c-Jun Contributes to Transcriptional Control of GNA12 Expression in Prostate Cancer Cells
Abstract
:1. Introduction
2. Results
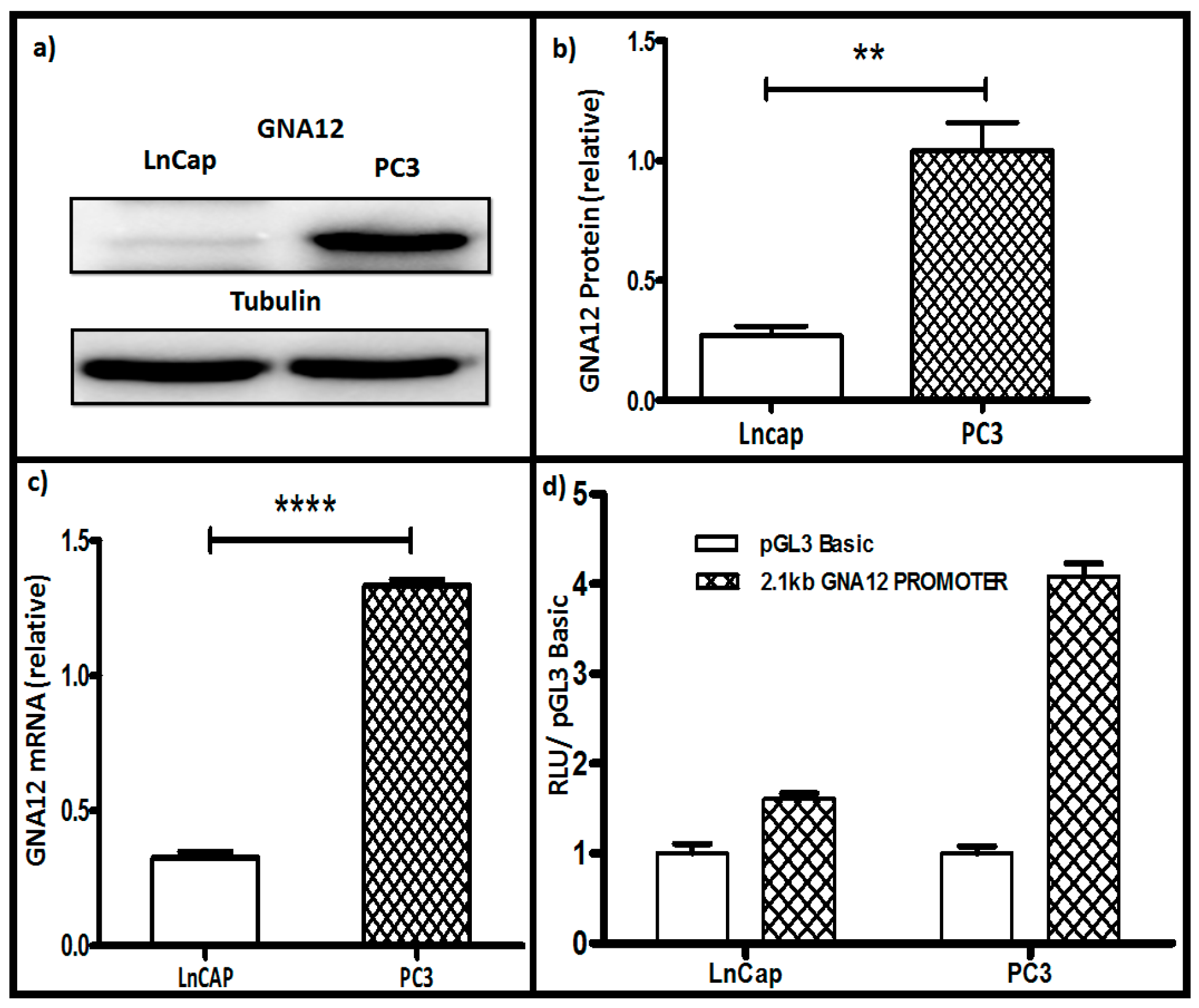
2.1. Correlation of GNA12 mRNA and Protein Levels in Prostate Cancer Cell Lines
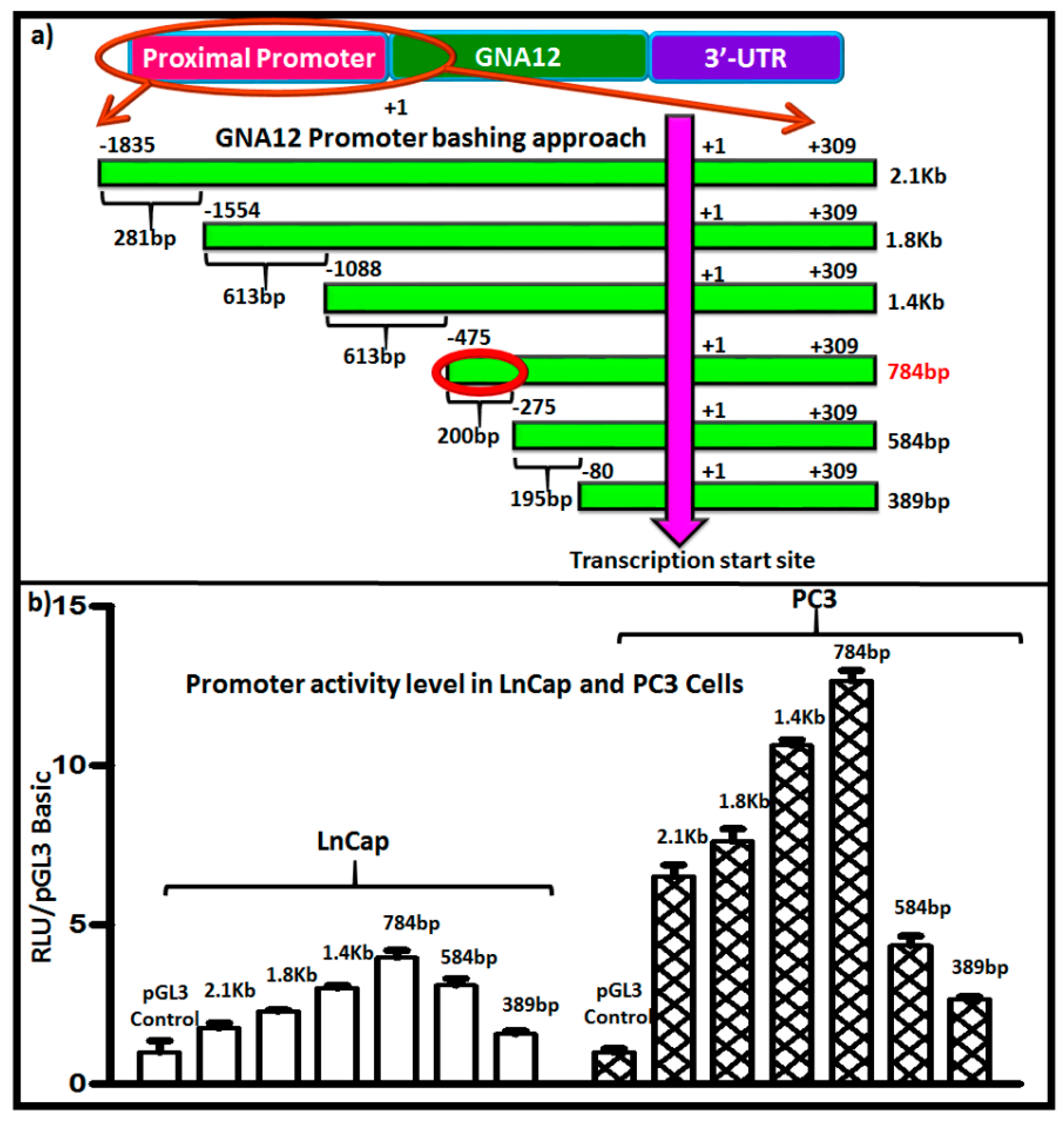
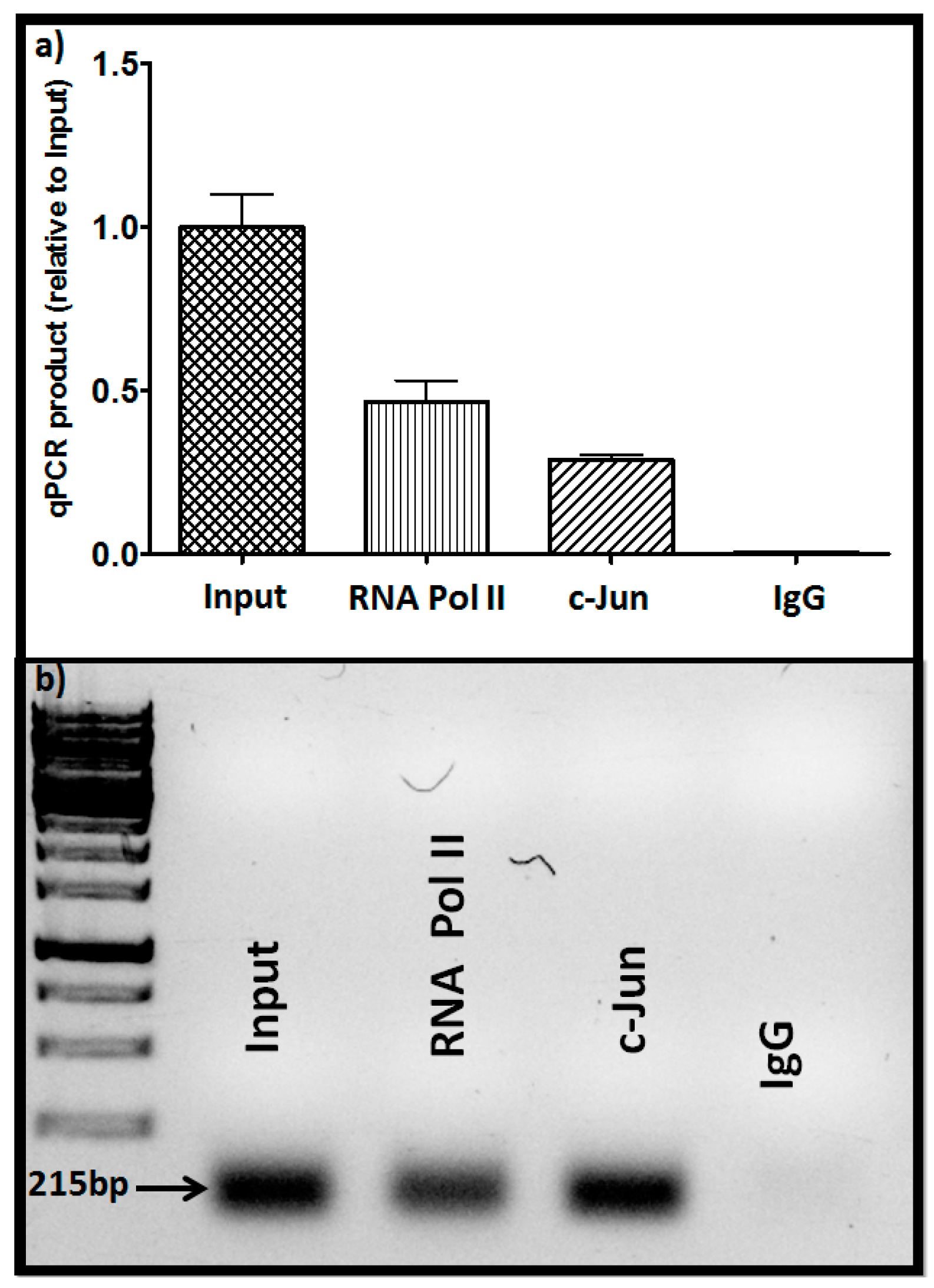
2.2. Identification of c-Jun as a Transcription Factor Impacting GNA12 Expression
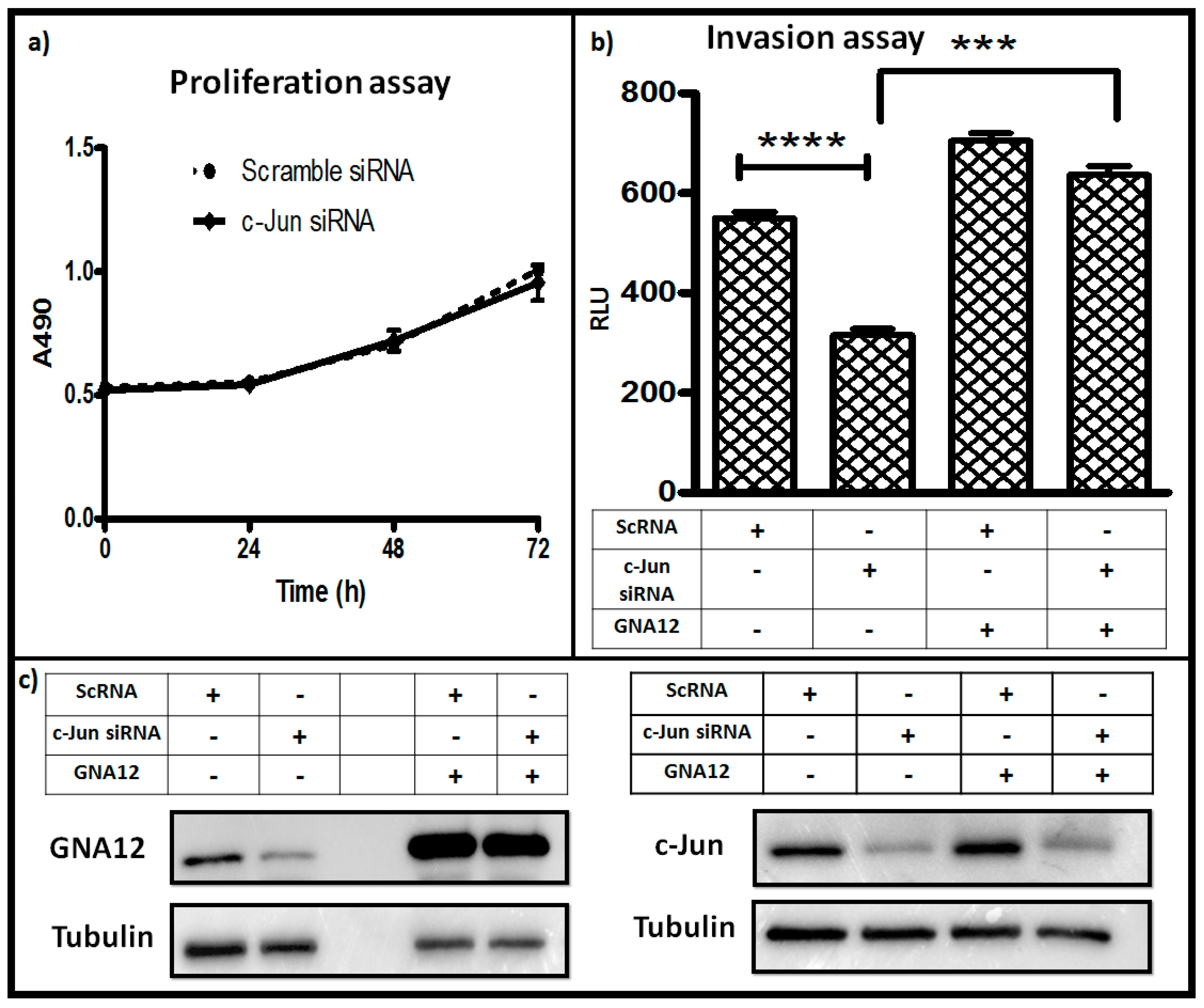
2.3. Silencing c-Jun Impacts GNA12-Dependent PC3 Cell Invasion
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Reagents
4.2. GNA12-5′ Regulatory Region Reporter Construct Cloning Strategies
4.3. Site Directed Mutagenesis
4.4. Luciferase Reporter Assay
4.5. Bioinformatics Analysis
4.6. RNA Extraction Quantitative Real-Time Reverse Transcription-PCR (qPCR)
4.7. Immunoblot Analysis
4.8. Proliferation and Invasion Assays
4.9. ChIP Assay
4.10. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| RLU | Relative light units |
| GPCR | G protein-coupled receptor |
| JNK | c-Jun N-terminal kinase |
References
- Kurose, H. Gα12 and Gα13 as key regulatory mediator in signal transduction. Life Sci. 2003, 74, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Kelly, P.; Casey, P.J.; Meigs, T.E. Biologic functions of the G12 subfamily of heterotrimeric G proteins: Growth, migration, and metastasis. Biochemistry 2007, 46, 6677–6687. [Google Scholar] [CrossRef] [PubMed]
- Juneja, J.; Casey, P.J. Role of G12 proteins in oncogenesis and metastasis. Br. J. Pharmacol. 2009, 158, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Kelly, P.; Moeller, B.J.; Juneja, J.; Booden, M.A.; Der, C.J.; Daaka, Y.; Dewhirst, M.W.; Fields, T.A.; Casey, P.J. The G12 family of heterotrimeric G proteins promotes breast cancer invasion and metastasis. Proc. Natl. Acad. Sci. USA 2006, 103, 8173–8178. [Google Scholar] [CrossRef] [PubMed]
- Kelly, P.; Stemmle, L.N.; Madden, J.F.; Fields, T.A.; Daaka, Y.; Casey, P.J. A role for the G12 family of heterotrimeric G proteins in prostate cancer invasion. J. Biol. Chem. 2006, 281, 26483–26490. [Google Scholar] [CrossRef] [PubMed]
- Juneja, J.; Cushman, I.; Casey, P.J. G12 signaling through c-Jun NH2-terminal kinase promotes breast cancer cell invasion. PLoS ONE 2011, 6, e26085. [Google Scholar] [CrossRef] [PubMed]
- Chia, C.Y.; Kumari, U.; Casey, P.J. Breast cancer cell invasion mediated by Gα12 signaling involves expression of interleukins-6 and -8, and matrix metalloproteinase-2. J. Mol. Signal. 2014, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Voyno-Yasenetskaya, T.A.; Pace, A.M.; Bourne, H.R. Mutant alpha subunits of G12 and G13 proteins induce neoplastic transformation of Rat-1 fibroblasts. Oncogene 1994, 9, 2559–2565. [Google Scholar] [PubMed]
- Nagao, M.; Kaziro, Y.; Itoh, H. The Src family tyrosine kinase is involved in Rho-dependent activation of c-Jun N-terminal kinase by Gα12. Oncogene 1999, 18, 4425–4434. [Google Scholar] [CrossRef] [PubMed]
- Jho, E.H.; Davis, R.J.; Malbon, C.C. c-Jun amino-terminal kinase is regulated by Gα12/Gα13 and obligate for differentiation of P19 embryonal carcinoma cells by retinoic acid. J. Biolol. Chem. 1997, 272, 24468–24474. [Google Scholar] [CrossRef]
- Arai, K.; Maruyama, Y.; Nishida, M.; Tanabe, S.; Takagahara, S.; Kozasa, T.; Mori, Y.; Nagao, T.; Kurose, H. Differential requirement of Gα12, Gα13, Gαq, and Gβγ for endothelin-1-induced c-Jun NH2-terminal kinase and extracellular signal-regulated kinase activation. Mol. Pharmacol. 2003, 63, 478–488. [Google Scholar] [CrossRef] [PubMed]
- Spinner, D.M.; Brandstetter, T.; Kiechle-Schwarz, M.; Du Bois, A.; Angel, P.; Bauknecht, T. c-Jun expression and growth stimulation in human ovarian carcinoma cell lines following exposure to cytokines. Int. J. Cancer 1995, 63, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Deyneko, I.V.; Kel, A.E.; Kel-Margoulis, O.V.; Deineko, E.V.; Wingender, E.; Weiss, S. Matrixcatch—A novel tool for the recognition of composite regulatory elements in promoters. BMC Bioinform. 2013, 14, 241. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.M.; Fleming, T.P.; McGovern, E.S.; Chedid, M.; Miki, T.; Aaronson, S.A. Expression cDNA cloning of a transforming gene encoding the wild-type Gα12 gene product. Mol. Cell. Biol. 1993, 13, 762–768. [Google Scholar] [CrossRef] [PubMed]
- Angel, P.; Hattori, K.; Smeal, T.; Karin, M. The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell 1988, 55, 875–885. [Google Scholar] [CrossRef]
- Rasheed, S.A.; Teo, C.R.; Beillard, E.J.; Voorhoeve, P.M.; Casey, P.J. MicroRNA-182 and microRNA-200a control gG-protein subunit α-13 (GNA13) expression and cell invasion synergistically in prostate cancer cells. J. Biolol. Chem. 2013, 288, 7986–7995. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, S.A.; Teo, C.R.; Beillard, E.J.; Voorhoeve, P.M.; Zhou, W.; Ghosh, S.; Casey, P.J. MicroRNA-31 controls G protein alpha-13 (GNA13) expression and cell invasion in breast cancer cells. Mol. Cancer 2015, 14, 67. [Google Scholar] [CrossRef] [PubMed]
- Shaulian, E.; Karin, M. AP-1 in cell proliferation and survival. Oncogene 2001, 20, 2390–2400. [Google Scholar] [CrossRef] [PubMed]
- Kitabayashi, I.; Kawakami, Z.; Chiu, R.; Ozawa, K.; Matsuoka, T.; Toyoshima, S.; Umesono, K.; Evans, R.M.; Gachelin, G.; Yokoyama, K. Transcriptional regulation of the c-jun gene by retinoic acid and E1A during differentiation of F9 cells. EMBO J. 1992, 11, 167–175. [Google Scholar] [PubMed]
- Sioletic, S.; Czaplinski, J.; Hu, L.; Fletcher, J.A.; Fletcher, C.D.; Wagner, A.J.; Loda, M.; Demetri, G.D.; Sicinska, E.T.; Snyder, E.L. c-Jun promotes cell migration and drives expression of the motility factor ENPP2 in soft tissue sarcomas. J. Pathol. 2014, 234, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Vleugel, M.M.; Greijer, A.E.; Bos, R.; van der Wall, E.; van Diest, P.J. c-Jun activation is associated with proliferation and angiogenesis in invasive breast cancer. Hum. Pathol. 2006, 37, 668–674. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.Y.; Cai, C.; Fisher, C.J.; Zheng, Z.; Omwancha, J.; Hsieh, C.L.; Shemshedini, L. c-Jun enhancement of androgen receptor transactivation is associated with prostate cancer cell proliferation. Oncogene 2006, 25, 7212–7223. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the reporter constructs generated in this study are available from the authors. |

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Udayappan, U.K.; Casey, P.J. c-Jun Contributes to Transcriptional Control of GNA12 Expression in Prostate Cancer Cells. Molecules 2017, 22, 612. https://doi.org/10.3390/molecules22040612
Udayappan UK, Casey PJ. c-Jun Contributes to Transcriptional Control of GNA12 Expression in Prostate Cancer Cells. Molecules. 2017; 22(4):612. https://doi.org/10.3390/molecules22040612
Chicago/Turabian StyleUdayappan, Udhaya Kumari, and Patrick J. Casey. 2017. "c-Jun Contributes to Transcriptional Control of GNA12 Expression in Prostate Cancer Cells" Molecules 22, no. 4: 612. https://doi.org/10.3390/molecules22040612




