an active constituent in cell biology. Chem. Rev. 2008, 108, 74–108. [Google Scholar] [CrossRef] [PubMed]
- Swanson, J.M.; Maupin, C.M.; Chen, H.; Petersen, M.K.; Xu, J.; Wu, Y.; Voth, G.A. Proton solvation and transport in aqueous and biomolecular systems: Insights from computer simulations. J. Phys. Chem. B 2007, 111, 4300–4314. [Google Scholar] [CrossRef] [PubMed]
- Taraphder, S.; Hummer, G. Protein side-chain motion and hydration in proton-transfer pathways. Results for cytochrome P450cam. J. Am. Chem. Soc. 2003, 125, 3931–3940. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Sharpe, M.A.; Qin, L.; Ferguson-Miller, S.; Voth, G.A. Storage of an excess proton in the hydrogen-bonded network of the D-pathway of cytochrome c oxidase: Identification of a protonated water cluster. J. Am. Chem.Soc. 2007, 129, 2910–2913. [Google Scholar] [CrossRef] [PubMed]
- McCullagh, M.; Saunders, M.G.; Voth, G.A. Unraveling the mystery of ATP hydrolysis in actin filaments. J. Am. Chem. Soc. 2014, 136, 13053–13058. [Google Scholar] [CrossRef] [PubMed]
- Martinez, S.E.; Huang, D.; Ponomarev, M.; Cramer, W.A.; Smith, J.L. The heme redox center of chloroplast cytochrome f is linked to a buried five-water chain. Protein Sci Publ. Protein Soc. 1996, 5, 1081–1092. [Google Scholar] [CrossRef] [PubMed]
- Schmeing, T.M.; Huang, K.S.; Strobel, S.A.; Steitz, T.A. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature 2005, 438, 520–524. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Gao, J.; Zhang, D.; Liu, C. A theoretical model investigation of peptide bond formation involving two water molecules in ribosome supports the two-step and eight membered ring mechanism. Chem. Phys. 2015, 111, 450–451. [Google Scholar] [CrossRef]
- Polikanov, Y.S.; Steitz, T.A.; Innis, C.A. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat. Struct. Mol. Biol. 2014, 21, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, J.Z.; Xiang, Y. Ab initio QM/MM free energy simulations of peptide bond formation in the ribosome support an eight-membered ring reaction mechanism. J. Am. Chem. Soci. 2012, 134, 16424–16429. [Google Scholar] [CrossRef] [PubMed]
- Świderek, K.; Marti, S.; Tuñón, I.; Moliner, V.; Bertran, J. Peptide Bond Formation Mechanism Catalyzed by Ribosome. J. Am. Chem. Soci. 2015, 137, 12024–12034. [Google Scholar] [CrossRef] [PubMed]
- Erlacher, M.D.; Lang, K.; Wotzel, B.; Rieder, R.; Micura, R.; Polacek, N. Efficient ribosomal peptidyl transfer critically relies on the presence of the ribose 2′-OH at A2451 of 23S rRNA. J. Am. Chem. Soci. 2006, 128, 4453–4459. [Google Scholar] [CrossRef] [PubMed]
- Lang, K.; Erlacher, M.; Wilson, D.N.; Micura, R.; Polacek, N. The role of 23S ribosomal RNA residue A2451 in peptide bond synthesis revealed by atomic mutagenesis. Chem. Biol. 2008, 15, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Sievers, A.; Beringer, M.; Rodnina, M.V.; Wolfenden, R. The ribosome as an entropy trap. Proc. Natl. Acad. Sci. USA 2004, 101, 7897–7901. [Google Scholar] [CrossRef] [PubMed]
- Wallin, G.; Aqvist, J. The transition state for peptide bond formation reveals the ribosome as a water trap. Proc. Natl. Acad. Sci. USA 2010, 107, 1888–1893. [Google Scholar] [CrossRef] [PubMed]
- Gindulyte, A.; Bashan, A.; Agmon, I.; Massa, L.; Yonath, A.; Karle, J. The transition state for formation of the peptide bond in the ribosome. Proc. Natl. Acad. Sci. USA 2006, 103, 13327–13332. [Google Scholar] [CrossRef] [PubMed]
- Thirumoorthy, K.; Nandi, N. Role of chirality of the sugar ring in the ribosomal peptide synthesis. J. Phys. Chem. B 2008, 112, 9187–9195. [Google Scholar] [CrossRef] [PubMed]
- Byun, B.J.; Kang, Y.K. A mechanistic study supports a two-step mechanism for peptide bond formation on the ribosome. Phys. Chem. Chem. Phys. 2013, 15, 14931–14935. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Gao, J.; Liu, Y.; Liu, C. Validating a new proton shuttle reaction pathway for formation of the peptide bond in ribosomes: A theoretical investigation. Chem. Phys. Lett. 2010, 501, 113–117. [Google Scholar] [CrossRef]
- Acosta-Silva, C.; Bertran, J.; Branchadell, V.; Oliva, A. Quantum-mechanical study on the mechanism of peptide bond formation in the ribosome. J. Am. Chem. Soci. 2012, 134, 5817–5831. [Google Scholar] [CrossRef] [PubMed]
- Hiller, D.A.; Singh, V.; Zhong, M.; Strobel, S.A. A two-step chemical mechanism for ribosome-catalysed peptide bond formation. Nature 2011, 476, 236–239. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Truhlar, D.G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: Two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor. Chem. Acc. 2008, 120, 215–241. [Google Scholar]
- Zhao, Y.; Truhlar, D.G. Density functionals with broad applicability in chemistry. Acc. Chem. Res. 2008, 41, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Trylska, J.; McCammon, A.J.; Brooks, C.L. Exploring Assembly Energetics of the 30S Ribosomal Subunit Using an Implicit Solvent Approach. J. Am. Chem. Soci. 2005, 127, 11125–11133. [Google Scholar] [CrossRef] [PubMed]
- Kazemi, M.; Himo, F.; Aqvist, J. Peptide Release on the Ribosome Involves Substrate-Assisted Base Catalysis. ACS Catal. 2016, 6, 8432–8439. [Google Scholar] [CrossRef]
- Foresman, J.; Ortiz, J.; Cioslowski, J.; Fox, D. Gaussian 09, Revision D. 01; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
Sample Availability: Coordinates of the reaction species are available from the authors. |
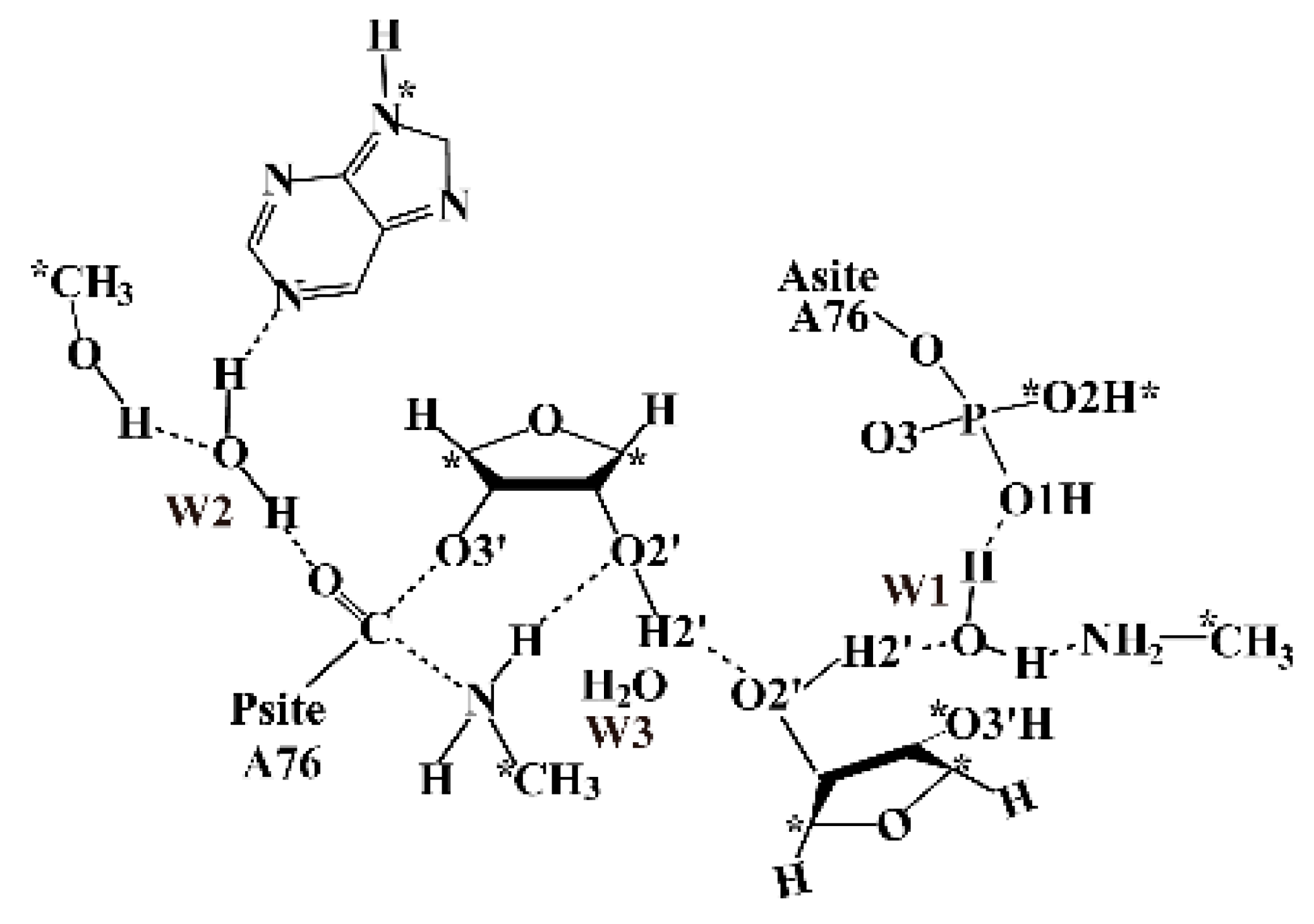
| Distal Proton Shuttle Mechanism | TS-8 Mechanism | |||||
|---|---|---|---|---|---|---|
| RC | TS1 | INT | TS2 | Pr | ||
| N-H | 0.775 | 0.242 | 0.203 | 0.035 | 0.003 | 0.140 |
| H-O2′(P-A76) | 0.027 | 0.434 | 0.474 | 0.451 | 0.007 | 0.525 |
| O2′-H2′(P-A76) | 0.633 | 0.234 | 0.179 | 0.490 | 0.646 | 0.336 |
| H2′(P-A76)-O2′(A2451) | 0.055 | 0.429 | 0.492 | 0.151 | 0.056 | 0.001 |
| O2′-H2′(A2451) | 0.603 | 0.302 | 0.207 | 0.549 | 0.605 | 0.619 |
| H2′(A2451)-OW1 | 0.090 | 0.362 | 0.460 | 0.130 | 0.085 | 0.081 |
| H-O3′(P-A76) | 0.002 | 0.003 | 0.003 | 0.151 | 0.665 | 0.001 |
| OW3-HW3 | 0.766 | 0.727 | 0.725 | 0.686 | 0.694 | 0.407 |
| C-N | 0.040 | 0.815 | 0.830 | 0.985 | 1.215 | 0.882 |
| C-O3′(P-A76) | 1.016 | 0.767 | 0.762 | 0.356 | 0.001 | 0.514 |
| C-O | 1.694 | 1.265 | 1.257 | 1.473 | 1.600 | 1.429 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Jiang, Y.; Mao, Q.; Tan, H.; Li, X.; Chen, G.; Jia, Z. Distal Proton Shuttle Mechanism of Ribosome Catalysed Peptide Bond Formation—A Theoretical Study. Molecules 2017, 22, 571. https://doi.org/10.3390/molecules22040571
Zhang X, Jiang Y, Mao Q, Tan H, Li X, Chen G, Jia Z. Distal Proton Shuttle Mechanism of Ribosome Catalysed Peptide Bond Formation—A Theoretical Study. Molecules. 2017; 22(4):571. https://doi.org/10.3390/molecules22040571
Chicago/Turabian StyleZhang, Xiaotong, Yafei Jiang, Qiuyun Mao, Hongwei Tan, Xichen Li, Guangju Chen, and Zongchao Jia. 2017. "Distal Proton Shuttle Mechanism of Ribosome Catalysed Peptide Bond Formation—A Theoretical Study" Molecules 22, no. 4: 571. https://doi.org/10.3390/molecules22040571





